Abstract
Interleukins (ILs), key cytokine family of inflammatory response, are closely associated with kidney function. However, the causal effect of various ILs on kidney function needs further investigation. Here we show two-sample summary-level Mendelian randomization (MR) analysis that examined the causality between serum IL levels and kidney function. Genetic variants with strong association with serum IL levels were obtained from a previous genome-wide association study meta-analysis. Summary-level data for estimated glomerular filtration rate (eGFR) were obtained from CKDGen database. As a main MR analysis, multiplicative random-effects inverse-variance weighted method was performed. Pleiotropy-robust MR analysis, including MR-Egger with bootstrapped error and weighted median methods, were also implemented. We tested the causal estimates from nine ILs on eGFR traits. Among the results, higher genetically predicted serum IL-1 receptor antagonist level was significantly associated with higher eGFR values in the meta-analysis of CKDGen and the UK Biobank data. In addition, the result was consistent towards eGFR decline phenotype of the outcome database. Otherwise, nonsignificant association was identified between other genetically predicted ILs and eGFR outcome. These findings support the clinical importance of IL-1 receptor antagonist-associated pathway in relation to kidney function in the general individuals, particularly highlighting the importance of IL-1 receptor antagonist.
Similar content being viewed by others
Introduction
The kidneys are vital organs that contribute to various biological processes. Various factors control kidney function, including cytokines1, neurohormonal responses2, and vascular regulations3. The substantial socioeconomic burden of kidney dysfunction4 warrants investigating the potential therapeutic or preventive effects of each factor on the impairment of kidney function.
Among the diverse regulators of glomerular filtration rates, inflammatory cytokines, including interleukins (ILs), directly affect kidney function5,6. Since the development and progression of major kidney diseases are linked to inflammation, the effect of these cytokines on kidney function has been shown in certain disease conditions. For instance, IL-1 and IL-6 are associated with kidney fibrosis and the decline of kidney function7,8, and IL-18 has been proposed to have detrimental effects on ischemic kidney diseases9,10. Several types of ILs are targeted as immunomodulating agents, including a recombinant IL-1 receptor antagonist (IL-1ra), which demonstrated rapid and sustained reduction of the inflammatory response in inflammation-mediated organ dysfunction (e.g., heart failure and stroke)11.
Mendelian randomization (MR) analysis is a method to estimate the causal effect of exposure on complex traits, overcoming the limitations of conventional observational studies by being less likely to be affected by confounding and reverse causation12. Furthermore, MR studies can provide valuable evidence to support the appropriate use of drugs in chronic kidney disease (CKD) patients. A previous MR study that did not identify a significant association between IL-6 inhibition and kidney function suggested MR evidence support the safe administration of tocilizumab, an IL-6 inhibitor, in patients with renal impairment13. Quantitative trait locus (QTL) studies enabled the discovery of genetic variants that affect a quantitative change of a particular trait, including the expression of one or more genes (eQTL) or protein (pQTL)14,15. Performing MR analysis with QTL instruments confers insight into identifying candidate genes and molecular targets for diseases by demonstrating the causal effect between gene and protein expression on traits16,17,18.
In this study, we performed MR analysis to examine the causal effects of certain ILs on kidney function. We selected genetic instruments that satisfied two criteria (cis-pQTL and cis-eQTL) and were robustly associated with serum IL levels. We tested the causal estimates in multiple population-scale genetic datasets of kidney function traits, hypothesizing that certain IL-associated molecules would significantly affect kidney function.
Results
Characteristics of the data sources
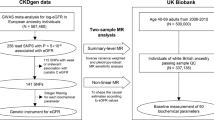
We performed a two-sample summary-level MR analysis of the serum IL levels and kidney function traits. The study flow diagram is presented in Fig. 1. The summary statistics of kidney function were obtained from the UK Biobank (UKB) and CKDGen genome-wide association study (GWAS) meta-analysis19,20,21. The mean age of the UKB participants was 57.6 ± 8.5 years and ~6.2% of the participants had CKD22. The median age of CKDGen GWAS meta-analysis participants was 54 years, and the median of mean estimated glomerular filtration rate (eGFR) values were 89 (interquartile range 81, 94) ml/min/1.73 m2 21. In the GWAS meta-analysis of eGFR decline, the median annual eGFR decline across studies was 1.32 ml/min/1.73 m2 per year19.
The study included summary-level Mendelian randomization analysis testing the causal effects between circulating interleukin (IL) level and kidney function. The cis-genetic instruments for genetically predicted serum IL levels, including cis-pQTL and cis-eQTL instruments, were developed from the previous genome-wide association study (GWAS) meta-analysis including 38,424 participants. Among 376 SNPs, 80 SNPs of nine ILs were utilized for the Mendelian randomization analysis. The summary statistics for kidney function traits were obtained from four respective GWAS meta-analysis datasets: the meta-analysis for creatinine-based log-eGFR values of CKDGen and UKB (n = 1,201,930), creatinine-based log-eGFR values from the phase 4 CKDGen study (n = 765,348), cystatin-C-based log-eGFR values from CKDGen and UKB (n = 460,826), and degree of annual eGFR decline from CKDGen and UKB (n = 343,339). Below the boxes of each dataset, it is indicated whether the datasets were analyzed as “Main analysis” or “Validation”. UKB UK Biobank, SNP single nucleotide polymorphism, GWAS genome-wide association study, eGFRcr estimated glomerular filtration rate by creatinine, eGFRcys estimated glomerular filtration rate by cystatin-C.
MR analysis of serum IL-1ra levels on kidney function
Among the ILs, genetically predicted serum IL-1ra levels showed a consistently significant association (P < 0.05) with eGFR, which was also supported by the significant (P < 0.05) pleiotropy-robust MR results (Table 1). Using 20 single nucleotide polymorphisms (SNPs; 18 pQTL and two eQTL), higher serum IL-1ra levels were significantly associated (P < 0.05) with higher eGFR in the multiplicative random-effects inverse-variance weighted (IVW) analysis (Fig. 2). In the MR analysis with log-transformed creatinine-based eGFR outcome statistics provided by CKDGen and UKB datasets, the eGFR increased by 0.28 % (standard error [SE], 0.11 %; P value, 0.009) as genetically predicted standard deviation increase in serum IL-1ra concentration. The association was also consistent when the MR analyses were performed against the other two eGFR outcome statistics: creatinine-based and cystatin-C-based eGFR. Pleiotropy-robust MR sensitivity analyses, including MR-Egger with bootstrapped error and the weighted median method, supported these findings. The nonsignificant P value of the intercept in MR-Egger indicated the absence of pleiotropic effects in the above findings. Furthermore, when we applied MR analysis toward the degree of annual eGFR decline, the result showed a significant finding, indicating that genetically predicted higher IL-1ra levels were significantly associated (P < 0.05) with a lower degree of annual eGFR decline. When we performed MR analysis with annual eGFR decline summary statistics from CKDGen and UKB datasets, the annual eGFR decline decreased by 2.18 % (SE, 1.09 %; P value, 0.043) a genetically predicted standard deviation increase in serum IL-1ra concentration. The leave-one-out analysis results demonstrate the absence of a notable outlier effect in the calculated causal estimates (Fig. 3).
The causal estimates were derived from the UK Biobank and CKDGen genome-wide association study meta-analysis for log-transformed creatinine-based eGFR. The shapes indicate beta coefficients per standard deviation change in corresponding serum interleukin level, and error bars indicate the 95% confidence interval. a The rhombus, square, and circle shapes indicate estimates calculated from weighted median, MR-IVW, and MR-Egger, respectively. b The triangle, rhombus, square, and circle shapes indicate estimates calculated from the weighted median, ratio of coefficient, MR-IVW, and MR-Egger, respectively. eGFR estimated glomerular filtration rate, IL interleukin, 95% CI 95% confidence interval, MR-IVW multiplicative random-effects inverse-variance weighted.
a Scatter plot of the effect sizes of the SNP-serum IL-1ra level association (x axis, SD units) against the SNP-log-eGFRcr associations (y axis, beta) with standard error bars. The slopes of the lines correspond to the causal estimates per method. b Leave-one-out sensitivity analysis. Each black point represents the MR-IVW method applied to estimate the causal effect of serum IL-1ra level on kidney function excluding that certain variant from the analysis. The red point indicates the IVW estimate using all SNPs. There are no instances where the exclusion of a single SNP leads to dramatic changes in the overall result. Outcome statistics were obtained from the UK Biobank and CKDGen genome-wide association study meta-analysis of log-eGFRcr. MR Mendelian randomization, IL interleukin, SNP single nucleotide polymorphism.
MR analysis of other serum IL levels on kidney function
The results of the summary-level MR analyses of the eight types of ILs (except IL-1ra) are summarized in Supplementary Table 1. There was a significant association (P < 0.05) between genetically predicted higher serum IL-2ra levels and higher eGFR in the multiplicative random-effects IVW model; however, this association was not replicated by pleiotropy-robust MR analyses (Fig. 2). MR analyses did not reveal causal association between genetically predicted serum IL levels (for IL-1α, IL-16, and IL-18) and eGFR. We performed a ratio of coefficient analysis with IL-6, IL-7, and IL-12p70, each provided with a single genetic instrument; however, no association between the serum IL concentrations with a single instrumented variant and eGFR were observed. The MR estimate for IL-6 was calculated using single genetic instrument (rs7808457), because rs57349960 was not included in the outcome GWAS database.
Sensitivity analysis of serum IL levels on kidney function
In the sensitivity analysis excluding cis-eQTLs (Supplementary Table 2) and palindromic SNPs (Supplementary Table 3), we found that the MR estimates for genetically predicted IL-1ra levels were consistent with the main analysis, which revealed significant associations with eGFR and eGFR decline.
Discussion
In this MR study, we investigated the causal association between genetically predicted serum IL concentrations and eGFR, using cis-QTL instruments and GWAS summary statistics. These results demonstrate a significant causal effect of serum IL-1ra levels on kidney function. Higher genetically predicted serum IL-1ra levels were significantly associated with a higher eGFR in the two population-scale genetic datasets, consistent with the results of the main MR analysis and various MR sensitivity analyses. Genetically predicted serum IL-1ra levels were also associated with the degree of annual eGFR decline, suggesting that higher IL-1ra levels ameliorate the rate of eGFR decline.
Because many kidney diseases are related to immune responses, understanding the mechanisms of circulating cytokines, including ILs, and their effect on the kidney microenvironment is crucial for preventing and treating kidney diseases23. The association between IL profiles and kidney function has been suggested in previous cross-sectional observational studies5,6. Moreover, several experimental and clinical studies are being conducted to find molecular targets that have immune-modulating effects in patients with kidney disease8,9,10,24. However, causality could not be inferred due to methodological limitations, and the roles of ILs related to general kidney function are still not fully understood. Our study found MR evidence of the protective effect of IL-1ra, a naturally occurring IL-1 receptor antagonist, demonstrating a significant association with eGFR.
Our study has several strengths. First, we performed MR analysis, which is less affected by bias from confounding effects or reverse causation and can provide evidence of a causal association between genetic predisposition to IL levels and kidney function. Second, we integrated the cis-pQTL and cis-eQTL information of ILs with summary-level outcome data from GWAS in the MR framework to estimate the causal effect of serum IL levels and the corresponding gene expression on kidney function traits. This further increased the power of MR estimates in identifying trait-associated loci, as well as reducing horizontal pleiotropy25,26. As genetic variants encoding a drug target have a similar effect as pharmacological agents that target this gene location, our findings may suggest a potential drug target for modifying kidney diseases27. Third, various types of ILs that are known to be associated with the pathogenesis of kidney diseases were included in the study5,6. Fourth, the MR results of IL-1ra were consistently validated even when we analyzed the association between IL-1ra and eGFR using four different outcome statistics on kidney traits, providing evidence for the protective effect of serum IL-1ra levels on kidney function.
IL-1 is a key pro-inflammatory cytokine that activates immune cells and promotes secretion of downstream cytokines including IL-6 and TNF-a, further driving amplification of innate immunity and inflammation in various organs and tissues28,29,30. Currently, drugs targeting IL-1, including recombinant IL-1 receptor antagonist (anakinra), monoclonal IL-1β antibodies (canakinumab), and IL-1 traps (rilonacept) are approved for the treatment of inflammation-mediated diseases, including arthritis, gout, type 2 diabetes, and heart failure11. In the kidney, activated IL-1 pathway causes cell stress, tissue damage, and fibrosis, eventually leading to loss of kidney function8. Kidney dysfunction may also be contributed by systemic release of IL-1 in lupus or diabetes that promotes leukocyte adhesion and vascular leakage in the glomeruli31. Experimental evidence suggests anti-hypertensive, anti-fibrotic effects of IL-1 receptor blockade in kidney diseases32,33. Our observations are in line with the findings of these experimental studies, elucidating the important role of IL-1 pathway in kidney function impairment. Furthermore, in a clinical trial of IL-1 inhibitors that primarily investigated the effect of an IL-1β inhibitor for lowering the risk of hypertension and major adverse cardiovascular events, significant benefits for preventing adverse cardiovascular events were observed in a subgroup of patients with mild CKD34. The current study’s findings provide MR evidence supporting the use of IL-1 blockade as a potential therapeutic target aimed at preserving kidney function or mitigating the adverse events associated with the deterioration of kidney function among individuals without pre-existing kidney function abnormalities.
Another valuable insight produced by implementing MR analysis between genetically proxied IL-1ra and kidney function, in addition to the identification of the underlying mechanisms of cytokine-associated inflammatory pathways and kidney function, is that the evidence for the safe administration of pharmacological agents in CKD patients could be provided by MR analysis. For example, in the previous MR study that investigated the association between genetically proxied IL-6 inhibition and kidney function, no significant causal association was found between IL-6 inhibition and kidney function. This finding supports that pharmacological IL-6 inhibition is unlikely to have a direct adverse effect on kidney function13. Given that patients with CKD are excluded from up to 75% of all randomized-controlled trials35, the findings from the MR study can provide reliable evidence for the appropriate use of IL inhibitors in CKD patients. According to studies on IL-1 inhibitors, including the CANTOS or IL-1ra Arthritis Study36,37, no dose adjustment has been suggested for patients with renal impairment. However, it should be noted that this drug has not been studied in patients with moderate or severe kidney dysfunction. Further clinical trials are necessary to investigate the safety of IL-1 inhibitors in patients with impaired kidney function, and the current study suggests the possible safe administration of IL-1 inhibitors in CKD patients by providing MR evidence.
Our study has some limitations. First, the number of genetic instruments was insufficient for some ILs, which restricted the type of sensitivity analysis that could be performed. Although the strengths of the instruments were evaluated using F-statistics and proportion of the explained variance (r2)38, there were weak-powered instruments with F-statistics <10. It should be noted that there is potential for false-negative bias, particularly when the true effect is below the minimum effect size (Supplementary Table 4). Thus, one may not preclude the possibility of a kidney function effect based on the null findings in this study. Specifically, we could expect that the IL-1α, which agonizes the IL-1 receptor, may produce an inverse effect on kidney function. However, the current study could not support the inverse association between serum IL-1α and kidney function. This may be explained by the broader biological function of IL-1ra than that of IL-1α, including the blockade of IL-1 receptors from binding with IL-1 agonists39. However, the difference in the statistical power may have caused the discrepancy. Second, the effect estimates of MR analysis are for lifetime exposure to genetic predisposition and does not consider temporal and spatial fluctuations in gene expression in tissues. Additionally, as the overall effect sizes in the results were small, a transient change with a higher degree of ILs may have different effects on kidney function. Third, the generalizability is limited as the analysis was mainly based on European ancestry samples.
In conclusion, we examined the causal effects of various ILs on kidney function using MR analysis and found a positive association between genetically predicted IL-1ra levels and kidney function traits. Based on our MR results, IL-1ra may be highlighted for its effects on eGFR changes in the general population. However, further studies are required to ascertain the specific biological pathways involving ILs, particularly IL-1ra, in kidney function.
Methods
Ethics and inclusion statement
This study was approved by the Institutional Review Board (IRB) of the Seoul National University Hospital (IRB number: E-2301-066-1394). The summary statistics of the kidney function traits are from the public domain (http://ckdgen.imbi.uni-freiburg.de/) of CKDGen GWAS meta-analysis database for kidney function traits. The requirement for informed consent was waived because the study analyzed anonymous public data and summary statistics.
Study setting
The current study is a two-sample summary-level MR analysis to examine the association between serum IL levels and kidney function. The genetic instruments for serum IL levels were developed from the previous study40. The outcome GWAS summary statistics for kidney function traits were obtained from the CKDGen consortium, which is widely used for MR analysis towards kidney function traits41,42,43. As a main analysis, we assessed the causal effects of circulating concentrations of nine types of IL, including IL-1α, IL-1ra, IL-2ra, IL-6, IL-7, IL-8, IL-12p70, IL-16, and IL-18, on kidney function using summary-level MR analyses. The examined ILs were previously reported to be associated with the pathogenesis of kidney inflammation, fibrosis, and injury8,31,39,44,45,46,47,48,49. Then, we also performed sensitivity analyses to ascertain the robustness of our findings50.
Genetic instrument for MR analysis
Genetic instruments were developed from a previous GWAS meta-analysis study including genetic information of 47 inflammatory cytokines40. Using these genetic instruments, a total of 80 cis-instruments (60 as cis-pQTL and 20 as cis-eQTL) that are strongly associated with the effect of circulating IL concentration were developed (Supplementary Tables 5, 6)40. Two cis-instrumental selection criteria were used to identify variants that reflected the effect of circulating cytokine levels and are described in Supplementary Note 1. To mitigate the potential loss of causal variants in a cis-region MR study due to a small correlation threshold, clumping was performed with a pairwise linkage disequilibrium threshold of r2 < 0.1.
Kidney function outcome in the MR analysis
The CKDGen consortium provides a publicly available database for GWAS and meta-analyses of kidney function (https://ckdgen.imbi.uni-freiburg.de). Because the genetic instruments were developed from individuals of European ancestry in the SCALLOP consortium and the Finnish population, we used the summary statistics of individuals of European ancestry and used the data as the outcome statistics in our two-sample MR analysis. We used two independent datasets, the CKDGen consortium data, and the UKB, as outcome datasets for log-transformed eGFR values because validation analysis is crucial for the validity of a genetic study.
We used three eGFR outcome datasets as eGFR values are a current standard parameter for the assessment of kidney function; (1) the meta-analysis for creatinine-based log-eGFR values of the CKDGen and UKB data from 2021 (n = 1,201,930) which has strength as this is the currently largest GWAS meta-analysis for eGFR trait20, (2) creatinine-based log-eGFR values from the phase 4 CKDGen study from 2019 (n = 765,348)21, and (3) cystatin-C-based log-eGFR values from the CKDGen and UKB (n = 460,826) from 202120, which has particular strength in that the dataset can be used to test the causal estimates towards cystatin-C-based eGFR values which is less affected from non-kidney factors than creatinine-based levels51. The main analysis was performed using the largest GWAS meta-analysis of creatinine-based log-eGFR from CKDGen and UKB because of the greater statistical power and lower possibility of weak-instrumental bias. Nevertheless, considering the potential for healthy volunteer bias resulting from the relatively lower prevalence of CKD in the UKB dataset22, validation analyses employing the other two GWAS meta-analysis datasets were conducted. Additionally, the UKB dataset was independent of the CKDGen data, fulfilling an independent validation analysis, and was independent of the samples included in the GWAS for genetic instrument development. Such a two-sample MR without participant overlap has statistical power even if the results are affected by instrumental power because the potential bias would be towards false-negative findings. Therefore, a positive finding from such an independent two-sample MR is more likely to reflect true causal effects52.
Furthermore, to test whether IL levels have causal effects on accelerated eGFR decline rather than on a static value, we implemented MR analysis using a GWAS meta-analysis of the degree of annual eGFR decline from CKDGen and UKB from 2022 (n = 343,339) as another outcome dataset19.
MR assumptions
In the context of selecting the genetic instruments for circulating IL concentration and performing MR analysis, we considered the three assumptions of MR analysis to demonstrate valid causal effects53. First, the genetic instrument is robustly associated with the exposure of interest (the relevance assumption), second, the genetic instrument is independent of the confounding factors of the association between the exposure and the outcome of interest (the independence assumption), and third, the genetic effects occur only through its association with the exposure of interest (the exclusion-restriction assumption).
For the relevance assumption, genetic instruments provided from the previous meta-analysis results of the Finnish and SCALLOP GWAS were strongly associated with circulating cytokine concentrations at genome-wide significance (P < 5 × 10−8) divided by the number of cytokines analyzed40,53,54. In addition, the F-statistic and r2 for each genetic variant based on the circulating protein concentration were calculated as a measure of the strength of the instrument40,53,54,55.
Horizontal pleiotropy occurs when a genetic variant affects the outcome outside the exposure pathway56. For the independence and exclusion-restriction assumption, both of which require the absence of horizontal pleiotropy and possible confounder phenotype, pleiotropy-robust MR analysis methods were performed as sensitivity analysis50. The directions of the variants' causal effects were ascertained using MR Steiger filtering to establish that there is no direct effect of the genetic instruments on kidney function traits, which may potentially violate the exclusion-restriction assumption57. Steiger filtering results for each instrumental variant demonstrated evidence of causality in the expected direction.
Statistics and reproducibility
Two-sample summary-level MR analysis was performed using two sets of genetic instruments (cis-pQTLs and cis-eQTLs) for each IL on the four outcome datasets. First, we implemented a multiplicative random-effects IVW method when more than one genetic instrument was available to construct an instrumental variable for a given IL. Provided with only a single genetic instrument, the ratio of coefficient, also referred to as the Wald ratio, was applied to measure MR estimates58. Multiplicative random-effects model is the current standard for main MR analysis as the method allows balanced pleiotropic effects50,59. In addition, leave-one-out analysis, leaving one SNP out of the analysis at a time, was performed to inspect whether there was bias from an extreme outlier suspected with pleiotropic effect. The MR estimates were converted into the degree of change (percentage [SE]) of log-transformed eGFR according to the genetically predicted standard deviation in serum IL concentration to facilitate interpretation.
Sensitivity analysis for summary-level MR analysis
To explore the possibility of horizontal pleiotropy, pleiotropy-robust MR analyses that hypothesized the inclusion of pleiotropic variants were implemented as sensitivity analyses when ≥3 instrument variants were available. First, MR-Egger regression analysis with bootstrapped standard error provides a pleiotropy-robust causal estimate that is valid even if all genetic variants are invalid due to a violation of the independence assumption60. A significant MR-Egger intercept P value would raise the suspicion of directional pleiotropy. Second, the weighted median method provides a valid causal estimate even up to 50% of the weighted instruments are invalid61, and this method was implemented as another sensitivity MR analysis.
Other statistical aspects of the MR analysis
We performed two additional sensitivity MR analyses: (1) excluding cis-eQTL instruments and (2) excluding palindromic SNPs to test whether differences in instrument development methods or uncertain harmonization might have influenced the results. As cis-eQTL instruments are near the genomic location of the influenced genes that can influence the expression of multiple genes, potential pleiotropy due to cis-eQTL instruments can occur62,63. Therefore, sensitivity analysis with the exclusion of cis-eQTL instruments was implemented to assess the validity of the causal inference.
We considered the possibility of false-positive findings from multiple comparisons as the causal estimates were tested for multiple types of ILs. To address this issue, we performed multiple sensitivity analyses and only considered an MR result as a true positive signal when the significance threshold of two-sided P was lower than 0.05, across all the sensitivity analyses performed per IL.
The minimum effect size of each IL for a power of 80% was calculated in the application for calculating the post hoc power (https://shiny.cnsgenomics.com/mRnd/) using the meta-analysis dataset for log-eGFRcr of the CKDGen and UKB datasets and r2 values of the instrumental variables of each IL at a type-I error rate of 0.25.
The summary-level MR analysis was performed using the “TwoSampleMR” package in R (version 0.4.26).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data used in this work are presented in the Supplementary information that accompanies the manuscript and is available in the original publications. The data used in this study is publicly available on the consortium website of The CKDGen (URL: https://ckdgen.imbi.uni-freiburg.de/)
Code availability
This manuscript used public software those are available online. The names of the software are presented in the Methods and the detailed statistical codes can be found in the related resources.
References
Ramesh, G. & Reeves, W. B. Inflammatory cytokines in acute renal failure. Kidney Int. Suppl. S56–S61. https://doi.org/10.1111/j.1523-1755.2004.09109.x (2004)
Ye, S., Ozgur, B. & Campese, V. M. Renal afferent impulses, the posterior hypothalamus, and hypertension in rats with chronic renal failure. Kidney Int. 51, 722–727 (1997).
Bidani, A. K. & Griffin, K. A. Pathophysiology of hypertensive renal damage: implications for therapy. Hypertension 44, 595–601 (2004).
Golestaneh, L. et al. All-cause costs increase exponentially with increased chronic kidney disease stage. Am. J. Manag. Care 23, S163–s172 (2017).
Amdur, R. L. et al. Inflammation and progression of CKD: the CRIC study. Clin. J. Am. Soc. Nephrol. 11, 1546–1556 (2016).
Gupta, J. et al. Association between albuminuria, kidney function, and inflammatory biomarker profile in CKD in CRIC. Clin. J. Am. Soc. Nephrol. 7, 1938–1946 (2012).
Akchurin, O. et al. Interleukin-6 contributes to the development of anemia in juvenile CKD. Kidney. Int. Rep. 4, 470–483 (2019).
Bandach, I., Segev, Y. & Landau, D. Experimental modulation of Interleukin 1 shows its key role in chronic kidney disease progression and anemia. Sci. Rep. 11, 6288 (2021).
Nozaki, Y. et al. Signaling through the interleukin-18 receptor α attenuates inflammation in cisplatin-induced acute kidney injury. Kidney. Int. 82, 892–902 (2012).
Wu, H. et al. IL-18 contributes to renal damage after ischemia-reperfusion. J. Am. Soc. Nephrol. 19, 2331–2341 (2008).
Dinarello, C. A., Simon, A. & van der Meer, J. W. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat. Rev. Drug. Discov. 11, 633–652 (2012).
Smith, G. D. & Ebrahim, S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 32, 1–22 (2003).
Ryan, D. K., Karhunen, V., Walker, D. J. & Gill, D. Inhibition of interleukin 6 signalling and renal function: a Mendelian randomization study. Br. J. Clin. Pharmacol. 87, 3000–3013 (2021).
Yao, D. W., O'Connor, L. J., Price, A. L. & Gusev, A. Quantifying genetic effects on disease mediated by assayed gene expression levels. Nat. Genet. 52, 626–633 (2020).
Võsa, U. et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 53, 1300–1310 (2021).
Lawrenson, K. et al. Cis-eQTL analysis and functional validation of candidate susceptibility genes for high-grade serous ovarian cancer. Nat. Commun. 6, 8234 (2015).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Karhunen V, G. D., et al. Genetic study of circulating cytokines offers insight into the determinants, cascades and effects of systemic inflammation. medRxiv https://doi.org/10.1101/2020.10.26.20219477 (2020).
Gorski, M. et al. Genetic loci and prioritization of genes for kidney function decline derived from a meta-analysis of 62 longitudinal genome-wide association studies. Kidney Int. 102, 624–639 (2022).
Stanzick, K. J. et al. Discovery and prioritization of variants and genes for kidney function in >1.2 million individuals. Nat. Commun. 12, 4350 (2021).
Wuttke, M. et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet. 51, 957–972 (2019).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Anders, H. J. & Ryu, M. Renal microenvironments and macrophage phenotypes determine progression or resolution of renal inflammation and fibrosis. Kidney Int. 80, 915–925 (2011).
Widjaja, A. A. et al. A neutralizing IL-11 antibody improves renal function and increases lifespan in a mouse model of alport syndrome. J. Am. Soc. Nephrol. 33, 718–730 (2022).
Porcu, E. et al. Mendelian randomization integrating GWAS and eQTL data reveals genetic determinants of complex and clinical traits. Nat. Commun. 10, 3300 (2019).
Davey Smith, G. & Hemani, G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23, R89–R98 (2014).
Swerdlow, D. I. et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet 379, 1214–1224 (2012).
Dinarello, C. A. Biologic basis for interleukin-1 in disease. Blood 87, 2095–2147 (1996).
Garlanda, C., Dinarello, C. A. & Mantovani, A. The interleukin-1 family: back to the future. Immunity 39, 1003–1018 (2013).
Sims, J. E. & Smith, D. E. The IL-1 family: regulators of immunity. Nat. Rev. Immunol. 10, 89–102 (2010).
Anders, H. J. Of inflammasomes and alarmins: IL-1β and IL-1α in kidney disease. J. Am. Soc. Nephrol. 27, 2564–2575 (2016).
Lemos, D. R. et al. Interleukin-1β activates a MYC-dependent metabolic switch in kidney stromal cells necessary for progressive tubulointerstitial fibrosis. J. Am. Soc. Nephrol. 29, 1690–1705 (2018).
Ling, Y. H. et al. Anakinra reduces blood pressure and renal fibrosis in one kidney/DOCA/salt-induced hypertension. Pharmacol. Res. 116, 77–86 (2017).
Rothman, A. M. et al. Effects of interleukin-1β inhibition on blood pressure, incident hypertension, and residual inflammatory risk: a secondary analysis of CANTOS. Hypertension 75, 477–482 (2020).
Charytan, D. & Kuntz, R. E. The exclusion of patients with chronic kidney disease from clinical trials in coronary artery disease. Kidney Int. 70, 2021–2030 (2006).
Ridker, P. M. et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl. J. Med. 377, 1119–1131 (2017).
Campion, G. V., Lebsack, M. E., Lookabaugh, J., Gordon, G. & Catalano, M. Dose-range and dose-frequency study of recombinant human interleukin-1 receptor antagonist in patients with rheumatoid arthritis. The IL-1Ra Arthritis Study Group. Arthritis Rheum. 39, 1092–1101 (1996).
Pierce, B. L., Ahsan, H. & Vanderweele, T. J. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int. J. Epidemiol. 40, 740–752 (2011).
Netea, M. G., van de Veerdonk, F. L., van der Meer, J. W., Dinarello, C. A. & Joosten, L. A. Inflammasome-independent regulation of IL-1-family cytokines. Annu. Rev. Immunol. 33, 49–77 (2015).
Bouras, E. et al. Circulating inflammatory cytokines and risk of five cancers: a Mendelian randomization analysis. BMC Med. 20, 3 (2022).
Park, S. et al. Short or long sleep duration and CKD: a mendelian randomization study. J. Am. Soc. Nephrol. 31, 2937–2947 (2020).
Park, S. et al. A Mendelian randomization study found causal linkage between telomere attrition and chronic kidney disease. Kidney Int. 100, 1063–1070 (2021).
Park, S. et al. Atrial fibrillation and kidney function: a bidirectional Mendelian randomization study. Eur. Heart J. 42, 2816–2823 (2021).
Lei, Y. et al. Interleukin-1β inhibition for chronic kidney disease in obese mice with type 2 diabetes. Front. Immunol. 10, 1223 (2019).
Yazdi, A. S. & Drexler, S. K. Regulation of interleukin 1α secretion by inflammasomes. Ann. Rheum. Dis. 72, ii96–ii99 (2013).
Rose, A. et al. IL-2 Therapy diminishes renal Inflammation and the activity of kidney-infiltrating CD4+ T cells in murine lupus nephritis. Cells 8, 1234 (2019).
Hsieh, P. F. et al. The role of IL-7 in renal proximal tubule epithelial cells fibrosis. Mol. Immunol. 50, 74–82 (2012).
Tucci, M., Lombardi, L., Richards, H. B., Dammacco, F. & Silvestris, F. Overexpression of interleukin-12 and T helper 1 predominance in lupus nephritis. Clin. Exp. Immunol. 154, 247–254 (2008).
Wang, S. et al. Decreased renal ischemia-reperfusion injury by IL-16 inactivation. Kidney Int. 73, 318–326 (2008).
Burgess, S. et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 4, 186 (2019).
Lees, J. S. et al. Glomerular filtration rate by differing measures, albuminuria and prediction of cardiovascular disease, mortality and end-stage kidney disease. Nat. Med. 25, 1753–1760 (2019).
Burgess, S., Davies, N. M. & Thompson, S. G. Bias due to participant overlap in two-sample Mendelian randomization. Genet. Epidemiol. 40, 597–608 (2016).
Davies, N. M., Holmes, M. V. & Davey Smith, G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362, k601 (2018).
Ahola-Olli, A. V. et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am. J. Hum. Genet. 100, 40–50 (2017).
Palmer, T. M. et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat. Methods Med. Res. 21, 223–242 (2012).
Verbanck, M., Chen, C. Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018).
Hemani, G., Tilling, K. & Davey Smith, G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 13, e1007081 (2017).
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013).
Bowden, J. et al. Improving the accuracy of two-sample summary-data Mendelian randomization: moving beyond the NOME assumption. Int. J. Epidemiol. 48, 728–742 (2019).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525 (2015).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314 (2016).
Tian, J. et al. The dissection of expression quantitative trait locus hotspots. Genetics 202, 1563–1574 (2016).
Liu, Y. et al. Genome-wide analysis of expression QTL (eQTL) and allele-specific expression (ASE) in pig muscle identifies candidate genes for meat quality traits. Genet. Sel. Evol. 52, 59 (2020).
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (no. 2021R1A2C2094586). This work was also supported by the Research Fund from Seoul National University Hospital (3020220160).
Author information
Authors and Affiliations
Contributions
The corresponding author attests that all of the listed authors meet the authorship criteria and that no others meeting the criteria have been omitted. J.M.C., J.H.K., S.G.K., S.L., Y.K., S.C., Y.C.K., S.S.H., H.L., and J.P.L. performed the main statistical analysis including data curation, formal analysis, and investigation. K.K. contributed to the investigation and methodology. K.W.J., C.S.L., Y.S.K., D.K.K., and S.P. contributed to the conceptualization and design of the study. S.P. advised on statistical aspects and interpreted the data. D.K.K. and S.P. offer advice regarding data interpretation and supervision. S.P. obtained funding and supervised the overall project. All of the authors participated in drafting the manuscript. All of the authors reviewed the manuscript and approved the final version to be published.
Corresponding author
Ethics declarations
Competing interests
All authors declare no competing interests
Peer review
Peer review information
Communications Biology thanks Tommy Hon Ting Wong, Jianbo Qing, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Eirini Marouli and Christina Karlsson Rosenthal. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cho, J.M., Koh, J.H., Kim, S.G. et al. Mendelian randomization uncovers a protective effect of interleukin-1 receptor antagonist on kidney function. Commun Biol 6, 722 (2023). https://doi.org/10.1038/s42003-023-05091-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-023-05091-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.