Abstract
Alport syndrome (AS) is a hereditary glomerulonephritis caused by COL4A3, COL4A4 or COL4A5 gene mutations and characterized by abnormalities of glomerular basement membranes (GBMs). Due to a lack of curative treatments, the condition proceeds to end-stage renal disease even in adolescents. Hampering drug discovery is the absence of effective in vitro methods for testing the restoration of normal GBMs. Here, we aimed to develop kidney organoid models from AS patient iPSCs for this purpose. We established iPSC-derived collagen α5(IV)-expressing kidney organoids and confirmed that kidney organoids from COL4A5 mutation-corrected iPSCs restore collagen α5(IV) protein expression. Importantly, our model recapitulates the differences in collagen composition between iPSC-derived kidney organoids from mild and severe AS cases. Furthermore, we demonstrate that a chemical chaperone, 4-phenyl butyric acid, has the potential to correct GBM abnormalities in kidney organoids showing mild AS phenotypes. This iPSC-derived kidney organoid model will contribute to drug discovery for AS.
Similar content being viewed by others
Introduction
Alport syndrome (AS) is the second most common hereditary glomerulonephritis that proceeds early in life to end-stage renal disease (ESRD)1,2,3,4. It is caused by mutations in COL4A3, COL4A4, or COL4A5 genes and defects in α3α4α5(IV) heterotrimers that compose glomerular basement membranes (GBMs)5,6,7. Some clinical studies show differences in the disease progression that depends on the mutation types, and there is a correlation between the detection of α5(IV) and clinical symptoms4,8. However, the disease mechanisms have not been fully elucidated, and there are no curative treatments9,10. Further, the lack of efficient in vitro tools to analyze the collagen composition of GBMs has made it difficult to develop curative therapeutic approaches that correct collagen abnormalities in AS.
Type IV collagen is a main component of basement membranes and functions as heterotrimers: α1α1α2(IV), α3α4α5(IV), or α5α5α6(IV). In particular, α3α4α5(IV) is mainly localized in GBMs and constitutes the glomerular filtration barrier7,11,12,13. In development, mature podocytes switch to produce α3α4α5(IV) from α1α1α2(IV) around the capillary loop stage11,13,14,15. These developmental changes are essential to construct a robust filtration system, and COL4A5 mutations disrupt normal switching and cause renal dysfunction11. Type IV collagens form heterotrimers from the NC1 domain in their C-terminus as a starting point16. Thus, mutation types that lead to NC1 domain anomalies or defective trimer formation cause severe AS clinical symptoms. In such cases, no α3/α4/α5(IV) subunits are detected in GBMs17,18. In contrast, some mutations do not hamper trimer formation but lead to the formation of misfolding structures. These mutations are correlated with mild AS symptoms18.
Previous studies reported the generation of various AS mouse models, such as Col4a5tm1Yseg (p. G5X) mice, which exhibit similar phenotypes to humans, including GBM anomalies, proteinuria, and renal fibrosis19,20,21. Although the mouse models have advantages in that renal function can be assessed, it is difficult to validate a direct approach to GBMs for the development of new therapies because of the time required to create and evaluate them. Higher-order human cell evaluation systems are useful considering species differences.
Kidney organoids are self-organizing 3D cellular aggregates that contain nephron-like structures and are differentiated from embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) by mimicking kidney developmental processes22,23,24,25. In embryonic kidneys, the GBM collagen composition changes from α1α1α2(IV) to α3α4α5(IV) heterotrimers during the transition from the S-shaped stage to capillary loop stage of nephrogenesis11,12,14,15,26. Although kidney organoids also develop avascular glomerulus structures containing podocyte-like cells in vitro, the collagen composition of the GBM-like structures is immature27. While a recent study revealed that kidney organoids conserve the temporal sequence of the basement assembly28, no reports have reproduced AS disease phenotypes using kidney organoids differentiated from patient iPSCs.
In this study, we successfully generated kidney organoids that contain GBM-like structures with α3α4α5(IV) collagen heterotrimers using agitated cultures with an orbital shaker to mature the collagen composition in a time-dependent manner. We established iPSCs from two male X-linked AS (XLAS) patients carrying COL4A5 mutations on the X chromosome, one mild case with a c.1634 G > A (p.G545D) missense mutation in exon 24 and one severe case with a c.1652_53 dupTC (p.T552Sfs*6) nonsense mutation in exon 24 that causes a truncated form of α5(IV) and lack of heterotrimer formation. We repaired the gene mutation in iPSCs from the mild AS patient, which led to the restoration of the collagen α5(IV) protein expression in the induced kidney organoids. Furthermore, we found that kidney organoids from the AS patient iPSCs showed different abnormal collagen expression patterns in the GBM-like structures depending on the disease severity, suggesting that these cells can reproduce the phenotypic differences among AS patients. Finally, we found that a chemical chaperone can potentially normalize misfolded proteins in kidney organoids from the mild AS patient iPSCs but not from the severe AS patient iPSCs, confirming the recovery of α5(IV) antigen conformation on the GBM-like structures. These results suggest that kidney organoids from AS patient iPSCs are a powerful tool for analyzing disease mechanisms and predicting the effectiveness of therapeutic agents for individual AS patients, enabling personalized medicine.
Results and discussion
Generation of AS patient iPSCs
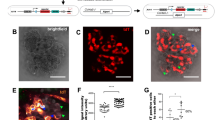
We generated iPSCs from the peripheral blood mononuclear cells (PBMCs) of two male XLAS patients with distinct symptomatic severity using episomal plasmids29,30. Although both patients were around 20 years old, the mild patient showed only slight proteinuria and hematuria as a symptom, while the severe patient proceeded to ESRD in his late teens. The mild patient’s mother had received a kidney biopsy test and was found α5(IV) negative. To establish disease models for the development of novel AS therapies by correcting the structural abnormalities of type IV collagen heterotrimers with low-molecular-weight compounds, we corrected the point mutation of the COL4A5 gene in mild AS patient iPSCs by CRISPR-Cas9 ribonucleoprotein and single-stranded DNA-mediated homologous recombination31. We confirmed that all iPSCs used in this study exhibited a normal morphological appearance, normal karyotype, and expression of the pluripotency marker NANOG (Fig. 1a, S1a, b). A genetic analysis of PBMCs revealed that both patients had COL4A5 mutations in exon 24, with a c.1634 G > A (p.G545D) missense mutation in the mild patient and a c.1652_53 dupTC (p.T552Sfs*6) nonsense mutation in the severe patient. These mutations were preserved in the patient’s iPSCs (Fig. 1b–d).
a The expression of a pluripotency marker, NANOG, in iPSCs. Scale bars, 100 µm. b, c The sequencing analysis of exon 24 of COL4A5 gene in 1383D2 cells, mild AS patient iPSCs, and mutation-corrected mild AS patient iPSCs (b) and in 1383D2 cells and severe AS patient iPSCs (c). Black and red bars and arrows indicate normal and mutated sequences, respectively. d A schematic showing COL4A5 mutations in mild and severe AS patients. e The kidney organoid differentiation protocol. f Immunostaining images of 1383D2 cell-derived day 13 + 28 kidney organoids for NEPHRIN and WT1 (podocytes) and LAMININ β1 (GBMs and tubular basement membranes) and for PODOCALYXIN (podocytes) and WT1. Scale bars, 50 µm.
AS patient iPSCs differentiate into kidney organoids
To develop a simple culture system suitable for drug screening for AS, we modified our previously reported kidney organoid differentiation method through nephron progenitor cells (NPCs) from iPSCs and adopted floating and agitated cultures with an orbital shaker (Fig. 1e). Culture day 9 cells induced from a healthy control iPSC line (1383D222) were positively stained with NPC markers, including WT1, SALL1, SIX2, PAX2 and HOXD11 (Figure S1c), as previously reported25. To quantitatively analyze the induction efficiency of NPCs among four iPSC lines (1383D2, mild AS patient iPSCs, mutation-corrected mild AS patient iPSCs, and severe AS patient iPSCs), we used WT1 and PAX2 as markers of NPCs on days 9 and 13, respectively, but found no significant differences in the differentiation efficiency among the lines (Figures S1d, e). During the 28 days of organoid culture of 1383D2 cells (from days 13 + 0 to 13 + 28), no obvious morphological changes in the bright field microscopy images were observed (Figure S2a). We confirmed that 1383D2-derived kidney organoids generated in the floating and agitated cultures show higher expressions of NPHS1, NPHS2, and the collagen IV genes COL4A3, COL4A4, and COL4A5 than those in the floating and non-agitated cultures, although the differences in NPHS1 and COL4A5 expression were not statistically significant (Figures S2b, c). Furthermore, the agitated cultures resulted in a significantly higher rate of PODOCALYXIN+ or EpCAM+ nephron-constituent cells than the non-agitated cultures (Figures S2d, S3). Although the mechanisms of these benefits of agitated cultures remain unknown, some possibilities including the diffusion-dependent maintenance of nutrients and oxygenation, waste removal, and mechanical stress were reported in cerebral organoids32,33.
In 1383D2-derived kidney organoids, the formation of glomerulus-like structures containing NEPHRIN+WT1+ or PODOCALYXIN+WT1+ podocyte-like cell clusters demarcated by LAMININ β1 was observed by immunostaining (Fig. 1f). We also confirmed that the kidney organoids from all four iPSC lines contained PODOCALYXIN+ podocyte-like cell clusters, EpCAM+Lotus tetragonolobus lectin (LTL)+ or EpCAM+CDH6+ proximal tubule-like structures and EpCAM+Dolichos biflorus agglutinin (DBA)+ or EpCAM+CDH1+ distal tubule-like structures (Figures S4a, S4b, S5a). Furthermore, we confirmed the development of nephron-constituent cells in 1383D2-derived day 13 + 28 kidney organoids by transmission electron microscopy (TEM), including podocyte-like cells exhibiting foot processes and surrounded by parietal epithelial cells, proximal tubule-like cells with brush borders and loop of Henle- or distal tubule-like cells that showed fewer microvilli and cilia and the morphology of a flat or cubic epithelium (Figures S5b-f). Immunostaining of a mild AS patient iPSC-derived day 13 + 28 kidney organoid for the vascular endothelial marker CD31 and the podocyte marker WT1 did not show the vascularization of glomerulus-like structures in kidney organoids (Figure S5g).
Time-dependent GBM maturation in kidney organoids
To develop in vitro AS kidney organoid models, the expression of α3α4α5(IV) heterotrimers is necessary. However, we found that the kidney organoids at early culture stages lacked α5(IV) expression in the GBM-like structures (Fig. 2). Therefore, we temporally examined GBM-associated gene expression by RNA sequencing and qRT-PCR analysis in 1383D2-derived kidney organoids on days 13 + 4, 13 + 7, 13 + 14, 13 + 21 and 13 + 28 (Figs. 2a, b). Mature GBM markers, including COL4A3, COL4A4, and COL4A5, were upregulated in a time-dependent manner, whereas early-stage GBM markers, such as COL4A1, COL4A2, LAMA1, and LAMAB1, were gradually downregulated. These results indicate a transition in the composition of the collagen IV chains in GBMs from α1α1α2(IV) to α3α4α5(IV). Consistently, immunostaining analysis revealed α5(IV) expression in the GBM-like structures after day 13 + 21 (Fig. 2c, S6a). Thus, long-term agitated culture with an orbital shaker generated kidney organoids containing GBM-like structures with α3α4α5(IV) collagen in vitro, indicating that the organoids can be used to create AS models.
a A heatmap showing log2FC gene expression relevant to GBMs in 1383D2 cell-derived kidney organoids on days 13 + 4 to 13 + 28 by RNA-seq analysis. b qRT-PCR analysis of 1383D2 cell-derived kidney organoids on days 13 + 4 to 13 + 28 for COL4A3, COL4A4, and COL4A5. The data from three independent experiments are represented as the means ± SEM (n = 3). c Immunostaining images of glomerulus-like structures in 1383D2 cell-derived kidney organoids on days 13 + 7, 13 + 14, and 13 + 21 for WT1, α5(IV) and α2(IV). Arrows and arrowheads indicate α5(IV)- and α2(IV)-expressing GBM-like structures, respectively. Scale bars, 50 µm. *p < 0.05, **p < 0.01, and ***p < 0.001 by Dunnett’s test, n.s., not statistically significant.
Kidney organoids containing GBMs with α3α4α5(IV) collagen recapitulate AS phenotypes in collagen composition
We examined whether α5(IV)-expressing kidney organoids have collagen α3α4α5(IV) in their GBM-like structures. Upon confirming α3(IV) protein expression in the NIDOGEN+ GBM-like structures in 1383D2 and mutation-corrected mild AS iPSC-derived kidney organoids by immunostaining (Figure S6b), we assumed that the kidney organoids contain GBMs with α3α4α5(IV) collagen chains. Then we tested whether kidney organoids from mild and severe AS patient iPSCs express α5(IV) in the GBM-like structures. Consistent with the clinical histopathology, the expression of α2(IV), but not α5(IV), was observed in kidney organoids on day 13 + 28 from both patient iPSCs, while kidney organoids from mutation-corrected mild AS patient iPSCs expressed α5(IV) protein (Fig. 3a), suggesting that kidney organoids from AS patient iPSCs recapitulate the α5(IV) depletion phenotypes.
a Immunostaining images of glomerulus-like structures in day 13 + 28 kidney organoids from mild AS patient iPSCs, mutation-corrected mild AS patient iPSCs and severe AS patient iPSCs for WT1, α5(IV) and α2(IV). Magnified images of the red-boxed areas are also shown. Arrows and arrowheads indicate α5(IV)- and α2(IV)-expressing glomerulus-like structures, respectively. Scale bars, 50 µm. b qRT-PCR analysis of day 13 + 28 kidney organoids from 1383D2 cells, mild AS patient iPSCs, mutation-corrected mild AS patient iPSCs, and severe AS patient iPSCs for COL4A5 and NPHS1. The data from three independent experiments are represented as the means ± SEM (n = 3). c Immunostaining images of glomerulus-like structures in day 13 + 28 kidney organoids from mild and severe AS patient iPSCs for WT1, α3(IV), and NIDOGEN. Magnified images of the red-boxed areas are also shown. Arrows and arrowheads indicate α3(IV)- and NIDOGEN-expressing glomerulus-like structures, respectively. Scale bars, 50 µm. d qRT-PCR analysis of day 13 + 28 kidney organoids from 1383D2 cells, mild AS patient iPSCs, mutation-corrected mild AS patient iPSCs, and severe AS patient iPSCs for COL4A3. The data from three independent experiments are represented as the means ± SEM (n = 3). *p < 0.05, by Tukey–Kramer test, n.s. not statistically significant.
Next, we conducted a TEM analysis to clarify whether kidney organoids show GBM ultrastructural abnormalities. We confirmed the GBM ultrastructure on day 13 + 28 organoids from all iPSC lines. However, kidney organoids from both mild and severe AS iPSCs failed to show several GBM ultrastructural abnormalities, such as thickening, splitting, and fragmenting of the lamina densa34,35,36 (Figure S6c). GBM-like structures in the organoids were ~100 nm thick and uniform (arrows in Figure S6c), but some GBM-like structures were divided into two basement membranes, which is consistent with observations of the early developmental stage of GBMs37, 38 (arrowheads in Figure S6c). Then, we analyzed the embryonic kidneys of AS model mice that harbor the G5X mutation in Col4a5 and develop the severe GBM lesions seen in adults, such as lamellation and splitting19,39,40. However, we did not detect any GBM ultrastructural abnormalities in E15.5 male col4a5Y/- mice (Figure S6d). Taken together with the finding that the GBM-like structures of our kidney organoids show isoform switching of type IV collagen (Figs. 2b, c), which occurs around the capillary loop stage in mouse embryonic kidneys11,12,14,15,26, we assumed that our kidney organoids are equivalent to around E15.5 mouse embryonic kidneys. These data suggest that our kidney organoids reproduce the switching of type IV collagen isoforms and AS phenotypes in collagen composition but not in ultrastructure.
Kidney organoid model recapitulates atypical AS phenotypes
Col4A5 expression is downregulated in AS model mice19,20. Consistently, the mRNA expression of COL4A5 was significantly decreased in our severe AS kidney organoids compared with 1383D2-derived organoids (Fig. 3b). However, COL4A5 expression did not change in mild AS kidney organoids, and no significant changes were observed in the mRNA expression of a podocyte marker, NPHS1, in all kidney organoids from all iPSC lines, although severe AS organoids showed a large batch variation (Fig. 3b). The difference in COL4A5 expression between mild and severe AS cases was explained by the types of COL4A5 mutations. Severe AS with a frameshift mutation (T552fs*6) could cause the nonsense-mediated mRNA decay (NMD) of COL4A5 mRNA, while mild AS with a point mutation (G545D) could not trigger NMD. Further analysis revealed that mild AS kidney organoids expressed α3(IV) protein, but severe AS organoids did not (Figs. 3c, d). Although both α3(IV) and α5(IV) are usually undetectable in typical AS patients, the expression of abnormal collagen proteins has been detected in GBMs by immunofluorescence in atypical cases18,41,42. It is known that type IV collagens form heterotrimers from the NC1 domain as a starting point6,16. Severe AS organoids showed a significant decrease in COL4A5 mRNA expression and lack α5(IV) C-terminal regions, including the NC1 domain, because of their mutations, whereas mild AS organoids carried a point mutation that does not significantly affect COL4A5 mRNA expression levels. These results suggest that α3α4α5(IV) heterotrimers were not present in the GBM-like structures of severe AS kidney organoids, but heterotrimers comprising mutated α5(IV) were expressed in the GBM-like structures of mild AS organoids. Previous clinical studies reported that AS patients with detectable α3(IV) or α5(IV) have mild symptoms, including the later onset of ESRD and hearing loss and fewer ocular abnormalities despite the genetic mutations8,17,18,42. Consistently, our results suggest that mild AS iPSCs are capable of differentiating into kidney organoids that recapitulate the clinical hallmark of α3(IV) expression. Thus, our kidney organoids are useful for reproducing individual differences in AS, indicating that iPSC-derived kidney organoids have advantages as a less invasive diagnostic tool for studying α3α4α5(IV) expression over other tests, such as renal biopsy.
Kidney organoids transplanted into renal subcapsules become vascularized and recapitulate AS phenotypes
In embryonic kidneys, GBMs mature by fusion of the basement membranes of podocytes and vascular endothelial cells37. Kidney organoids are vascularized when transplanted into mouse renal subcapsules and develop higher-order renal tissue-like structures25,43. Therefore, to more accurately characterize GBM-like structures in kidney organoids, we transplanted day 13 NPCs under the renal capsules of NOD/SCID mice. In the grafts 21 days after transplantation, we found glomerulus-like structures with WT1+ podocyte-like cells and CD31+ vascular endothelial cells from host mice (Fig. 4a, b). Moreover, α5(IV) expression was observed between CD31+ endothelia and WT1+ podocyte-like structures in the 1383D2-derived grafts (Fig. 4b). In contrast, although well-vascularized glomerulus-like structures were observed, no α5(IV) expression was found in mild or severe AS grafts (Fig. 4b). These results further indicate that our kidney organoids recapitulate AS phenotypes in GBMs.
a Immunostaining images of glomerulus-like structures in day 21 grafts for WT1 (podocytes), α5(IV), and CD31 (mouse endothelial cells). Mouse kidney and grafts from NPCs are divided by the yellow dotted lines. Scale bars, 100 µm. b Magnified images of the yellow boxed areas in a. Arrows indicate α5(IV)-expressing GBM-like structures. G glomerulus. Scale bars, 50 µm.
Chemical chaperone treatment restores α5(IV) conformation in mild AS kidney organoids
While α3α4α5(IV) heterotrimers are assembled in the endoplasmic reticulum (ER) and secreted to the extracellular space via the Golgi apparatus, unassembled or misassembled α3α4α5(IV) heterotrimers cause ER stress and autophagy activation, which leads to their degradation44,45. A previous study reported that chemical chaperone treatment in COL4A2-mutated patient primary dermal fibroblasts reduces the intracellular accumulation of collagen IV mutants and attenuated ER stress, although the study did not reveal whether chemical chaperone treatment rescues basement membrane integrity46. Chemical chaperones potentially normalize misfolded proteins and are candidate AS drugs, accelerating the extracellular secretion of mutated α3α4α5(IV) or ameliorating ER stress by modulating collagen secretion that stacks in the ER44,45. Because our results suggested that α3α4α5(IV) heterotrimers with structural abnormalities can form GBM-like structures in mild AS (Fig. 3c), we selected mild AS kidney organoids to examine chemical modifications in the α3α4α5(IV) formation. We treated mild AS kidney organoids with three representative low-molecular-weight chemical chaperones: 4-phenyl butyric acid (4-PBA), trimethylamine N-oxide (TMAO) and mannitol (Fig. 5a) for 1 day, and confirmed α5(IV) expression in the GBM-like structures after 4-PBA exposure (Fig. 5b). However, 4-PBA treatment did not restore α5(IV) expression in severe AS kidney organoids (Fig. 5c). These data suggest that 4-PBA has the potential to correct α5(IV) abnormalities and/or restore α3α4α5(IV) heterotrimer folding. Therefore, because the chaperone effects of collagen trimerization differ depending on the type of COL4A5 mutation44,45, our AS patient iPSC-based kidney organoids are useful for predicting the efficacy of chemical chaperones as personalized medicine.
a Experimental protocol. b Immunostaining images of glomerulus-like structures in mild AS patient iPSC-derived kidney organoids treated with medium, 4-PBA, TMAO, and mannitol for WT1, α5(IV), and α2(IV). c Immunostaining images of glomerulus-like structures in severe AS patient iPSC-derived kidney organoids treated with medium and 4-PBA for WT1, α5(IV), and α2(IV). Arrows and arrowheads indicate α5(IV)- and α2(IV)-expressing GBM-like structures, respectively. Scale bars, 50 µm.
It was reported that laminin α2-defective basement membrane in skeletal muscle can be rescued by the expression of laminin α1 chain47. Similarly, AS symptoms may be attenuated by augmenting GBM strength with other collagen subunits. In AS patients and mouse models, the compensatory expression of α1α1α2(IV) in GBM was reported48, 49. Although a compensatory increase in α1(IV) may contribute to increased GBM strength, previous studies using a Col4a3 and Ddr double knockout AS mouse model suggested that the Ddr pathway is crucial for the pathogenesis50,51. Since α1(IV) stimulates Ddr, the compensatory expression of α1(IV) may activate Ddr signaling and exacerbate the symptoms. Another study showed that the overexpression of Col4a3 in vascular endothelial cells does not restore the loss of α3α4α5(IV)52. Thus, it remains unclear whether the expression of only a single subunit has therapeutic effects. Epigenetic regulatory treatments may increase the expression of mutated α3α4α5(IV) or α1α1α2(IV). However, no studies have attempted to epigenetically regulate chromatin structures and induce the expression of α3α4α5(IV) or α1α2α2(IV).
In conclusion, we succeeded in creating an in vitro disease model using iPSC-based kidney organoids that recapitulate the characteristics of early-stage AS. Our model reproduces type IV collagen switching in GBMs during embryonic development and patient-specific α5(IV) and α3(IV) defects in GBM-like structures. A low-molecular-weight compound, 4-PBA, improved α5(IV) protein expression in the GBM-like structures of our kidney organoids, indicating that these organoids are potentially applicable to drug screening for restoring GBM abnormalities in AS towards patient-specific personalized medicine. Thus, these findings demonstrate the resourcefulness of patient iPSC-based organoids for the study of AS disease mechanisms and related drug discovery.
Methods
Inclusion and ethics
Experiments using iPSCs were approved by the Ethics Committees of Kyoto University, Kobe University, and Taisho Pharmaceutical Co., Ltd. All patients and donors from whom iPSCs were derived provided written informed consent. Animal experiments were approved by the CiRA Animal Experiment Committee and conducted in accordance with institutional guidelines.
Generation of patient iPSC lines
The iPSC lines were generated from two male XLAS patients based on a previously reported protocol29,30. PBMCs were obtained from the patients at the Department of Pediatrics, Kobe University Graduate School of Medicine, and reprogrammed by introducing pCEhOct4, pCEhSK, pCEhUL, pCEmp53DD, and pCXB-EBNA1, which encoded OCT3/4, SOX2, KLF4, L-MYC, LIN28, and mouse p53DD under feeder-free culture conditions.
Animal experiments
12-20-week-old female G5X AS model mice (B6.Cg-Col4a5tm1Yseg/J), 10–30-week-old male C57BL/6 mice, and 8–12-week-old male NOD/SCID mice were maintained at an SPF animal facility in CiRA. E15.5 and E18.5 male embryos from G5X AS model mice were obtained by crossing female G5X AS model mice and male C57BL/6 mice. For genotyping, genomic DNA was extracted from the ears or tails using the KANEKA Easy DNA Extraction Kit version 2 (KANEKA), and sequencing analysis was performed. The primers used are described in Supplementary Table 2. The renal subcapsular transplantation of NPCs was conducted based on our previously established protocol25. Induced NPCs on day 9 were seeded in low-binding 96U-plates at 1 × 104 cells/well and cultured for 4 days. Then, approximately 120 NPC aggregates were transplanted into renal subcapsules of NOD/SCID mice under systemic anesthesia. Day 21 grafts were used for the analysis.
Cell culture and karyotype analysis of iPSCs
All iPSC lines were cultured in StemFit® AK02N (Ajinomoto) and iMatrix-511 silk (Matrixome) under feeder-free conditions30. 4 × 104 cells were seeded on a six-well plate (Greiner) and passaged with the EDTA method53 every 4 days. The cells were routinely tested for mycoplasma contamination. All iPSC lines generated in this study were examined for their karyotypes by LSI Medience (Tokyo, Japan).
Sequencing analysis
For Sanger sequencing, genomic DNA was extracted from iPSCs using Cell Lysis Solution and Protein Precipitation Solution in Gentra Puregene Kits (QIAGEN). Extracted crude genomic DNA was precipitated with isopropanol and washed with ethanol for purification. The sequence around exon 24 of COL4A5 was amplified with the standard polymerase chain reaction (PCR) protocol using the primer sequences listed in Supplementary Table 2. After purification of the PCR products using the MinElute Reaction Cleanup Kit (QIAGEN), sequencing was performed using BigDye™ and analyzed using an Applied BiosystemsTM 3500/3500xL Genetic Analyzer.
iPSC differentiation into NPCs and kidney organoids
We slightly modified our previously published protocol for NPC induction from iPSCs25. iPSCs cultured in StemFit® AK02N and iMatrix-511 silk were differentiated into NPCs using DMEM/F12 GlutaMAXTM (Thermo Fisher Scientific) supplemented with B27 supplement minus vitamin A (Thermo Fisher Scientific) on a 24-well plate (Greiner) from days 0 to 9 and on a low-binding 96-well U plate (Sumitomo Bakelite) from days 9 to 13. Then, the basal culture medium was switched to KR5 medium, which is DMEM/F12 GlutaMAXTM containing 0.1 mM non-essential amino acids (Thermo Fisher Scientific), 55 µM 2-mercaptoethanol (Thermo Fisher Scientific), and 5% Knockout serum replacement (Thermo Fisher Scientific). NPC aggregates on day 13 were incubated for 2 days in a 96-well U plate using KR5 medium containing 200 ng/mL fibroblast growth factor (FGF)9 (Peprotech), 1 µM CHIR99021 (Axon) and 2% Matrigel (BD) for the organoid formation. Further organoid maturation steps were performed in KR5 medium containing 0.05% polyvinyl alcohol (Sigma-Aldrich) in a low-binding six-well plate (Iwaki) using an orbital shaker (CS-LR; Taitec).
Chemical chaperone treatment
Low-molecular-weight chemical chaperone compounds were purchased from the suppliers listed in Supplementary Table 1. Kidney organoids were treated for 24 hours with 4-PBA (10 mM), TMAO (150 mM), or mannitol (150 mM), which were dissolved in KR5 medium and filtrated before use.
Immunofluorescence analysis
Immunofluorescence analysis was conducted based on our previously established protocol25. Cells were fixed with 4% PFA/phosphate buffer (PB) for 15 min at 4 °C. Kidney organoids were fixed with 4% PFA/PB for 30 min at 4 °C or ice-cold acetone for 15 min, embedded into OCT compounds (Sakura Finetek Japan), and frozen quickly at −80 °C. Then, 4-µm frozen sections were prepared using a CM1520 cryostat (Leica). Primary antibodies were diluted in 5% donkey serum/PBST (PBS/0.3% Triton-X100) and incubated overnight at 4 °C. After washing with PBST, secondary antibodies were incubated for 1 hour at room temperature (RT). Stained samples were observed using a confocal microscope, FV-3000 (Olympus). The immunofluorescence reagents used in this study are listed in Supplementary Table 1.
Induction rates
We adopted a quantitative immunofluorescence analysis to verify the NPC induction rate. NPCs induced on day 9 or 13 were seeded in 96-well plates at 3–5 × 104 cells/well and incubated for three hours. Then, fixed NPCs were stained by anti-WT1 or anti-PAX2 antibodies listed in Supplementary Table 1. Immunofluorescence data were obtained using a BZ-X700 and analyzed by a BZ-X Analyzer (KEYENCE).
Preparation of CRISPR sgRNA by in vitro transcription (IVT)
We designed CRISPR-Cas9 gRNA for the c.1634 G > A (p.G545D) missense mutation on the COL4A5 gene and performed ssODN-mediated homologous recombination based on a previously published protocol54,55. The PCR template for IVT reaction to prepare gRNA was prepared using the following two primers: forward, 5′-GAAATTAATACGACTCACTATAGGCCTGGATCTAAAGATGAACCGTTTTAGAGCTAGAAATAGCAAG-3′; and reverse, 5′-AAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3′. The purified PCR product was used for the IVT reaction using the MEGAshortscript™ T7 Transcription Kit (Thermo Fisher Scientific) for 6 hours at 37 °C. Then, 2 µL of TURBOTM DNase (Thermo Fisher Scientific) was added and incubated for 30 min at 37 °C to remove the DNA template. The sgRNA produced by the IVT reaction was purified using the QIAGEN RNeasy MinElute Cleanup Kit (QIAGEN).
CRISPR-Cas9- and ssODN-mediated correction of COL4A5 mutation in iPSCs
For electroporation, semi-confluent iPSCs (mild AS patient) cultured with StemFit AK02N media in iMatrix-511-coated six-well plates were washed with 2 mL PBS and incubated with 0.5 mL TrypLE Select for 10 min at 37 °C. The cells were then detached from the plate by pipetting and transferred to a 1.5 mL tube containing StemFit AK02N media with 10 µM Y-27632 (Wako). After counting the number of cells, 3 × 105 cells were transferred to a 1.5 mL tube per sample and centrifuged for 5 min at 120 × g. The cell pellet was resuspended in 20 µL P4 Primary Cell Nucleofector Solution from a P4 Primary Cell 4D-Nucleofector Kit (Lonza). For RNP electroporation, 5 µg of recombinant SpCas9 protein (Thermo Fisher Scientific) and 1.25 µg of sgRNA prepared by the IVT reaction (as described above) were incubated for 5 min at RT and then added to the cell suspension with 6 µg of ssODN (5′-TCAGGGCATTCCAGGAGCTCCAGGTGCTCCAGGCTTTCCTGGATCTAAAGgTGAACCTGGTGATATCCTCACTTTTCCAGGAATGAAGGGTGACAAAGGA-3′). The cell suspension was transferred to a Nucleocuvette Strip and electroporated using the CA-137 protocol of 4D-Nucleofector. Then, the cells were cultured in iMatrix-511-coated six-well plates containing 2 mL of AK02N with 10 µM Y-27632 per well. Three to 7 days post-transfection, some cells were expanded for further culture, and the other cells were harvested for genomic DNA extraction. The target region on the COL4A5 gene was PCR amplified from the genomic DNA using the forward primer 5′-CATGCCTCACTTGATTCAGCC-3′ and reverse primer 5′-CAGCATCAGTCCCATCCTTTG-3′ for Sanger sequencing. After confirmation of the desired genetic correction of the bulk cells by Sanger sequencing, we isolated subclones by limiting dilution and established several corrected subclones confirmed by Sanger sequencing.
Transmission electron microscopy
After washing with PBS, day 13 + 28 kidney organoids and mouse embryonic kidneys were fixed with 1.4% PFA/1% glutaraldehyde/0.1 M PB overnight at 4 °C. Then, the organoids were washed with isotonic phosphate-buffered sucrose, refixed with phosphate-buffered 1% OsO4, dehydrated through a graded series of ethanol solutions, and embedded in Luveak 812 (Nacalai Tesque). Thin sections (70–90 nm thick) were cut with a diamond knife on an EM UC7 ultramicrotome (Leica), stained with uranyl acetate and lead citrate, and observed using a JEM-1400Flash electron microscope (JEOL).
Real-time quantitative RT-PCR (qRT-PCR)
Kidney organoids were harvested and lysed in buffer RLT using BioMasher II (Nippi). Total RNA was extracted using an RNeasy Kit (QIAGEN) according to the manufacturer’s protocol. cDNA was synthesized using ReverTra Ace (TOYOBO), and qPCR was performed with SYBR Green PCR Master Mix (Takara) using QuantStudio 3 (Thermo Fisher Scientific). The PCR reactions were performed in triplicate for each sample. The primer sequences used in this study are listed in Supplementary Table 2.
RNA sequencing
For RNA sequencing, total RNA was extracted as described above. The samples preserved at −80 °C were shipped and analyzed by DNAFORM. The quality of total RNA was evaluated by a Bioanalyzer (Agilent) to ensure over 8.0 RIN (RNA integrity number) or by electrophoresis waveforms. Double-stranded cDNA libraries (RNA-seq libraries) were prepared using a SMART Seq Stranded Kit (Clontech) according to the manufacturer’s protocols. RNA-seq libraries were sequenced using paired-end reads (50 nt of read 1 and 25 nt of read 2) on a NextSeq 500 (Illumina). Obtained reads were mapped to the human GRCh38 genome analyzed by STAR (version 2.7.3a). Annotated reads were counted using featureCounts (version 2.0.1) and RSEM (version 1.3.1). FPKM values were calculated from mapped reads by normalizing to total counts. The gene expression heatmap was drawn based on the log2 fold change (log2FC) compared with day 13 + 4 kidney organoid samples using FPKM.
Flow cytometry
Kidney organoids were dissociated in TrypLE Select Enzyme (Thermo Fisher Scientific) for approximately 30 min at 37 °C. The cell suspension was incubated with primary antibodies in 2% fetal bovine serum (FBS; Wako)/PBS for 30 min on ice. After washing, the cell suspension was incubated with secondary antibodies for 30 min on ice. Antibody-labeled cells were resuspended with 2% FBS/PBS containing 4’,6-diamidino-2-phenylindole (DAPI; Sigma). Flow cytometry was performed using a BD FACSAria II (BD Biosciences). The FACS Diva (BD) software program was used to analyze the data. The cells stained with isotype control and secondary antibodies were used as a negative control. Gating was set such that DAPI(−) live negative control cells had a positive fraction of less than 1%.
Statistics and reproducibility
All quantitative data are represented as the mean ± SE. Student’s t test was used to compare the means of two groups, and Dunnett’s test or Tukey–Kramer test was used for multiple comparisons among groups. p < 0.05 was considered statistically significant for all analyses. *p < 0.05, **p < 0.01, and ***p < 0.001 in the figures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The NCBI GEO accession number for the RNA sequencing data in this paper is GSE236314. All other data are available from the corresponding author upon reasonable request. The source data for the graphs are available in Supplementary Data 1.
References
Barker, D. et al. Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science 248, 1224–1227 (1990).
Lemmink, H. H. et al. Mutations in the type IV collagen α3 (COL4A3) gene in autosomal recessive Alport syndrome. Hum. Mol. Genet. 3, 1269–1273 (1994).
Jefferson, J. A. et al. Autosomal dominant Alport syndrome linked to the type IV collage a3 and a4 genes (COL4A3 and COL4A4). Nephrol. Dial. Transplant. 12, 1595–1599 (1997).
Bekheirnia, M. R. et al. Genotype–phenotype correlation in X-linked Alport syndrome. JASN 21, 876–883 (2010).
Harvey S.J., et al. Transfer of the α5(IV) collagen chain gene to smooth muscle restores in vivo expression of the α6(IV) collagen chain in a canine model of alport syndrome. Am. J. Pathol. 162, 13 (2003).
Kobayashi, T. & Uchiyama, M. Characterization of assembly of recombinant type IV collagen α3, α4, and α5 chains in transfected cell strains. Kidney Int. 64, 1986–1996 (2003).
Lennon, R. et al. Global analysis reveals the complexity of the human glomerular extracellular matrix. JASN 25, 939–951 (2014).
Hashimura, Y. et al. Milder clinical aspects of X-linked Alport syndrome in men positive for the collagen IV α5 chain. Kidney Int. 85, 1208–1213 (2014).
Savva, I., Pierides, A. & Deltas, C. RAAS inhibition and the course of Alport syndrome. Pharmacol. Res. 107, 205–210 (2016).
Stock, J. et al. Prospective study on the potential of RAAS blockade to halt renal disease in Alport syndrome patients with heterozygous mutations. Pediatr. Nephrol. 32, 131–137 (2017).
Kalluri, R., Shield, C. F., Todd, P., Hudson, B. G. & Neilson, E. G. Isoform switching of type IV collagen is developmentally arrested in X-linked Alport syndrome leading to increased susceptibility of renal basement membranes to endoproteolysis. J. Clin. Invest. 99, 2470–2478 (1997).
Miner, J. H. Renal basement membrane components. Kidney Int. 56, 2016–2024 (1999).
Naylor et al. of the glomerular basement membrane. Nat. Rev. Nephrol. 17, 112–127 (2021).
Miner, J. H. & Sanes, J. R. Collagen IV α3, α4, and α5 chains in rodent basal laminae: sequence, distribution, association with laminins, and developmental switches. J. Cell Biol. 127, 13 (1994).
Kuroda, N. et al. Expression of type IV collagen in the developing human kidney. Pediatr. Nephrol. 12, 554–558 (1998).
Söder, S. & Pöschl, E. The NC1 domain of human collagen IV is necessary to initiate triple helix formation. Biochem. Biophys. Res. Commun. 325, 276–280 (2004).
Kashtan, C. E. Alport syndromes: phenotypic heterogeneity of progressive hereditary nephritis. Pediatr. Nephrol. 14, 0502–0512 (2000).
Wei, G. et al. Spectrum of clinical features and type IV collagen -chain distribution in Chinese patients with Alport syndrome. Nephrol. Dialysis Transplant. 21, 3146–3154 (2006).
Rheault M. N. et al. Mouse model of X-linked Alport syndrome. J. Am. Soc. Nephrol. 15, 1466–74 (2004).
Hashikami, K. et al. Establishment of X-linked Alport syndrome model mice with a Col4a5 R471X mutation. Biochem. Biophys. Rep. 17, 81–86 (2019).
Nikolaou, S. & Deltas, C. A comparative presentation of mouse models that recapitulate most features of alport syndrome. Genes 13, 1893 (2022).
Taguchi, A. et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14, 53–67 (2014).
Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015).
Morizane, R. et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200 (2015).
Tsujimoto, H. et al. A modular differentiation system maps multiple human kidney lineages from pluripotent stem cells. Cell Rep. 31, 107476 (2020).
Lohi, J. et al. Expression of type IV collagen α1(IV)–α6(IV) polypeptides in normal and developing human kidney and in renal cell carcinomas and oncocytomas. Int. J. Cancer 72, 43–49 (1997).
Hale, L. J. et al. 3D organoid-derived human glomeruli for personalised podocyte disease modelling and drug screening. Nat. Commun. 9, 5167 (2018).
Morais, M. R. et al. Kidney organoids recapitulate human basement membrane assembly in health and disease. eLife 11, e73486 (2022).
Okita, K. et al. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 31, 458–466 (2013).
Nakagawa, M. et al. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci. Rep. 4, 3594 (2014).
Xu, H. et al. Targeted disruption of HLA genes via CRISPR-Cas9 generates iPSCs with enhanced immune compatibility. Cell Stem Cell 24, 566–578.e7 (2019).
Qian, X. et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254 (2016).
Giandomenico, S. L., Sutcliffe, M. & Lancaster, M. A. Generation and long-term culture of advanced cerebral organoids for studying later stages of neural development. Nat. Protoc. 16, 579–602 (2021).
Hinglais, N., Grünfeld, J. P. & Bois, E. Characteristic ultrastructural lesion of the glomerular basement membrane in progressive hereditary nephritis (Alport’s syndrome). Lab. Invest. 27, 473–487 (1972).
Spear, G. S. & Slusser, R. J. Alport’s syndrome. Emphasizing electron microscopic studies of the glomerulus. Am. J. Pathol. 69, 213–224 (1972).
Rumpelt, H. J., Langer, K. H., Schärer, K., Straub, E. & Thoenes, W. Split and extremely thin glomerular basement membranes in hereditary nephropathy (Alport’s syndrome). Virchows Arch. A Pathol. Anat. Histol. 364, 225–233 (1974).
Abrahamson, D. R. Origin of the glomerular basement membrane visualized after in vivo labeling of laminin in newborn rat kidneys. J. Cell Biol. 100, 1988–2000 (1985).
Morrissey, M. A. & Sherwood, D. R. An active role for basement membrane assembly and modification in tissue sculpting. J. Cell Sci. 128, 1661–1668 (2015).
Fukuda, R. et al. Podocyte p53 limits the severity of experimental alport syndrome. JASN 27, 144–157 (2016).
Ning, L., Suleiman, H. Y. & Miner, J. H. Synaptopodin deficiency exacerbates kidney disease in a mouse model of Alport syndrome. Am. J. Physiol. Ren. Physiol. 321, F12–F25 (2021).
Wongtrakul P. et al. Immunohistochemical study for the diagnosis of Alport’s syndrome. J. Med. Assoc. Thai. 89, 11 (2006).
Yamamura, T. et al. Genotype-phenotype correlations influence the response to angiotensin-targeting drugs in Japanese patients with male X-linked Alport syndrome. Kidney Int. 98, 1605–1614 (2020).
Bantounas, I. et al. Generation of functioning nephrons by implanting human pluripotent stem cell-derived kidney progenitors. Stem Cell Rep. 10, 766–779 (2018).
Wang, D. et al. The chemical chaperone, PBA, reduces ER stress and autophagy and increases collagen IV α5 expression in cultured fibroblasts from men with X-linked alport syndrome and missense mutations. Kidney Int. Rep. 2, 739–748 (2017).
Omachi, K. et al. A split-luciferase-based trimer formation assay as a high-throughput screening platform for therapeutics in alport syndrome. Cell Chem. Biol. 25, 634–643.e4 (2018).
Murray, L. S. et al. Chemical chaperone treatment reduces intracellular accumulation of mutant collagen IV and ameliorates the cellular phenotype of a COL4A2 mutation that causes haemorrhagic stroke. Hum. Mol. Genet. 23, 283–292 (2014).
Gawlik, K. I. & Durbeej, M. Skeletal muscle laminin and MDC1A: pathogenesis and treatment strategies. Skelet. Muscle 1, 9 (2011).
Kashtan, C. E. & Kim, Y. Distribution of the α1 and α2 chains of collagen IV and of collagens V and VI in Alport syndrome. Kidney Int. 42, 115–126 (1992).
Miner, JeffreyH., Sanes & Joshua, R. Molecular and functional defects in kidneys of mice lacking collagen ot3(IV): implications for alport syndrome. J. Cell Biol. 135, 1403–1413 (1996).
Gross, O. et al. Loss of collagen-receptor DDR1 delays renal fibrosis in hereditary type IV collagen disease. Matrix Biol. 29, 346–356 (2010).
Kim, J.-J. et al. Discoidin domain receptor 1 activation links extracellular matrix to podocyte lipotoxicity in Alport syndrome. EBioMedicine 63, 103162 (2021).
Funk, S. D., Bayer, R. H. & Miner, J. H. Endothelial cell-specific collagen type IV-α 3 expression does not rescue Alport syndrome in Col4a3 −/− mice. Am. J. Physiol. Ren. Physiol. 316, F830–F837 (2019).
Beers, J. et al. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat. Protoc. 7, 2029–2040 (2012).
Kagita, A. et al. Efficient ssODN-mediated targeting by avoiding cellular inhibitory RNAs through precomplexed CRISPR-Cas9/sgRNA ribonucleoprotein. Stem Cell Rep. 16, 985–996 (2021).
Xu, H., Kita, Y., Bang, U., Gee, P. & Hotta, A. Optimized electroporation of CRISPR-Cas9/gRNA ribonucleoprotein complex for selection-free homologous recombination in human pluripotent stem cells. STAR Protoc. 2, 100965 (2021).
Acknowledgements
The authors thank Drs. Keiko Okamoto-Furuta and Haruyasu Kohda, Division of Electron Microscopic Study, Center for Anatomical Studies, Graduate School of Medicine, Kyoto University, for technical support with the electron microscopy, Dr. Kanae Mitsunaga, CiRA, Kyoto University, for the flow cytometry analysis, Dr. Naoki Kojima, Taisho Pharmaceutical Co., Ltd., for helpful discussions, and Dr. Peter Karagiannis for critically reading and revising the manuscript. This study was supported by Taisho Pharmaceutical Co., Ltd., the Japan Agency for Medical Research and Development (AMED) under Grant numbers JP23bm1123002 and JP22bm0804013 to K. Osafune, and the iPS Cell Research Fund.
Author information
Authors and Affiliations
Contributions
R.H. carried out all primary analyses and wrote and edited the paper. K.W. and A.H. generated the mutation-corrected iPSC line and wrote parts of the paper. K.T., T.O., T.A., and S.I.M. performed some experiments and analyzed the data. T.H., T.Y., K.I., and K.N. provided PBMCs to the AS patients. K. Okita established the patient’s iPSCs. K. Osafune supervised the project and wrote and edited the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
R.H. is an employee of Taisho Pharmaceutical Co., Ltd. T.O. is an employee of Ono Pharmaceutical Co., Ltd. K. Osafune is a founder and member of the scientific advisory boards of iPS Portal, Inc., and a founder and chief scientific advisor of RegeNephro Co., Ltd. T.A. is a founder and scientific advisor of RegeNephro Co., Ltd. S.I.M. is a scientific advisor of RegeNephro Co., Ltd. R.H., T.A. and K. Osafune are the inventors of the iPSC-based disease model for AS (patent application 2021–184043). The other authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Ryuji Morizane and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Simona Chera and Manuel Breuer.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hirayama, R., Toyohara, K., Watanabe, K. et al. iPSC-derived type IV collagen α5-expressing kidney organoids model Alport syndrome. Commun Biol 6, 854 (2023). https://doi.org/10.1038/s42003-023-05203-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-023-05203-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.