Abstract
Maintaining stable and transient quiescence in differentiated and stem cells, respectively, requires repression of the cell cycle. The plant RETINOBLASTOMA-RELATED (RBR) has been implicated in stem cell maintenance, presumably by forming repressor complexes with E2F transcription factors. Surprisingly we find that mutations in all three canonical E2Fs do not hinder the cell cycle, but similarly to RBR silencing, result in hyperplasia. Contrary to the growth arrest that occurs when exit from proliferation to differentiation is inhibited upon RBR silencing, the e2fabc mutant develops enlarged organs with supernumerary stem and differentiated cells as quiescence is compromised. While E2F, RBR and the M-phase regulatory MYB3Rs are part of the DREAM repressor complexes, and recruited to overlapping groups of targets, they regulate distinct sets of genes. Only the loss of E2Fs but not the MYB3Rs interferes with quiescence, which might be due to the ability of E2Fs to control both G1-S and some key G2-M targets. We conclude that collectively the three canonical E2Fs in complex with RBR have central roles in establishing cellular quiescence during organ development, leading to enhanced plant growth.
Similar content being viewed by others
Introduction
In multicellular organisms, both the entry into and the exit from cell division cycle is developmentally regulated. There is a tight balance among cells proliferating in meristematic regions of emerging young organs, stem cells that are in a reversible quiescent status with a capacity to re-entry into cell division upon stimulation, and the most common cellular state of terminally differentiated cells formed after permanent exit from the cell cycle1,2,3. The principal regulation of cell proliferation by highly conserved molecular mechanisms is well established4, but how the cell cycle machinery is controlled in quiescent cells to maintain transient and stable repression of mitosis remains unclear.
Evidence from animal model organisms indicates that the RETINOBLASTOMA (Rb)-E2F transcriptional regulatory pathway plays a central role in coordinating proliferation and quiescence2,5. Binding of Rb to the E2F-dimerisation partner (DP) heterodimers can convert these complexes to transcriptional repressors, while phosphorylation of Rb by Cdk-cyclin complexes releases E2Fs and their ability to activate transcription. Accordingly, animal E2Fs can function as bistable switches on their target genes, determining whether the cells enter into the cell cycle or exit to establish quiescence6.
The Rb-E2F pathway is remarkably conserved in plants, and it is well established that the plant RETINOBLASTOMA-RELATED (RBR) coordinates cell proliferation and differentiation by regulating the activity of three canonical E2F transcription factors (E2FA, E2FB and E2FC) that form dimers with one of the two DP proteins (DPA and DPB)7,8,9,10. The three non-canonical E2Fs (E2FD, E and F) function independently of DP and RBR11,12. Canonical E2Fs are structurally related to their animal counterparts, and consensus E2F-binding sites were identified in many cell cycle regulatory genes11,13. Their C-termini contain the transactivation domain overlapping with the RBR-binding region14,15 and thus function as transcriptional activators or repressors depending on whether they are in complex with RBR16,17. Overexpression studies suggest that E2FA and E2FB function as activators, but recent findings showed that they could also repress genes specifically involved in embryo and seed development18. Surprisingly, genetic studies showed that E2F mutations either in single, double or even triple mutants did not universally compromise cell proliferation17,18,19,20.
It has been established that loss of function rbr mutants are gametophytic lethal due to abnormalities during meiosis and gametophyte development21,22,23. Manipulation of RBR level during sporophyte development severely affect the balance between proliferation and differentiation; ectopic overexpression of RBR led to diminishment of root and shoot meristems, while the consequence of RBR silencing is the inhibition of differentiation and increased number of stem cells7,24. Conditional RBR silencing strongly represses leaf growth by stimulating the excessive proliferation of stem cells in the stomatal lineage and inhibiting terminal differentiation8,25, and triggers cell death7,26. It was suggested that upon RBR silencing the E2Fs are de-repressed, which is responsible for the above described developmental defects. However, the recent description of a fully viable, although partially sterile mutant plant, in which all three canonical E2Fs carry a T-DNA insertion (a triple e2fabc mutant) questions whether E2Fs are indeed essential for the activation of cell proliferation20,27. This is surprising because there is ample evidence that plant E2Fs are the primary effectors of RBR just like in animals28,29.
To understand the role of E2Fs for the developmental regulation of cell cycle, in this work we analysed the e2fabc triple mutant in detail for cell proliferation defects and for the deregulation of E2F targets. Surprisingly, just as upon RBR silencing, we observed hyperplasia in the e2fabc triple mutant, suggesting that E2Fs are not required for maintaining cell proliferation, but rather have a repressive role. RBR silencing leads to growth arrest during seedling establishment due to the inhibition of this developmental transition and of cellular differentiation9. This is not the case for the e2fabc mutant, which shows hyperplasia during embryonic and post-embryonic organ development with delayed cell cycle exit and unstable quiescence both in stem and in terminally differentiated cells. The production of excess number of cells in a developmental setting naturally does not mean that all cells uncontrollably proliferate, as these cells become incorporated into an enlarged organ. Some of these extra cells are the result of continued proliferation, e.g., more rounds of amplifying divisions of stomata meristemoids. In other cases, we observed differentiated pavement cells to form cell division planes. It is equally possible that these differentiated pavements cells retain their ability to proliferate or they re-enter cell division after they attained quiescence. We also established that the three E2Fs and RBR have largely overlapping gene targets and cell cycle genes were markedly upregulated in e2fabc mutant seedlings. We conclude that the three canonical E2Fs are collectively involved in cell cycle exit and in establishing transient and stable cellular quiescence by forming repressor complexes with RBR.
Results
The triple e2fabc mutant displays hyperplasia, leading to enlarged organs and plant stature
Several alleles of T-DNA insertion lines have been described for canonical E2Fs, all of which disrupt the carboxyl terminus containing the entire transactivation and RBR-binding domains and they, except e2fa-2 also lost the MARKED BOX (MB) region, that mediates the interactions between E2Fs and various other transcription factors in animals (Supplementary Fig. 1a)18,30. By using specific antibodies in immunoblot assays, we confirmed the loss of the corresponding E2F proteins as well as the accumulation of a truncated E2FA protein in the e2fab double and in the e2fabc triple mutants (Supplementary Fig. 1b, c). These E2F mutations, however, did not affect DPA, DPB or RBR protein level in the triple e2fabc mutant seedlings (Supplementary Fig. 1d). As described previously, the triple e2fabc mutant is partially sterile, mostly due to abnormal gametophyte development (Fig. 1a), but the reason for this is not yet known27. In addition, at a very low frequency, there is a so far unnoticed anomaly of supernumerary nuclei in the e2fabc mutant embryo sacs (2%; n = 240), a characteristic phenotype for rbr mutant ovules (Fig. 1a)21,31. Further similarities to rbr mutant phenotypes are the twin embryo sacs31, that may originate from two megagametophyte mother cells (Fig. 1a)27. These excessive proliferation phenotypes suggest that the canonical E2Fs may also have common repressor functions with RBR at some stages of gametophytic development.
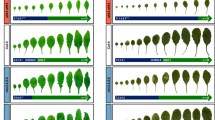
a Gametophytic defects observed in e2fabc triple mutants (highlighted in the lower panel of pictures). DIC images of cleared ovules showing embryo sacs with non-distinguishable nuclei (96% of images), a minority of normal-looking embryo sacs that account for the few viable seeds produced by the mutant (2.9%), but also embryo sacs with supernumerary nuclei (2,0%) or even twin embryo sacs (1,1%). The latter two are excessive proliferation phenotypes that were also reported in rbr mutants. Scale bars = 50 µm. b A representative picture of mature plants showing that the e2fabc mutant has shorter siliques and is taller than the WT. Pictures were taken 30 DAG. Bar is 1,5 cm. c Graph showing average height of WT and e2fabc plants (N = 10 samples in each). ***P ≤ 0.001 indicates statistically relevant differences between the mutant and the WT (two-tailed paired t test between the mutant and the WT). Root length is increased in e2fabc mutants as shown by representative pictures of plantlets at 7 DAG (d), and quantification of root length (e), data are average +/− standard deviation (n = 3 biological replicates; N = 10 samples in each), ***P ≤ 0.001 indicates statistically relevant differences between the mutant and the WT (two-tailed paired t test between the mutant and the WT). f Seed weight of e2fabc mutant is increased (weight of seeds is per 100 seeds, n = 10/line). ***P ≤ 0.001 was considered significant between the WT and the mutant (two-tailed paired t test between the mutant and the WT). g–j e2fabc embryos are larger than those of WT and consist of more and smaller cells. g representative pictures of dissected embryos from mature seeds (Bar: 200 µm), (h) cotyledon area, (i) epidermal cell size, (j) epidermal cell number in embryonic cotyledon. Cell size was calculated using ImageJ. Sample size N ≥ 200 cells/image. For all graphs, data are average +/− standard deviation (n = 3 biological replicates; N = 8 sample in each) and ***P ≤ 0.001 (two-tailed paired t test between the mutant and the WT) indicate statistically relevant differences.
To our surprise, the embryo and plant that came through the viable gametes of e2fabc triple mutant was overall normal but showed an increased plant size at the adult stage (Fig. 1b, c) as well as longer roots (Fig. 1d, e) and larger plantlet and rosette sizes (Supplementary Fig. 2a–e). Importantly, another triple e2fabc mutant obtained by crossing the e2fab double with the e2fc-2 T-DNA insertion mutant (Supplementary Fig. 1a), gave identical phenotypes with the original e2fabc mutant line (Supplementary Fig. 2f–h).
Due to the above-described sterility, the e2fabc mutant only produced few seeds, but these were significantly larger and heavier than WT (Fig. 1f)20, containing enlarged embryos (Fig. 1g and Supplementary Fig. 3) that consisted of smaller and more numerous cells than WT and double e2fab mutant (Fig. 1h–j and Supplementary Fig. 3a–e)17. Contrary to the triple e2fabc mutant embryo, double e2f mutant embryos (e2fab, e2fbc or e2fac) did not show hyperplasia (Supplementary Fig. 3e, f). Likewise, developing leaves of the e2fabc mutant seedlings became gradually larger than those of WT, until they reached 1.5 times the size of WT leaves at 20 DAG (Fig. 2a, b). Cellular analysis revealed that cell size was comparable with WT at early leaf development but failed to reach WT values at 16 DAG (Fig. 2c). Consequently, epidermal cell number was significantly higher in e2fabc mutants than in the WT (Fig. 2d). In addition, we observed some fully differentiated lobbed pavement cells with straight division planes in the e2fabc mutant, but not in WT, indicating their re-entry into cell division or continue proliferation while simultaneously differentiating (Fig. 2e, f and Supplementary Fig. 4a). Introducing a nuclear targeted CYCB1;2-YFP, that marks mitotic cells until telophase, into the triple e2fabc mutant further supported that enlarged, lobbed pavement cells divide, while in the control WT, CYCB1;2-YFP signal is never present in these type of cells (Supplementary Fig. 4b). Similar divisions of differentiated leaf epidermal cells was reported when regulators of this pathway like CYCD3;1 or E2FB was ectopically expressed in tobacco leaves32,33. The number of these extra divisions continuously rose and became the highest in the most advanced leaves at 16 DAG, resulting in smaller sized cells than in the WT (Fig. 2d–f), further suggesting that differentiated cells undergo extra cell divisions. As a result, the e2fabc mutant leaves contained almost twice as many cells (48%) as the control WT at 16 DAG (Fig. 2d). In some large pavement cells multiple cell division planes could be detected, indicating repeated cell division cycles (Supplementary Fig. 4a). Leaf cells dispersed among lobbed pavement cells, round and smaller than 60 µm2 are considered to be stomatal meristemoids34. The number of these cells was significantly increased in the expanding e2fabc mutant leaf at 12 and 16 DAG (Fig. 2e, g), suggesting that also the cell cycle exit is compromised in stomata stem cells in the e2fabc mutant. Consistent with the excessive proliferation the expression levels of the S-phase-related MCM3 (Supplementary Fig. 4c) and the mitotic CDKB1;1 (Fig. 2h), continuously increased in the e2fabc mutant during leaf development whereas these were strongly reduced or only detectable at the youngest stage of 9 DAG in the WT. To see whether the elevated expression of S-phase genes is related to endocycle or overproliferation in the triple e2fabc mutant compared to WT, we measured the nuclear DNA content of the first leaf pairs at early, when normally there is no endocycle, and at a later developmental stages, when control WT leaves enter endocycle (8 and 14 DAG, respectively), as well as in cotyledons, which is known to have high degree of endoreduplication at 10DAG (Supplementary Fig. 4d). As expected, in WT the 8C DNA content started to appear in true leaves at 14 DAG and reached 16C in the cotyledons, while in the triple mutant the 4C DNA level was the highest both in 14 DAG leaf and in the cotyledon, indicating a G2 arrest and block of endocycle. Similar effect on the leaf nuclear DNA content was reported when RBR level was silenced, further supporting that RBR functions together with E2Fs7. The elevated expression of two genes associated with the stem cell lineage of stomatal meristemoids: TOO MANY MOUTH (TMM; Fig. 2i), and SPEECHLESS (SPCH; Supplementary Fig. 4c) was also in agreement with the observed phenotype. Moreover, both RBR protein amount and its phosphorylation increased and remained at high levels in the e2fabc mutant leaf during its development in comparison to WT, supporting that quiescence was not stable in the triple e2fabc (Supplementary Fig. 4e). In contrast, the leaf size of e2fab double mutant was comparable to WT (Supplementary Fig. 5a, b). Additionally, the expression pattern of the mitotic CYCB1;1 in the developing e2fab double mutant leaf was similar to WT, while it was significantly upregulated in the triple e2fabc mutant (Supplementary Fig. 5c). Taken together, these data suggest that the three canonical E2Fs together are essential to allow cell cycle arrest to establish both stable and transient quiescence.
a–d First leaves of e2fabc mutant are enlarged due to prolonged proliferation, such as in stomata meristemoids and due to possible re-entry into cell division, such as in differentiated puzzle shaped pavement cells. a representative pictures of the first leaves of WT plants or e2fabc mutants at 20 DAG. b quantification of leaf area, (c) quantification of cell area, and (d) quantification of cell number in the leaf epidermis (first leaf pair). For all graphs, data are average +/− standard deviation n = 3 biological replicates, N = 10 samples in each. **P ≤ 0.001, ***P ≤ 0.0001 indicates statistically relevant differences between the mutant and the WT (two-tailed paired t-test between the mutant and the WT). e–g The increased cell number is derived from prolonged proliferation of stomata meristemoids and re-entry into cell division in differentiated puzzle shaped pavement cells in the e2fabc mutant. e Confocal microscope image of leaf epidermis of WT and e2fabc plants stained with propidium iodide (PI). Clustered stomata meristemoids (*) and division of differentiated puzzle-shaped cells (arrowheads) are observed in the e2fabc mutant. White dashed boxes outline the epidermal region of leaf in WT and e2fabc and magnified below. (Bar:75 µm and 25 µm). f Quantification of extra cell division events (n = 3 biological replicates, N = 6 samples in each), (g) proportion of small cells corresponding to stomata meristemoids. Data are mean +/− sd., n = 3 biological replicates, N = 5 samples in each and sample size ≥600 cells *P < 0.05, ***P < 0.001 (two-tailed, paired t test between the mutant and the WT at a given developmental time point). h, i The cell cycle gene CDKB1;1 and the stomata development gene TMM are elevated in e2fabc mutant compared to the wild-type. Expression of the two genes was monitored by qRT-PCR. Inset shows CDKB1;1 expression in the WT. Values represent fold changes normalised to the value of the relevant transcript of the wild type at 9DAG, which was set arbitrarily at 1. Data are means +/− sd., n = 3 biological repeats. **P < 0.01; ***P < 0.001 (two-tailed, paired t test between the WT and the mutant at a given time point). Abbreviations and primer sequences are listed in Supplementary Table 1.
Canonical E2Fs maintain quiescence through the recruitment of RBR to repress cell cycle-related genes
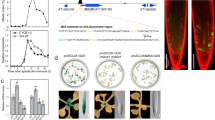
To further understand the underlying mechanism for the repressor role of E2Fs, we investigated global gene expression changes in the triple e2fabc mutant seedlings (Supplementary Data 1). 1766 genes were significantly upregulated and only 647 were downregulated in e2fabc compared to the WT (adj P value < 0.01). Additionally, Gene Ontology (GO) terms associated with upregulated, but not with downregulated genes, were highly enriched with functions linked to cell cycle control (Fig. 3a). To go further, we analysed the expression changes of two gene categories: the G1/S transition specific genes consisting of the canonical E2F targets, that were defined as genes misregulated in E2FA overexpressing lines, and harbouring a consensus E2F binding motif in their promoter13, and the G2/M-specific genes, respectively35. Among the genes upregulated in e2fabc, we found significant enrichment for canonical E2F target genes (P-value < 2.2e-16), but interestingly G2/M-specific genes were also affected (P-value = 0.0001401, Fig. 3b). On the other hand, neither categories were enriched among the downregulated genes in e2fabc mutant (Fig. 3b, p = 0.3336 and 1 respectively). Similarly, the scatter plot comparison of transcript levels in e2fabc and wild type seedlings showed general upregulation of the canonical E2F target genes (Fig. 3c).
a–c Cell cycle genes are globally upregulated in e2fabc triple mutant. a GO term enrichment found amongst genes up- or downregulated in e2fabc triple mutant. The colour scale corresponds to the -log10 of the FDR. b Venn diagram showing the overlap between up- or downregulated genes in e2fabc mutant and G1/S or G2/M cell cycle regulators. c Scatterplot highlighting the expression of G2/M specific (orange) and canonical E2F target (blue) genes in e2fabc mutant compared to the WT. d–f E2FA, B, C and RBR have largely overlapping target sites genome-wide. d Pearson correlation of binding sites identified by ChIP-seq for E2FA, B, C and RBR. e plots showing 2 by 2 comparison of the position of binding sites for E2FA, B, C and RBR on their targets. Plots are centred on the TSS of target genes. f Example ChIP-seq profiles to illustrating the similarities among E2FA, B, C and RBR.
Strong upregulation of cell cycle genes is also observed when RBR function is inhibited8,9. To test whether E2FA, B, C and RBR actually target the same loci, we performed chromatin immunoprecipitation (ChIP) followed by sequencing (ChIP-seq) using plants expressing translational GFP fusions under the control of the native promoters16,17,36,37. Binding of all four proteins occurs mostly just upstream from transcriptional start sites of genes (Supplementary Fig. 6a), and targets identified here for E2FA and RBR largely overlapped with previous reports (Supplementary Fig. 6b, c)38, validating the reliability of our data. Pearson correlation analysis (Fig. 3d), as well as plots representing 2 by 2 comparisons of distances separating defined peaks from the closest TSS (Fig. 3e) revealed that the four proteins have very similar binding profiles genome-wide, as further illustrated by a representative chromatin enrichment profile in Fig. 3f.
The ability of canonical E2Fs and RBR to target the same set of genes, together with the observation that the simultaneous loss of E2FA, B and C triggers excessive proliferation like RBR silencing, suggest that one key function of E2F proteins during vegetative development could be the recruitment of RBR to impose cell proliferation arrest. We therefore monitored the presence of RBR-DPB complexes in the e2fabc mutant, compared to e2fab mutants in which E2FC is still intact and its heterodimeric E2FC-DPB form is able to form complexes with RBR. As expected, RBR was in complex with DPB in the e2fab double mutant (Fig. 4a), but strongly reduced in the e2fabc mutant lacking E2FC, supporting our hypothesis that RBR recruitment to its targets is affected in these mutants. The residual RBR binding in the e2fabc mutant might be mediated by the MB region retained on E2FA in the case of e2fa-2 mutant allele.
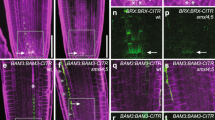
a RBR-DPB association is strongly reduced in the e2fabc mutant. Protein complexes were immunoprecipitated from protein extracts obtained from WT, e2fab or e2fabc mutant seedlings grown at 5 DAG using anti-DPB antibody. Comparable levels of both RBR and DPB can be detected in all genotypes (left, Input). However, although RBR could be co-IP-ed with DPB using protein extracts of WT or e2fab seedlings, amount of RBR immune-precipitated from e2fabc protein extracts were dramatically reduced (see also in Supplementary Fig. 14). b Comparison of the cellular phenotypes observed in the cotyledon epidermis of e2fabc triple mutant and RBRcs line. Asterisks indicate clustered stomata meristemoids, arrowheads point to extra cell division events in puzzle-shaped differentiated pavement cells found only in e2fabc. In RBRcs line all epidermal cells are small embryonic-like. White and blue boxes outline the epidermal region of cotyledon in e2fabc and RBRcs magnified on the right side. Arrowhead in RBRcs shows a dividing guard cell, while white arrows mark dead epidermal cells. Western blot in the left corner shows the lack of RBR in the RBRcs in comparison to the WT seedlings at 6DAG, while DPA and CDKA;1 amounts were comparable between them. Arrowhead shows RBR protein, and molecular weight markers are indicated on the left side (see also in Supplementary Fig. 14). c, d e2fabc and amiRBR lines display enlarged root meristems. Roots of the indicated genotypes were labelled with EdU for 60 min. c EdU staining of root tips of WT, e2fabc and amiRBR lines. White arrowheads mark the position of QC and the end of the meristematic zone indicated by enlarged nuclear size in the root epidermal cells. Bar: 100 µm. d Percentage of EdU-positive S-phase cells relative to DAPI-stained nuclei in the root meristems of WT, amiRBR and e2fabc lines. Average of 10 roots were imaged for each genotype. Data are average +/− standard deviation (n = 3 biological replicates, N = 10 samples in each). **P < 0.01 (two-tailed, paired t test between the mutant and the WT). e, f e2fabc triple mutant with supernumerary columella initials. e microscopy images of lugol-stained root tips of WT and e2fabc mutants (Bar: 25 µm). Differentiated root cap cells can be identified by amyloplast accumulation. The red arrowhead points to the QC and blue arrowheads indicate layers of columella initials. f: quantification of the proportion of roots with 1, 2, 3, 4, 5 or 6 layers of columella initials in WT, e2fabc or e2fabc-2 mutants. g Cell proliferation is reactivated in the QC of e2fabc and amiRBR lines. Plants were incubated in EdU for 24 h. Representative images are shown on the top panel and the white arrows point at the QC (Bar: 25 and 50 µm). White boxes outline the QC region of root meristems and magnified in below (Bar: 20 µm). White arrowheads mark the QC. The graph below shows the proportion of roots showing EdU labelling in the QC. Average of 35 roots were imaged for each genotype. Data are average +/− standard deviation (n = 3 biological replicates, N = 35 samples in each). ***P < 0.001 (two-tailed, paired t test between each mutant and the WT).
Because the strong RBR co-suppression line (RBRcs) is arrested during seedling growth9 (Supplementary Fig. 7a), we analysed the cotyledons for excessive proliferation and compared them to the e2fabc mutant (Fig. 4b, Supplementary Fig. 7). As in developing leaves, puzzle-formed pavement cells and guard cells in their regular bicellular forms could form in the cotyledon of e2fabc mutants, but meristemoids appeared clustered and differentiated pavement cells showed large number of straight cross walls, while neither of these ectopic proliferations were present in the WT (Fig. 4b, Supplementary Figs. 7b and 4a, Supplementary 8c, d). In the e2fabc cotyledon epidermis, straight cross walls could be detected in nearly all pavement cells supporting that quiescence was neither efficiently established nor maintained in these otherwise committed epidermal cells (Fig. 4b and Supplementary Fig. 8d). By contrast, most of the epidermal cells in RBRcs were small embryonic-like (Fig. 4b; Supplementary Fig. 7c, d), and many dead cells could be visualised (Fig. 4b; Supplementary Fig. 7c, d)8,9. Cell size was markedly shifted from large cells towards smaller cells, and cell number was increased in both RBRcs and e2fabc, although much more in RBRcs (Supplementary Fig. 8a–c). Consistently, the expression of key cell cycle regulators, DNA repair genes and embryonic genes was markedly increased in RBRcs line, and to a lesser extent in e2fabc mutant (Supplementary Fig. 9a). Extra cell divisions could also be visualised both in cotyledons and leaves of e2fabc triple mutant through the accumulation of the CYCB1;2 protein labelled with YFP (Supplementary Figs. 4b and 10). By contrast, photosynthesis-related genes that fail to be activated in RBRcs line due to defects in cell differentiation, were expressed at comparable levels in e2fabc and WT plants (Supplementary Fig. 9a). These findings match the observation that cell differentiation is delayed but not compromised in e2fabc mutant whereas the RBRcs line is severely growth arrested because cell differentiation is prevented9. Interestingly, late embryonic LEC2 and ABI3 genes were expressed at a comparable high level in both RBRcs and e2fabc mutants, and additionally seed storage 12S globulin was accumulated in the e2fabc seedlings suggesting that embryonic genes are repressed by RBR after germination probably in complex with E2Fs (Supplementary Fig. 9b).
We also compared cell proliferation defects in the root meristem of e2fabc mutants and in a weaker RBR loss of function line, where RBR levels are reduced by an artificial microRNA (amiRBR)39. Pulse labelling with 5-ethynyl 20-deoxyuridine (EdU) revealed a similar increase of S-phase cell number in the e2fabc mutant and the amiRBR line in comparison to the WT (Fig. 4c, d). Furthermore, we observed supernumerary undifferentiated columella stem cells in the e2fabc root cap, as well as supernumerary cortex/endodermis initials (Fig. 4e, f and Supplementary Fig. 11a), as previously shown in an RBR silenced line7. Mitosis, as followed by the YFP-tagged CYCB1;2 marker was clearly present in small dividing epidermal cells and the supernumerary columella initials, providing further evidence for their sustained ability to divide (Supplementary Fig. 10–11b). The lack of signal in quiescent centre (QC) compared to the surrounding cells when using prolonged EdU labelling allows to visualise the low frequency of S-phase in cells in QC, as shown in WT, while both in the e2fabc triple mutant and in amiRBR the QC cells were positive for EdU (Fig. 4g). Quiescence of stem cells was shown to protect them against DNA damage39. As it was shown before for amiRBR, we found an increased sensitivity of root stem cells of the e2fabc line compared to WT to the DNA damaging agent cisplatin (Supplementary Fig. 12c). The RBR silenced lines also display spontaneous cell death within the root meristem, but this is not the case for the e2fabc line (Supplementary Fig. 12a, b). Together, these data indicate that proliferation activities of the root meristem and specifically of the stem cells increased in the triple e2fabc mutant root, as is the case in amiRBR line, providing evidence for the role of E2F/RBR complexes in the maintenance of the quiescent state.
E2Fs and MYB3Rs participate in common multiprotein DREAM-complexes but have unique regulatory roles in the cell cycle
MYB3Rs together with E2FB, E2FC, and RBR, but not E2FA have been implicated in the repression of cell cycle genes as part of large multiprotein complexes, known as DREAM26,37,40. We therefore investigated the interplay among E2Fs, RBR and repressive MYB3Rs in relation to their target gene regulation. We defined target genes as those detected with the highest confidence (q-value < 0.01 enrichment > 3; Supplementary Data 2). As expected, lists of genes bound by each E2F were largely overlapping, irrespective of the combination of isoforms looked at (Supplementary Fig. 13a). To collectively deal with target genes of E2FA, E2FB and E2FC, we defined a gene category representing general E2F targets, that is characterised by being amongst the most significant targets of at least two E2Fs (hereafter called E2F(2)). E2F(2)-, RBR- and MYB3R3-bound genes showed dramatic overlap in any combinations (Supplementary Fig. 13b), defining 7 gene categories depending on the combination of factors that bind to them (Supplementary Data 3). EMR (bound by E2F, MYB3R3, and RBR) corresponded to 637 genes (Fig. 5a). For most of these genes, peaks of ChIP signals exactly coincide for E2FA, E2FB, E2FC, RBR and MYB3R3 at the site of transcriptional initiation (Supplementary Fig. 13c). GO terms associated with EMR genes were highly and exclusively enriched with categories related to cell cycle (Fig. 5a). Enrichment of cell cycle-related GO terms was observed both for EMR and EM, and less prominently in M, but not at all in other categories (Fig. 5a). Both the G1/S E2F targets and the G2/M-specific genes comprise a significant fraction of EMR and EM genes (Fig. 5b). This suggests that genes with cell cycle-related functions are preferentially regulated under the dual control of E2F and MYB3R transcription factors.
a GO term enrichment analysis shows that genes bound by E2Fs, RBR, and MYB3Rs are significantly overrepresented for cell-cycle related genes, whereas this enrichment is not as obvious for genes targeted by 2 or only one of these factors. b G1/S and G2/M-related genes are highly enriched in the common targets of E2Fs, MYB3Rs and RBR (EMR). This graph represents the percentage of G1/S (yellow), or G2/M (blue) genes amongst the six categories of genes defined according to their binding by E2F (E), MYB3Rs (M) RBR (R). c Genes targeted by E2Fs, RBR and/or MYB3R are preferentially regulated either by E2Fs or by MYB3Rs. This graph represents the percentage of genes in each category that are upregulated either in e2fabc or myb3R1,3,5 mutants, or upregulated in both. d–f Cell-cycle related genes targeted by E2Fs, RBR and/or MYB3R are preferentially regulated either by E2Fs or by MYB3Rs. d graph representing the percentage of G1/S or G2/M genes that are upregulated either in e2fabc or myb3r1,3,5 mutants, or upregulated in both. e Graph representing the percentage of G2/M and G1/S genes harbouring canonical E2F- or MYB3R-binding motifs in their promoters. f Graph representing the percentage of genes harbouring a canonical E2F or MYB3R binding motif in their promoters within the 6 categories defined according to the binding profiles of E2Fs, MYB3Rs and RBR. g The activator MYB3R4 but not the repressor MYB3R3 is upregulated in e2fabc mutant. Expression of MYB3R4, MYB3R3 and the MYB3R4 target and G2/M marker KNOLLE was monitored by qRT-PCR in the first leaf pairs of WT, e2fab and e2fabc mutants at 9, 12, 16 and 20 DAG. Values represent fold changes normalised to the value of the relevant transcript of the WT at 9DAG, which was set arbitrarily at 1. Data are means +/− sd., n = 3 biological repeats. **P < 0.01, ***P < 0.001 (two-tailed, paired t test between the WT and the mutant at a given time point). Abbreviations and primer sequences are listed in Supplementary Table 1.
We then analysed the effects of E2F and MYB3R mutations on the expression of genes in each category as defined by our ChIP-seq data. For this purpose, RNA-seq gene expression datasets of e2fabc and myb3r135 triple mutant were utilised. Genes within the EMR and EM groups were frequently upregulated in either e2fabc or myb3r135 mutants, but very rarely in both (Fig. 5c). Therefore, we concluded that in spite of the fact that MYB3Rs and E2Fs can be present in the same protein complex, and they are recruited onto the same promoter site, on a particular target gene only one or the other predominantly regulates the expression. This unique mode of regulation can be also confirmed for canonical E2F target and G2/M-specific genes, which are preferentially influenced by E2Fs and MYB3Rs, respectively, but much less frequently by both factors (Fig. 5d). Interestingly, the propension of genes to be influenced either by E2Fs or MYB3Rs correlated with the presence of consensus motifs for E2F or MYB3R binding, respectively (Fig. 5e, f). Altogether, these data indicate that E2F and MYB3R transcription factors could bind to common cell cycle genes, but they specifically regulate the expressions of genes holding their characteristic E2F or MSA binding sequences, respectively.
While MYB3Rs are specifically targeted and regulate mitotic genes containing MSA elements, E2Fs can regulate cell-cycle genes involved both in G1-S and G2-M transitions (Fig. 3b), an example is CDKB1:1 (Fig. 2h), a critical controller of G2-M and the switch from mitosis to endoreduplication41. Interestingly, the activating MYB3R, MYB3R4 is also a direct target of canonical E2Fs and is upregulated in the e2fabc mutant leaves at all analysed time points, whereas expression of the repressor MYB3R3 remained unchanged (Fig. 5g). Accordingly, the MYB3R4 target, KNOLLE with a role in cytokinesis, was also upregulated (Fig. 5g). These suggest that E2Fs may also regulate mitosis indirectly through the activating MYB3Rs. The ability of RBR and the three canonical E2Fs to regulate both G1/S and G2/M transitions may explain why only upon their loss the quiescence is broken, but not upon MYB3Rs that only act on mitotic genes.
Discussion
The E2F-Rb regulatory pathway is a pivotal conserved regulator of cell proliferation both in animal and plant cells, acting at the critical transition point of G1 to S phase, known as restriction point. The textbook model is that E2Fs, when Rb-bound, restrict cell proliferation by preventing the expression of key cell cycle genes. This repression is relieved through Rb phosphorylation by CDKs, the activity of which is regulated by a plethora of signalling inputs. Therefore, it is surprising that genetic knockout of all three canonical E2Fs in Arabidopsis can produce viable plants with a functional cell cycle. One possibility could be that the distantly related DP and E2F-like DELs (DEL1-3 or E2FD-F) might replace canonical E2Fs to activate cell cycle genes, but they considered to be repressors without the ability to induce gene expression42. Here we show that while none of the canonical E2Fs are fully required for activation of cell cycle genes, but collectively they are responsible for their repression and thereby to establish both transient quiescence in stem cells and stable quiescence in differentiated cells leading to supernumerary cell numbers in all organs examined. This scenario resembles to what has been established for the mammalian activator E2Fs (E2F1-3), as on the one hand they were also found dispensable for cell proliferation while on the other hand they form complexes with Rb in differentiating cells to repress cell cycle genes43.
Much of the previous studies have focused on the functional differences among the three canonical E2Fs16,44,45,46,47. E2FA and E2FB are portrayed as transcriptional activators while E2FC as a repressor. Moreover, it appears that they work through different mechanisms, only E2FB and E2FC but not E2FA are part of DREAM-like multi-subunit protein complexes37,40. Cell proliferation in single e2fa, e2fb or e2fc mutants is largely normal and these plants only display mild phenotypes, e.g., the compromised meristem maintenance and reduced formation of lateral root primordia in e2fa mutant, and a slight increase in cell number during leaf development in e2fb mutant and an e2fc silenced line17,18,47,48, suggesting that the distinction between activator and repressor classes of E2Fs is more nuanced. Interestingly, full loss of function e2fab double knock-out line could not be obtained49, but this appears to be allele specific, as combining the e2fb mutant with e2fa-2 is viable and did not show proliferation defects either during embryo or leaf development18,19. The hyperplasia in the e2fabc triple was not characteristic for any of the e2fc double mutant combinations with e2fa-2 (e2fc-1/e2fa-2) or e2fb-1 (e2fc-1/e2fb-1), suggesting that it is not E2FC, but the action of the three E2Fs together that is responsible for the repression. This was also the case when combining the e2fa/e2fb double mutant with two different e2fc knockout lines, both displayed hyperplasia, mostly due to the inability of maintaining quiescence. This suggests that the three canonical E2Fs are collectively required for the repression of cell cycle genes even if they act through different molecular mechanisms17,18. The principal component of repressor complexes on all canonical E2Fs is RBR and it was demonstrated that all three canonical Arabidopsis E2Fs strongly interact with it16,17,37. However, we have seen a stunning difference between the phenotypes of RBR silencing and the triple e2fabc mutant, both showing hyperplasia during organ development, but only upon RBR silencing and not in the e2fabc mutant the cell differentiation is repressed. A similar inhibition of differentiation occurs when CYCD3;1, the regulatory component of the major RBR kinase is overexpressed50. Transient overexpression of CYCD3;1 and CYCD4;2 in nonproliferating tobacco epidermal leaf cells was shown to induce the re-entry into cell division, demonstrating that quiescent leaf cells maintain their competence to divide32. These suggest that the de-repression of active E2Fs upon RBR silencing or phosphorylation interferes with differentiation and maintenance of quiescence. Because through the dimerisation partner, DPB, we still could detect a tiny amount of RBR association in the triple e2fabc mutant, presumably through the conserved MB region that was retained on E2FA in the e2fa-2 allele, we cannot rule out that a small residual E2F-RBR complex is able to repress cell proliferation to an extent that lead to cell cycle exit and differentiation51. However, the RBR association in the e2fabc is far less than in the double e2fab mutant, supporting that the main RBR binding activity of E2Fs is at the C-terminal RBR-binding domain, which in the case of e2fab, is presented on E2FC16.
On the one hand, RBR repression on E2Fs is the pivotal regulator of the restriction point in G1 and in response to developmental cues can determine the G1 length and whether cells can exit to differentiation. In this scenario tuning E2F activation of cell cycle genes by RBR is the critical mechanism. On the other hand, mutation of all three E2Fs impairs both the RBR-E2F corepressor function and the transcriptional activation of cell cycle genes. The comparison of RBR silencing and e2fabc mutant lines indicates that E2F activator function is important to maintain proliferation through preventing cell cycle exit and the onset of differentiation, while maintaining quiescence is primarily regulated by the RBR-E2F corepression. However, quiescence was not disrupted in each of the studied organs and cell types the same way in the e2fabc mutant, reflecting that different cell types have various levels of quiescence dependent on the developmental context, developmental regulation, and positional organisation of stem cells and cell files, as it was demonstrated in animals3. Leaves and roots for example have different temporal and spatial proliferation patterns under the control of the E2F-RBR regulation6,16 suggesting that this pathway is involved to establish and maintain quiescence in both organs but whether alone or in combination with other regulatory pathway is not known yet. In addition, our results show that plant E2Fs, like their animal counterparts, are the primary effectors of RBR in nearly every process where RBR is involved (Fig. 6a). The transient stem cell quiescence is most sensitively abrogated in the stomata lineage when RBR is silenced or the canonical E2Fs are mutated, as indicated by the abundance of small cells characterised as stomata stem cells and by the continuous upregulation of stomata stem cell factors, TMM and SPCH8. Interestingly, RBR, in an E2F-independent manner, by direct interaction with the stomata terminal differentiation transcription factor, FAMA, was also shown to regulate the late steps of differentiation within the stomata cell linage52. However, in the e2fabc mutant we have not seen the characteristic phenotype of the fama-1 mutant, called fama-tumours, clusters of small, narrow epidermal cells that are unable to go through the final stages of stomata differentiation. RBR and E2Fs can also directly regulate genes involved in late embryonic differentiation programs, e.g., LEC2 and ABI3, which are direct E2F and RBR targets and are upregulated both when RBR is silenced or in the e2fabc mutant, but the developmental transition of seedling establishment is blocked only in the former 9. Therefore, it is likely that as during leaf emergence, the developmental arrest in seedlings, when RBR is silenced, is due to the deregulated cell cycle.
a Canonical E2Fs function as co-repressors of cell cycle genes with RBR. Left: in the wild-type, E2Fs form complexes with RBR to repress cell cycle genes and allow cell division arrest in quiescent and differentiated cells. Middle: in the e2fabc mutant, lack of RBR-E2F complexes results in de-repression of cell cycle genes leading to a delay in cell differentiation as well as division reactivation in quiescent cells. Right: in RBR-deficient lines, E2F/RBR complexes are also lacking, but E2Fs are fully active, leading to a more pronounced hyperactivation of cell cycle genes and thus completely preventing cell differentiation, and inducing cell death. b Distinct molecular roles of E2Fs and MYB3Rs in the control of cell cycle arrest. Top: in the wild-type, Repressor complexes containing RBR, E2Fs and MYB3Rs target G1/S and G2/M genes to repress their expression either via the MSA or the E2F motifs. Middle: in the myb3r1,3,5 mutant that lacks repressor MYB3Rs, E2Fs are still present and able to repress G1/S and some G2/M genes bound via the E2F motif. Cell proliferation is therefore not induced in differentiated cells. Bottom: in the e2fabc mutant, repressor MYB3Rs can still repress some G2/M genes with or without RBR, but repression of both G1/S and G2/M activators such as MYB3R4 is lost, resulting in enhanced cell proliferation. Model was created by using Biorender.
It was shown both in animals and plants that quiescence protects long-lived stem cells against stress and toxicities39,53,54. In agreement, we found that stem cells are sensitive to genotoxic stress both upon RBR silencing and in the e2fabc mutant. RBR and E2Fs were also implicated to directly regulate the expression of DNA damage genes and cell death. In this respect we observed a much more pronounced spontaneous cell death with RBR silenced cells than in the e2fabc mutant, which correlated with the level of induction of DNA damage genes, suggesting that both co-repression and E2F transactivation function contributes to this regulation.
RBR, E2FB and E2FC together with the mitotic MYB3Rs can be part of large repressor complexes, called DREAM37. Here we show that RBR, E2Fs and MYB3Rs are also recruited to largely overlapping sets of cell cycle target genes, but still maintain a specific regulatory function on E2F targets or mitotic genes dependent on whether the MSA or E2F target sequence is present in these promoters. We have shown previously that mutation of the three repressor MYB3R (myb3r1/3/5) in Arabidopsis resulted in excess cells in developing organs but exclusively within the meristems40. Interestingly, mitotic genes like CYCB1;2 were found to be ectopically expressed also in differentiated leaf cells, but this did not result in mitotic reactivation, possibly because E2F-RBR complexes are still present, and capable of repressing genes involved in the G1/S transition, which may explain why quiescence is efficiently maintained in the myb3r1/3/5 mutant (Fig. 6b). In contrast, in the e2fabc mutant, where the E2F target genes are de-repressed, we find that both the transient quiescence in stem cells and stable quiescence in differentiated cells are broken. Although E2Fs primarily regulate genes with G1-S function, they were also shown to control the expression of key G2-M regulatory genes, like the plant specific CDKBs, supporting that E2Fs regulate both cell cycle transitions45. Moreover, E2Fs and RBR were specifically enriched on the promoter of the activator MYB3R4, a key regulator of mitosis, and indeed it was activated in the e2fabc mutant leaf, which might explain the central role of E2Fs to maintain cellular quiescence by controlling both G1/S and G2/M transitions (Fig. 6b). Recently a similar feed forward regulatory loop has been discovered in the mammalian cell cycle, where the activity of G1/S regulator CDK4/6 was also responsible to maintain CDK2 throughout G2/M and thus the cell cycle re-activation and restriction point remains sensitive to mitogenic signals until mitosis55.
Taken together, we have shown that the canonical E2Fs together with RBR form a repressor complex on a cohort of cell cycle genes that is pivotal for maintaining quiescence. Through this repression mechanism it is possible to influence plant growth, which could open novel ways for improving crop biomass and productivity.
Methods
Plant material and growth conditions
Arabidopsis thaliana Col-0 ecotype was the WT and background of all transgenic lines used in this study. In vitro-cultured plants were grown on a half-strength germination medium under continuous light at 22 °C. Soil grown plants were cultivated in a greenhouse at 22 °C under long-day conditions (16 h light/8 h dark). Most of the transgenic and T-DNA insertion mutant lines used in this study have been previously published: E2FA-GFP, RBR-GFP16, E2FB-GFP17, E2FC-GFP36, CYCB1;2-YFP56, e2fb-1 (SALK_103138), e2fa-2 (GABI-348E09), e2fc-1, (GK-718E12,20,48; the double e2fab (e2fa-2/e2fb-1) was reported by Heyman et al.19, and the triple e2fabc (e2fa-2/e2fb-1/e2fc-1) was generated by Wang et al.20. The second T-DNA insertion line for E2FC, named as e2fc-2 (SAIL-1216G10) was obtained from the SALK Institute and the e2fabc-2 triple mutant was generated by crossing the e2fab double (e2fa-2/e2fb-1) with the e2fc-2. By crossing the corresponding E2F mutants we also generated e2fc-1/e2fa-2 and e2fc-1/e2fb-1 double mutants as well as we also generated an e2fabc triple mutant line expressing the CYCB1;2-YFP marker. RBR co-suppression (RBRcs) seedlings were identified in transgenic lines expressing either the RBR-3xCFP18 or the RBR-RFP57 in the WT background under the control of the native RBR promoter showing strong growth arrested phenotype identical with previously reported9.
Young seedlings (5, 7, or 14 days after germination, DAG) or the first true leaf pairs of the wild-type and mutant Arabidopsis lines (e2fabc and e2fab) grown in vitro were harvested 9, 12, 16 and 20 DAG, flash frozen and stored at −80 °C.
Microscopy, EdU labelling and flow cytometry analysis
Mature dried seeds were imbibed for 1 h and dissected under the stereo-microscope. Isolated embryos were stained with propidium iodide (PI) and photographed under confocal laser microscopy (SP5, Leica). Organ and epidermal cell sizes were measured using ImageJ software18,58. Cotyledons and the first true leaf pairs of wild-type and mutant lines were dissected from seedlings at 4–8 DAG and 8-16 DAG, respectively. Leaves were stained with propidium iodide (PI; 20 μg/mL) and images on the abaxial side of the cotyledon or the leaf were taken and analysed by confocal laser microscopy. 600–300 cells were counted and measured per cotyledon or leaf samples (n = 3 biological replicates and N = 5 samples in each case were studied for the transgenic lines and the control WT) using ImageJ software. Average cell size was calculated, and the total cell number was extrapolated to the whole cotyledon and the first real leaf pair17,59. To visualise the distributions of the cell area, only non-guard epidermal cells were used for calculation59. The number of elongated pavement cells with newly formed cell wall (described as extra cell division) was counted in three different zones (the basal, middle and tip parts) and extrapolated to the whole leaf. Roots were also visualised after PI staining (20 μg/mL) under confocal laser microscopy17.
For DIC microscopy of cleared tissues, plant tissues were fixed in a 9:1 of ethanol and acetic acid solution and cleared with Hoyer’s solution (a mixture of 100 g chloral hydrate, 10 g glycerol, 15 g gum arabic, and 25 mL water). They were mounted onto glass slides and used for microscopic observations with differential interference contrast (DIC) microscope (BX51, Olympus). Images were captured with a CCD camera (DP74, Olympus) and an image capture software (CellSens Standard, Olympus). For visualisation of starch granules in columella cells, roots were incubated for 3 min in Lugol solution (Sigma) before clearing.
For EdU incorporation assay WT and the mutant seedlings were grown in half strength liquid MS containing 10 µM EdU (Click-iT Alexa Fluor 647 Imaging Kit; Invitrogene) and incubated either for an hour or for 24 h. Afterward, seedlings were treated according to60, and the root samples were also stained by DAPI solution and the observations were done under the confocal laser microscope.
For flow cytometry measurement, the first leaf pair and the cotyledons were collected and chopped by razor blades in nuclei extraction buffer and stained with DAPI with the CyStain UV Precise Kit (Partec, Magyar et al., 2005). Nuclear DNA content was determined by using Partec PAS2 Particle Analysing system (Partec, Germany).
cDNA preparation and RT-qPCR
RNA was extracted from the plant material using a CTAB-LiCl method of61. RNA samples were treated with DNase1 (ThermoScientific #EN0521) according to the manufacturer’s protocol. cDNA was synthesised using 1 µg of RNA using the ThermoScientific Reverse Transcription Kit (#K1691) with random hexamers based on the manufacturer’s prescription. Mock reaction without RevertAid enzyme was also prepared to ensure that there is no contaminating genomic DNA in the samples. RT-qPCR in the presence of SYBR Green (TaKaRa TB Green Primer Ex TaqII, #RR820Q) was carried out according to the manufacturer’s instructions in a BioRAD CFX 384 Thermal Cycler (BioRAD) with the following setup: 50 °C 2 min, 95 °C 10 min, 95 °C 15 s, 60 °C 1 min, 40 cycles followed by melting point analysis. Each reaction was carried out in tree technical replicates and reaction specificity was confirmed by the presence of a single peak in the melting curve. All the data were normalised to the average Ct value of two housekeeping genes (ACTIN and UBIQUITIN) unless otherwise mentioned and the calculated efficiency was added to the analysis. Amplification efficiencies were derived from the slope of amplification curves at the exponential phase. Primer sequences are summarised in Supplementary Table 1.
Immunoprecipitation and immunoblotting
Total proteins were extracted from dissected leaves or young seedlings in extraction buffer (25 mM Tris-HCl, pH 7.5, 75 mM NaCl, 15 mM MgCl2, 15 mM EGTA, 15 mM p-nitrophenylphosphate, 60 mM β-glycerophosphate, 1 mM dithiothreitol, 0.1% (v/v) IGEPAL CA-630, 0.5 mM NaF, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail for plant tissue [P9599, Sigma]). For immunoprecipitation (IP), equal amounts of protein samples (800 µg) in the extraction buffer17 were incubated with anti-DPB antibodies for 1 h at 4 °C on a rotary shaker. Dynabeads Protein A (Invitrogen) was used to pull down polyclonal antibodies, and after washing the beads proteins were eluted by adding SDS sample buffer followed by 5 min boiling. Eluted proteins were loaded on SDS-PAGE gels (10%) and after protein gel electrophoresis proteins were transferred to polyvinylidene difluoride membrane (PVDF, Milipore) and afterwards immunoblotting assay was carried out. For immunoblotting assay17, equal amounts of proteins were loaded to SDS-PAGE gel (10%), and proteins were transferred onto PVDF membranes. The membranes were blocked in 5% (w/v) milk powder with 0.05% (v/v) Tween 20 in Tris-buffered saline (TBS; 25 mM Tris-Cl, pH 8.0, and 150 mM NaCl; TBS plus Tween 20 [TBST]) buffer for 1 h at room temperature. The membrane was incubated with 5% (w/v) milk-powder TBST containing the primary antibodies and agitated overnight at 4 °C.
Primary antibodies used in immunoblotting experiments were chicken anti-RBR antibody (1:2000 dilution; Agrisera), mouse monoclonal anti-PSTAIRE (1:40,000 dilution, CDKA;1 specific; Sigma), anti-phospho-specific Rb (Ser-807/811) rabbit polyclonal antibody (1:500 dilution; Cell Signalling Tech), anti-DPA, anti-E2FB polyclonal rabbit antibodies (both 1:400 dilution, Magyar et al., 2005)45, anti-DPB polyclonal rabbit antibody62, and anti-E2FA rat polyclonal antibody (1:400 dilution,18, anti-12S globulin rabbit polyclonal antibody (1:10,000;63. After the primary antibody reaction, the membrane was washed (TBST) and incubated with the appropriate secondary antibody conjugated with horseradish peroxidase at room temperature. Afterward, chemiluminescence substrate was applied according to the manufacturer description (SuperSignal West Pico Plus, Thermo Fisher Scientific) or Immobilon western horseradish peroxidase (Millipore).
Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) assay
ChIP-seq assays were performed on 14-d-old seedlings using anti-GFP antibody (Abcam). Plant material was cross-linked in 1% (v/v) formaldehyde for 15 min under vacuum at room temperature and cross-linking was then quenched with 0.125 M glycine for 5 min. Cross-linked plantlets were ground in liquid nitrogen, and nuclei were lysed in Nuclei Lysis Buffer (0.1% SDS, 50 mm Tris-HCl at pH 8, 10 mm EDTA, pH 8). Chromatin was then sonicated for 5 min using a Covaris S220 (Peak Power: 175, cycles/burst: 200. Duty Factory: 20). Immunoprecipitation was performed overnight at 4 °C with gentle shaking, and immuno-complexes were next incubated for 1 h at 4 °C with 40 μL of Dynabeads Protein A (Thermo Fisher Scientific). The beads were washed 2 × 5 min in ChIP Wash Buffer 1 (0.1% SDS, 1% Triton X-100, 20 mM Tris−HCl at pH 8, 2 mM EDTA at pH 8, 150 mM NaCl), 2 × 5 min in ChIP Wash Buffer 2 (0.1% SDS, 1% Triton X-100, 20 mM Tris−HCl at pH 8, 2 mM EDTA at pH 8, 500 mM NaCl), 2 × 5 min in ChIP Wash Buffer 3 (0.25 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 10 mM Tris−HCl at pH 8, 1 mM EDTA at pH 8) and twice in TE (10 mm Tris-HCl at pH 8, 1 mM EDTA at pH 8). ChIPed DNA was eluted by two 15-min incubations at 65 °C with 250 μL of Elution Buffer (1% SDS, 0.1 m NaHCO3). Chromatin was reverse cross-linked by adding 20 μL of NaCl 5 M and incubated overnight at 65 °C. Reverse cross-linked DNA was treated with RNase and Proteinase K and extracted with phenol−chloroform. DNA was ethanol precipitated in the presence of 20 μg of glycogen and resuspended in 10 μL of nuclease-free water in a DNA low-bind tube. Libraries were then generated using 10 ng of DNA and NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB), following the manufacturer’s instructions. The quality of libraries was assessed with an Agilent 2100 Bioanalyzer (Agilent), prior to 1 × 75 bp high-throughput sequencing by NextSeq 500 (Illumina).
Analysis of ChIP-seq data
Trimmomatic-0.38 was used for quality trimming64. Parameters for read quality filtering were set as follows: minimum length of 36 bp; mean Phred quality score greater than 30; leading and trailing bases removal with base quality <5. The reads were mapped onto the TAIR10 assembly using Bowtie 265 with mismatch permission of 1 bp. To identify significantly enriched regions, we used MACS266. Parameters for peaks detection were set as follows: number of duplicate reads at a location:1; mfold of 5:50; Q-value cutoff: 0.05; extsize 200; sharp peak. Average scores across genomic regions were extracted using the multiBigwigSummary command of the DeepTools package after data normalisation with the S3norm software67. Data were visualised using WashU and the plot Heatmap tool of the DeepTools package68 was used to generate heatmaps while the ggplot2 package was used to draw metaplots.
Motif enrichment analysis
Position weight matrix (PWM) data for binding motifs of E2FA and MYB3R5, determined by DNA affinity purification sequencing (DAP-Seq), were obtained from a website of Plant Cistrome Database at http://neomorph.salk.edu/PlantCistromeDB69, and used as E2F- and MYB3R-binding motifs. Motif search based on PWM was conducted using the matchPWM function implementing in R Biostrings package. Forward and reverse of each motif were used as PWM with minimum score of 50%.
Transcriptome analysis
Total RNAs were extracted using the RNeasy Plant Mini Kit (QIAGEN, Germany) from whole seedlings according to the manufacturer’s instructions. Total RNAs from WT and e2fabc at 9 DAG were used for construction of cDNA libraries using the TruSeq RNA Library Preparation Kit v2 (Illumina, United States) according to the manufacturer’s protocol. For transcriptome profiling in WT and e2fabc plants, we analysed three biological replicates for statistical analysis. The libraries were sequenced using the NextSeq500 sequencer (Illumina, United States). Raw reads containing adaptor sequences were trimmed using bcl2fastq (Illumina, United States), and nucleotides with low-quality (QV < 25) were masked by N using the original script. Reads shorter than 50 bp were discarded, and the remaining reads were mapped to the cDNA reference using Bowtie with the following parameters: “–all–best–strata”70. The reads were counted by transcript models. Differentially expressed genes were selected based on the adjusted P-value calculated using edgeR (version 3.20.9) with default settings71.
Other methods
Gene ontology analysis was carried out using the singular enrichment analysis tool offered by agriGO with the default settings72.
Statistics and reproducibility
Each experiments were performed with three biologically independent samples unless stated otherwise. Statistical analysis between two groups was analysed by a paired t test in Microsoft Excel. The p values were all two-tailed, and p < 0.05 was considered significant: *p < 0.05, **p < 0.01, ***p < 0.001. All experiments in this study were repeated three times.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
RNA-Seq data for WT and e2fabc can be accessed from the DDBJ database under accession number DRA016858. ChIP-Seq data of RBR, E2FA, E2FB and E2FC can be accessed at Gene Expression Omnibus database under accession number GSE218481. The source data are provided with this paper including raw western blots (Supplementary Data 4 and Supplementary Fig. 14). Other source data supporting the findings of this study and Arabidopsis mutants and transgenic lines generated and used in this study are available from the corresponding author upon reasonable request.
References
Scheres, B. Stem-cell niches: nursery rhymes across kingdoms. Nat. Rev. Mol. Cell Biol. 8, 345–354 (2007).
Marescal, O. & Cheeseman, I. M. Cellular mechanisms and regulation of quiescence. Dev. Cell 55, 259–271 (2020).
Johnson, M. S. & Cook, J. G. Cell cycle exits and U-turns: quiescence as multiple reversible forms of arrest. Fac. Rev. 12, 5 (2023).
Morgan D. O. The Cell Cycle: Principles of Control. (New Science Press, 2007).
Yao, G. Modelling mammalian cellular quiescence. Interface Focus 4, 20130074 (2014).
Yao, G., Tan, C., West, M., Nevins, J. R. & You, L. Origin of bistability underlying mammalian cell cycle entry. Mol. Syst. Biol. 7, 485 (2011).
Wildwater, M. et al. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123, 1337–1349 (2005).
Borghi, L. et al. Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22, 1792–1811 (2010).
Gutzat, R. et al. RETINOBLASTOMA-RELATED PROTEIN controls the transition to autotrophic plant development. Development 138, 2977–2986 (2011).
Magyar, Z., Bogre, L. & Ito, M. DREAMs make plant cells to cycle or to become quiescent. Curr. Opin. Plant Biol. 34, 100–106 (2016).
Mariconti, L. et al. The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J. Biol. Chem. 277, 9911–9919 (2002).
Lammens, T., Li, J., Leone, G. & De Veylder, L. Atypical E2Fs: new players in the E2F transcription factor family. Trends Cell Biol. 19, 111–118 (2009).
Vandepoele, K. et al. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14, 903–916 (2002).
Magyar, Z., Atanassova, A., De Veylder, L., Rombauts, S. & Inze, D. Characterization of two distinct DP-related genes from Arabidopsis thaliana. FEBS Lett. 486, 79–87 (2000).
Kosugi, S. & Ohashi, Y. Interaction of the Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant Physiol. 128, 833–843 (2002).
Magyar, Z. et al. Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J. 31, 1480–1493 (2012).
Oszi, E. et al. E2FB Interacts with RETINOBLASTOMA RELATED and regulates cell proliferation during leaf development. Plant Physiol. 182, 518–533 (2020).
Leviczky, T. et al. E2FA and E2FB transcription factors coordinate cell proliferation with seed maturation. Development 146, dev179333 (2019).
Heyman, J. et al. Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome. Plant Cell 23, 4394–4410 (2011).
Wang, S. et al. A noncanonical role for the CKI-RB-E2F cell-cycle signaling pathway in plant effector-triggered immunity. Cell Host Microbe 16, 787–794 (2014).
Ebel, C., Mariconti, L. & Gruissem, W. Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429, 776–780 (2004).
Chen, Z., Hafidh, S., Poh, S. H., Twell, D. & Berger, F. Proliferation and cell fate establishment during Arabidopsis male gametogenesis depends on the Retinoblastoma protein. Proc. Natl. Acad. Sci. USA 106, 7257–7262 (2009).
Chen, Z. et al. Retinoblastoma protein is essential for early meiotic events in Arabidopsis. EMBO J. 30, 744–755 (2011).
Wyrzykowska, J., Schorderet, M., Pien, S., Gruissem, W. & Fleming, A. J. Induction of differentiation in the shoot apical meristem by transient overexpression of a retinoblastoma-related protein. Plant Physiol. 141, 1338–1348 (2006).
Desvoyes, B., Ramirez-Parra, E., Xie, Q., Chua, N. H. & Gutierrez, C. Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 140, 67–80 (2006).
Horvath, B. M. et al. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J. 36, 1261–1278 (2017).
Yao, X. et al. The canonical E2Fs are required for germline development in Arabidopsis. Front. Plant Sci. 9, 638 (2018).
Desvoyes, B. & Gutierrez, C. Roles of plant retinoblastoma protein: cell cycle and beyond. EMBO J. 39, e105802 (2020).
Kent, L. N. & Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 19, 326–338 (2019).
Black, E. P., Hallstrom, T., Dressman, H. K., West, M. & Nevins, J. R. Distinctions in the specificity of E2F function revealed by gene expression signatures. Proc. Natl. Acad. Sci. USA 102, 15948–15953 (2005).
Zhao, X. et al. RETINOBLASTOMA RELATED1 mediates germline entry in Arabidopsis. Science 356, eaaf6532 (2017).
Xu, J., Lee, Y. J. & Liu, B. Establishment of a mitotic model system by transient expression of the D-type cyclin in differentiated leaf cells of tobacco (Nicotiana benthamiana). N. Phytol. 226, 1213–1220 (2020).
Jimenez-Gomicronngora, T., Tan, H. & Lozano-Duran, R. Transient overexpression of E2Fb triggers cell divisions in pavement cells of Nicotiana benthamiana leaves. Plant Cell Rep. 38, 1465–1471 (2019).
Dong, J., MacAlister, C. A. & Bergmann, D. C. BASL controls asymmetric cell division in Arabidopsis. Cell 137, 1320–1330 (2009).
Haga, N. et al. Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 157, 706–717 (2011).
Kallai, B. M. et al. gamma-Tubulin interacts with E2F transcription factors to regulate proliferation and endocycling in Arabidopsis. J. Exp. Bot. 71, 1265–1277 (2020).
Lang, L. et al. The DREAM complex represses growth in response to DNA damage in Arabidopsis. Life Sci. Alliance 4, e202101141 (2021).
Verkest, A. et al. A generic tool for transcription factor target gene discovery in Arabidopsis cell suspension cultures based on tandem chromatin affinity purification. Plant Physiol. 164, 1122–1133 (2014).
Cruz-Ramirez, A. et al. A SCARECROW-RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol. 11, e1001724 (2013).
Kobayashi, K. et al. Transcriptional repression by MYB3R proteins regulates plant organ growth. EMBO J. 34, 1992–2007 (2015).
Boudolf, V. et al. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16, 2683–2692 (2004).
Kosugi, S. & Ohashi, Y. E2Ls, E2F-like repressors of Arabidopsis that bind to E2F sites in a monomeric form. J. Biol. Chem. 277, 16553–16558 (2002).
Chong, J. L. et al. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature 462, 930–934 (2009).
De Veylder, L. et al. Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21, 1360–1368 (2002).
Magyar, Z. et al. The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17, 2527–2541 (2005).
Sozzani, R. et al. Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol. 140, 1355–1366 (2006).
del Pozo, J. C., Diaz-Trivino, S., Cisneros, N. & Gutierrez, C. The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18, 2224–2235 (2006).
Berckmans, B. et al. Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 23, 3671–3683 (2011).
Li, X. et al. Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc. Natl. Acad. Sci. USA 114, 2765–2770 (2017).
Dewitte, W. et al. Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15, 79–92 (2003).
Rubin, S. M., Gall, A. L., Zheng, N. & Pavletich, N. P. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release. Cell 123, 1093–1106 (2005).
Matos, J. L. et al. Irreversible fate commitment in the Arabidopsis stomatal lineage requires a FAMA and RETINOBLASTOMA-RELATED module. Elife 3, e03271 (2014).
Cheung, T. H. & Rando, T. A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 14, 329–340 (2013).
Legesse-Miller, A. et al. Quiescent fibroblasts are protected from proteasome inhibition-mediated toxicity. Mol. Biol. Cell 23, 3566–3581 (2012).
Cornwell, J. A. et al. Loss of CDK4/6 activity in S/G2 phase leads to cell cycle reversal. Nature 619, 363–370 (2023).
Iwata, E. et al. GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell 23, 4382–4393 (2011).
Biedermann, S. et al. The retinoblastoma homolog RBR1 mediates localization of the repair protein RAD51 to DNA lesions in Arabidopsis. EMBO J. 36, 1279–1297 (2017).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Asl, L. K. et al. Model-based analysis of Arabidopsis leaf epidermal cells reveals distinct division and expansion patterns for pavement and guard cells. Plant Physiol. 156, 2172–2183 (2011).
Vanstraelen, M. et al. APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 106, 11806–11811 (2009).
Jaakola, L., Pirttila, A. M., Halonen, M. & Hohtola, A. Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol. Biotechnol. 19, 201–203 (2001).
Umbrasaite, J. et al. MAPK phosphatase AP2C3 induces ectopic proliferation of epidermal cells leading to stomata development in Arabidopsis. PLoS One 5, e15357 (2010).
Shimada, T. et al. Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100, 16095–16100 (2003).
Martins, R. P., Gandini, L. G. Jr., Martins, I. P. & Martins, L. P. Crimpable double tubes for segmental retraction. Orthodontics (Chic.) 12, 400–403 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Gaspar J. M. Improved peak-calling with MACS2. bioRxiv (2018).
Xiang, G. et al. S3norm: simultaneous normalization of sequencing depth and signal-to-noise ratio in epigenomic data. Nucleic Acids Res. 48, e43 (2020).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–165 (2016).
O’Malley, R. C. et al. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292 (2016).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Du, Z., Zhou, X., Ling, Y., Zhang, Z. & Su, Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–70 (2010).
Acknowledgements
We thank Xinnian Dong for the e2fabc mutant line, Arp Schnittger for the RBR-RFP line. This work was supported by a grant from the Hungarian National Research Funding (NKFI-139202) to Z. Magyar, the Japan Society for the Promotion of Science KAKENHI (22K06261 and 22H04714) to M. Ito, Japan Science and Technology Agency (JST grant number JPMJPF2102) to M. Ito, Biotechnology and Biological Sciences Research Council BBSRC-NSF grant BB/M025047/1 to L. Bogre and Cs Papdi. We are grateful for Attila Fehér for the critical reading and helpful discussion.
Funding
Open access funding provided by ELKH Biological Research Center.
Author information
Authors and Affiliations
Contributions
Z.M., L.B., M.I., C.R. and M.B. conceived the study; M.G., C.R, Z.M., M.I. designed the experiments; M.G., C.R., Z.M., Y.N., E.M., R.B.-C., H.T., A.Z., D.B., D.L., K.M., F.N., H.I., E.Ő. and C.P. carried out the experiments; X.H., J.A. and T.S. analysed the data, C.B. did the model; Z.M., C.R., M.G., L.B., M.I. and M.B. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
This manuscript has been previously reviewed at another Nature Portfolio journal. Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: David Favero. A peer review file is available. This document only contains reviewer comments and rebuttal letters for versions considered at Communications Biology.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gombos, M., Raynaud, C., Nomoto, Y. et al. The canonical E2Fs together with RETINOBLASTOMA-RELATED are required to establish quiescence during plant development. Commun Biol 6, 903 (2023). https://doi.org/10.1038/s42003-023-05259-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-023-05259-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.