Abstract
Tuberculosis (TB) caused by Mycobacterium tuberculosis (Mtb) is the second-deadliest infectious disease worldwide. Emerging evidence shows that the elongation factor EF-Tu could be an excellent target for treating Mtb infection. Here, we report the crystal structures of Mtb EF-Tu•EF-Ts and EF-Tu•GDP complexes, showing the molecular basis of EF-Tu’s representative recycling and inactive forms in protein translation. Mtb EF-Tu binds with EF-Ts at a 1:1 ratio in solution and crystal packing. Mutation and SAXS analysis show that EF-Ts residues Arg13, Asn82, and His149 are indispensable for the EF-Tu/EF-Ts complex formation. The GDP binding pocket of EF-Tu dramatically changes conformations upon binding with EF-Ts, sharing a similar GDP-exchange mechanism in E. coli and T. ther. Also, the FDA-approved drug Osimertinib inhibits the growth of M. smegmatis, H37Ra, and M. bovis BCG strains by directly binding with EF-Tu. Thus, our work reveals the structural basis of Mtb EF-Tu in polypeptide synthesis and may provide a promising candidate for TB treatment.
Similar content being viewed by others
Introduction
Tuberculosis, caused by Mycobacterium tuberculosis (Mtb), is one of the deadliest infectious diseases and has posed a severe threat to human health for thousands of years, accounting for about one million-death worldwide each year1. There are currently four types of drugs that can partially inhibit the growth of Mtb1. Isoniazid and its derivatives represent one category inhibiting cell wall formation, disturbing the synthesis of mycolic acid by producing nitric oxide in vivo, and affecting the type II fatty acid synthase (FAS) system2. Pyrazinamide, a prodrug of the active form pyrazinoic acid, can inhibit the FAS I enzyme and disrupt membrane potential, therefore interfering with Mtb’s survival at an acidic site of infection3. The third category of medicine, such as quinolone, acts on inhibiting DNA replication. Various fluoroquinolone-based drugs have shown promising effects against the DNA gyrase enzyme and, in turn, are successful in combating multidrug-resistant tuberculosis (MDR-TB)4. The last one, including the third-line anti-TB drug linezolid, targets ribosomes to inhibit protein synthesis by binding to 23S rRNA in the catalytic site of the 50S ribosome5,6. However, Mtb exhibits latency during infection, making antibacterial drug development challenging. In addition, the increasing prevalence of drug-resistant Mtb has further accentuated the need for novel antimycobacterial drugs7.
The ribosome is the protein translation machine in cells, and the protein synthesis needs the help of many accessory proteins during the initiation, elongation, and recycling processes8,9. The most common EFs in prokaryotes include EF-G, EF-Ts, and EF-Tu10. Although EFs in bacteria are homologous, they possess distinct structures and exert variant functions11,12. EF-Tu, one member of the GTPase family and the most abundant protein in bacteria, is responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome by hydrolyzing GTP to GDP13,14,15. This process provides amino acids to elongate the newly synthesized protein via transferring the correct amino acid-binding in the A-site according to the codon-anticodon pairing. Upon binding with GTP, EF-Tu becomes an active form to bind with aa-tRNAs; then, EF-Tu-GTP recognizes the aminoacyl bond and has a high affinity for all aa-tRNAs by binding to the receptor motif16. Next, EF-Tu delivers aa-tRNAs to the mRNA-programmed ribosome, where the mRNA codon in the ribosomal A-site is recognized by aa-tRNA in a ternary complex with EF-Tu and GTP17. Also, the GTP hydrolysis by EF-Tu is activated by the cognate codon-anticodon interaction18. The additional rounds of ternary complex formation can occur on the ribosome during proofreading, to increase the rate and fidelity of aa-tRNA selection at the expense of GTP hydrolysis19. EF-Tu then hydrolyzes GTP to GDP, resulting in the EF-Tu•GDP complex having a low affinity to the ribosome, therefore releasing from the ribosome20. To be reactivated from the inactive form (EF-Tu•GDP) to the active form, EF-Tu needs to bind with EF-Ts (elongation factor thermo-stable), a GTPase-activating protein (GAP), in turn releasing the active EF-Tu•GTP12.
Many prokaryotic species encode two gene copies of EF-Tu, named tufA and tufB, which share high amino acid sequence similarity21. Reports showed that about two-thirds of EF-Tu was expressed from the tufA gene, and the other was expressed from the tufB gene in E. coli and Salmonella typhimurium21,22. Unlike other species, there is only one gene encoding EF-Tu in Mtb. Emerging evidence showed that EF-Tu could be an excellent target for treating infection from bacteria23,24. Currently, four compounds, including GE2270A, enactloxin lla, kirromycin, and pulvomycin, are used for targeting EF-Tu and inhibiting its activity23,25,26. The compounds inhibit either the dissociation between EF-Tu•GDP and ribosome, or the ternary complex formation. However, the compounds do not work well for Mtb25,26. Most importantly, although the homologic structure of E. coli has been reported27, the absence of structural information on Mtb EF-Tu and EF-Ts dampens the discovery of pharmacological intervention for treating Mtb.
Here, we report the crystal structures of Mtb EF-Tu•EF-Ts and EF-Tu•GDP complexes, elucidating the molecular basis of the representative recycling or inactive form of EF-Tu in protein translation in Mycobacterium, similar to reported in E. coli and T. ther. The GDP-binding pocket residues of EF-Tu dramatically changed conformations upon binding with EF-Ts, illustrating how EF-Ts reactivate the inactivated EF-Tu. Also, we identified that Osimertinib could inhibit different mycobacterium strains by direct targeting EF-Tu.
Results
Crystallization and structure determination of the Mtb EF-Tu•EF-Ts complex
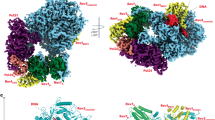
To reveal the molecular basis of the EF-Tu reactivating mechanism in Mtb, we investigated the crystal structure of the full-length EF-Tu and EF-Ts complex. We expressed and purified EF-Tu and EF-Ts in Escherichia coli cells. EF-Tu and EF-Ts formed a 1:1 complex in solution, evidenced by the fact that the complex came out at the elution peak of 76 mL on a Superdex200 16/600 GL column, which corresponds to a molecular mass of 71 kDa (Fig. 1a, b). Also, the EF-Tu or EF-Ts was eluted out at the peaks of 84 mL or 86 mL on the gel-filtration column, corresponding to a mass of 43 kDa and 30 kDa, respectively (Fig. 1a, b). The Mtb EF-Tu•EF-Ts crystals were obtained after several rounds of optimization, and the complex structure was solved at a resolution of 2.8 Å by molecular replacement method using the EF-Tu•GDP (PDB: 7VOK) and the AlphaFold-predicted EF-Ts (https://alphafold.ebi.ac.uk/entry/P9WNM1) as the searching models.
a The size-exclusion chromatography (SEC) profiles of Mtb EF-Tu, EF-Ts, and EF-Tu•EF-Ts complex on a Superdex200 16/600 column. b The SDS-PAGE results of EF-Tu, EF-Ts, and EF-Tu•EF-Ts complex, corresponding with (a). M protein marker, L protein loading sample. c Schematic domains of Mtb EF-Tu and EF-Ts. EF-Tu consisted of three domains, including Domain I (gray), Domain II (blue), and Domain III (purple). EF-Ts consisted of three domains, including the N-terminal domain (wheat), the C-terminal domain (green), and the core domain in the middle (yellow). d A ribbon representation of Mtb EF-Tu•EF-Ts complex structure, colored with that in (c). EF-Tu and EF-Ts formed a 1:1 symmetric complex. The binding interface was labeled with a black rectangle for key residues depicted with stick models, and the cartoon is shown with a 50% transparency to highlight the key residues. e A ribbon representation of the detailed binding interface between EF-Tu and EF-Ts. The involved key residues were labeled and shown as sticks. f The electrostatic potential surfaces of Mtb EF-Tu•EF-Ts complex (left), EF-Tu (middle), and EF-Ts (right). Red denoted negative potential, and blue denoted positive potential.
The structure of Mtb EF-Tu•EF-Ts complex
The Mtb EF-Tu contains three domains, named Domain I (residues 1–200), II (residues 213–296), and III (residues 306–396) (Fig. 1c and Supplementary Fig. 1a). Domain I, also called the GTP-binding domain or Ras-like domain, hydrolyzes GTP to GDP in an Mg2+-dependent manner. Domains II and III are oligonucleotide-binding domains, which bind to both charged tRNA and EF-Ts in T. thermophilus28. The Mtb EF-Tu structure showed three domains forming a flattened triangular shape with a hole in the middle, arranged like other homolog structures (Fig. 1d and Supplementary Fig. 1a). Moreover, Domains I and III were closer than Domain I and II, making face-to-face contact and resulting the side-chain interactions. Also, the flattened triangular shape permits a high degree of inter-domain flexibility, which benefits EF-Tu to bind with kinds of substrates in the polypeptide synthesis29,30. The EF-Ts comprises 13 α-helices and 6 β-strands, containing three domains, the N-terminal domain, the core domain, and the C-terminal domain (Fig. 1c and Supplementary Fig. 1b). The core domain comprises two subdomains, denoted as subdomain N (residues 61–65, 68–73, and 133–139) and subdomain C (residues 144–149, 157–166, and 259–267), which is composed of 6 β-strands surrounding by the α-helices (Supplementary Fig. 1b).
The Mtb EF-Tu•EF-Ts complex contains one EF-Tu and one EF-Ts per asymmetric unit (Fig. 1d), different from that in E. coli and T. thermophilus that is composed of two EF-Tu and two EF-Ts molecules27,28,31. In E. coli, the amino-terminal region of EF-Ts interacts with the nucleotide-binding Domain I of EF-Tu, and the other half binds to Domain III. In addition, residues K51, G54, D80, F81, I125, and G126 of E. coli EF-Ts are vital for the interaction with EF-Tu; however, the structure and sequence alignment shows that only D80 (corresponding to D77 in Mtb EF-Ts) is the conserved active site27. In T. thermophilus, EF-Ts is a dimer, in which both subunits contribute to the bipartite interface. Also, the conserved TDFV sequence of EF-Ts contacts with EF-Tu Domain I in E. coli, T. thermophilus, and B.mitochondrial32. Similar to homologs in other species, the Mtb EF-Tu bound with the N-terminal domain and the core domain of EF-Ts through Domains I and III (Fig. 1d, e). We chose the pair of amino acids with a relative distance within 3 Å as interface residues. Twenty residues were involved in the interface with hydrogen bonds or salt bridges (Fig. 1d, e). Also, the electrostatic potential surfaces analysis showed that the EF-Tu•EF-Ts complex was full of negative potential surfaces (Fig. 1f). Notably, the binding interface of EF-Tu was full of negative potential, while that in EF-Ts was positive potential, indicating the charge-charge interaction promoting the complex formation, which is critical for protein stacking33,34.
Mtb EF-Ts exhibits a high binding affinity to EF-Tu
The interface-involved residues of EF-Tu were located in Domain I and III, while the related residues of EF-Ts were mainly located in the N-terminal domain and the core domain (Fig. 1d). Briefly, residue K24 on the α2 helix of EF-Ts formed salt bridges with residues D144 and D145 of EF-Tu, and the K24 side chain formed a hydrogen bond with D145. Residue R13 on the α1 helix of EF-Ts formed hydrogen bonds with P114 and E155 of EF-Tu, respectively. Moreover, residue D77 of EF-Ts formed a hydrogen bond with residue H87, and a salt bridge with A88 of EF-Tu. Residue N82 of EF-Ts formed a hydrogen bond with the side chain of EF-Tu E120. In addition, the N atom on the side chain of EF-Ts-H149 formed a salt bridge with the O atom of the D357 side chain of EF-Tu. Also, the side chain of EF-Ts R151 formed a hydrogen bond with N358 of EF-Tu, and D154 formed a salt bridge with EF-Tu Q127 (Fig. 2a). Next, we investigated the interaction between EF-Tu and EF-Ts using the isothermal titration calorimetry (ITC) method. The result showed that EF-Tu bound with EF-Ts at a 1:1 ratio with a Kd value around 1.36 µM (Fig. 2b), different from the Kd values for EF-Tu and EF-Ts from E. coli and Bovine mitochondria (2 nM and 5.5 nM, respectively)35,36. Reports showed that the equilibrium dissociation constants would change among different species despite the high sequence similarity37. Furthermore, Mtb EF-Tu shared 74% and 71% similarities in amino acid sequence with that in E. coli and T. thermophilus, and the interfaced-related residues were highly conserved (Supplementary Fig. 2). However, EF-Ts’ amino acid sequence similarity between Mtb and E. coli, or T. thermophilus was 39% or less, except that residues R13, K24, and D77 in Mtb were conserved (Supplementary Fig. 3).
a Ribbon representations of crucial residues in the interface between EF-Tu (cyan) and EF-Ts (light pink). The residues were labeled and shown as sticks. The hydrogen bonds among interacted residues are shown in black dashed lines. b The isothermal titration calorimetry (ITC) result showed that the Kd value between EF-Tu and EF-Ts was 1.36 µM. The binding molar ratio was around 1:1. c The SEC results of the EF-Tu•EF-Ts complex and different mutants. The mutants Tu/Ts-R13, Tu/Ts-N82, and Tu/Ts-H149 dramatically changed their solution status. Tu/Ts is abbreviated for EF-Tu/EF-Ts. d The SDS-PAGE result of the EF-Tu•EF-Ts complex and different mutants, corresponding to (c). M protein marker, L protein loading sample.
To identify the critical residues for the EF-Tu/EF-Ts complex’s interaction, we mutated seven residues of EF-Ts into alanine, including R13, L21, K24, D77, N82, H149, D154, and EF-Tu N358. When EF-Tu was co-purified with the wild-type or mutant of EF-Ts on the size exclusion chromatography, it clearly showed that mutants R13A, N82A, and H149A dramatically disrupted the interaction profiles with EF-Tu, indicating the three residues are indispensable for the EF-Tu/EF-Ts formation (Fig. 2c, d).
EF-Ts exhibits a diverse binding conformation among different species
EF-Ts functions as a guanine nucleotide exchange factor and catalyzes the reaction of EF-Tu from the inactive form (GDP-bound) to the active form (GTP-bound)38. As EF-Ts shared low similarities among different species (Supplementary Fig. 3), and preferred to bind with and reactivate EF-Tu at different ratios between Mtb (1: 1, EF-Ts: EF-Tu) and T. thermophilus, (2:1, EF-Ts: EF-Tu)28,31, we compared the EF-Tu•EF-Ts complex structures using the ChimeraX and PymoL software. Superimposing the Mtb EF-Tu•EF-Ts complex with that of E. coli (PDB ID:1EFU) or T. thermophilus (PDB ID: 1AIP) revealed similar features of overall folds, which showed high similarities among the Cα atoms with RMSD values of 1.214 Å or 1.309 Å, respectively (Fig. 3a). In brief, the structural differences existed mainly in EF-Ts, while EF-Tu had similar architecture (Fig. 3a, b). Different from the Mtb EF-Ts, the EF-Ts of E. coli was composed of 13 α-helices and 6 β-strands, which assembled in four critical domains, the N-terminal domain (residues 1–54), the core domain (residues 55–179), the dimerization domain (residues 180–228), and the C-terminal domain (residues 264–282)27. The C-terminal domain was stretched out to interact with Domain I in EF-Tu of E. coli, whereas it was absent in Mtb EF-Ts (Fig. 3c, d and Supplementary Fig. 3)27,28, implying a distinct binding conformation in different bacteria.
a Superimposition of EF-Tu•EF-Ts complex structures among Mtb (marine, PDB: 7VMX), E. coli (light pink, PDB: 1EFU), and T. thermophilus (green, PDB: 1AIP). The RMSD values between Mtb and E. coli or T. thermophilus were 1.214 Å or 1.309 Å, respectively. b Superimposition of EF-Tu structures among Mtb, E. coli, and T. thermophilus. c Superimposition of EF-Ts structures among Mtb, E. coli, and T. thermophilus. The numbers denoted significant differences among the three structures. d The detailed difference of EF-Ts structures among three species, according to (c). ‘1’, the extra helix of E. coli EF-Ts for binding with EF-Tu Domain I. ‘2’ and ‘4’, the helices in the T. thermophilus EF-Ts, absent in Mtb and E. coli, which were not involved in binding with EF-Tu. ‘3’, the α10 and α11 helices that shifted about 4.7 Å or 6.0 Å within three species, respectively.
The second significant difference was that the T. thermophilus EF-Ts had one extra helix between the β1 and β2 strands. As the EF-Tu•EF-Ts complex is an asymmetrical heterotetramer in T. thermophilus, of which one EF-Tu interacts with two subunits of EF-Ts, forming a bipartite interface; the extra helix could explain why one EF-Tu molecule needed to be reactivated by two EF-Ts molecules in T. thermophilus (Fig. 3d). Mtb and E. coli EF-Ts is a monomer with a structural repeat that mimics the dimeric interface in T. thermophilus EF-Ts28. Also, the helices, α10 and α11, of Mtb EF-Ts dramatically shifted with distances around 4.7 Å and 6.0 Å to that of E. coli or T. thermophilus (Fig. 3d).
The Mtb EF-Tu•GDP complex structure
To elucidate the molecular basis of the inactive form of Mtb EF-Tu during the elongation cycle, we expressed and purified the full-length EF-Tu protein, and successfully obtained the EF-Tu•GDP complex structure at a resolution of 3.4 Å using the E. coli EF-Tu (PDB ID: 1EFC) as a searching model. The final structure contains four EF-Tu molecules per asymmetric unit (Table 1). Each EF-Tu molecule was bound with a GDP and an Mg2+ ion (Fig. 4a). Domain I of Mtb EF-Tu underwent a structural rearrangement of approximately 90° rotation to the other domains (Fig. 4a), similar to that in E. coli which occurred around switches 1 and 2 when GTP replaced GDP. This movement formed a binding site of aminoacyl-tRNA to interact with all three domains of EF-Tu. Domain II was composed of 84 residues from 213 to 296, with 6 antiparallel β-strands forming a β-barrel (Supplementary Fig. 1a), different from that in E. coli EF-Tu. Residues of Domain II took responsibility for discriminating between the charged and uncharged tRNA in the binding pocket. Moreover, Domain III of Mtb was composed of 91 residues from 306 to 396, with 6 antiparallel β-strands forming a β-barrel. In Mtb, Domains II and III were connected by a shorter peptide, but in E. coli, there was one more β-strand between the two domains20. Also, Domain II and III formed antiparallel β-barrels to regulate the activity of Domain I, increasing the affinity of GDP over GTP (Fig. 4)38.
a A ribbon representation of the EF-Tu•GDP structure, which is colored as Fig. 1a. GDP and Mg2+ were located in Domain I. GDP is shown as sticks, and Mg2+ is shown as a green sphere. b Representation of GDP-binding sites in the EF-Tu•GDP complex. The broken black lines represented the hydrogen bonds formed between EF-Tu and GDP. The related residues were labeled and shown as sticks. c The electrostatic potential surface of the EF-Tu•GDP complex. Red indicated the negative potential, and blue indicated the positive potential. GDP is shown as sticks, and Mg2+ is shown as a green sphere. d Superimposition of Mtb EF-Tu between the EF-Tu•GDP (yellow) and EF-Tu•EF-Ts (marine) complexes. The RMSD value for the Cα alignment between the two structures was 0.711 Å. e The detailed shift of GDP-bound residues between the EF-Tu•GDP and EF-Tu•EF-Ts complexes. The broken black lines represented the shifting of labeled residues.
The GDP-binding sites are conserved among different species
The GDP molecule was bound with Domain I of EF-Tu, which consisted of five loops, named G-1 to G-5, similar to other nucleotide-binding proteins (Fig. 4b and Supplementary Fig. 2)28,31,39. G-1 was referred to as the P-loop due to the conserved amino acid sequence like GX4GK(ST) from residues G19 to K25, which connected the β1 strand and α1 helix responsible for phosphate binding. Also, residues K25, T26, and T27, located on the α1 helix, forming hydrogen bonds with the α- and β-phosphate of GDP. G-2 contained the second β-strand and its preceding loop, which was bound with the γ-phosphate of GTP described in other homologous structures. G-3 contained the conserved sequence of DX4G, residues from 83 to 86 in Mtb EF-Tu, which connected the β5 strand and α3 helix. Especially, residue D83 was bound with the Mg2+ ion through several water molecules as described before. G-4 contained β5 strand and its preceding loop, and had the conserved amino acids sequence of (N/T) (K/Q) XD, which was NKAD in Mtb EF-Tu spanned from residues 138 to 145 that formed a loop to connect β7 strand and α5 helix. Also, residues D141 and A176 formed hydrogen bonds with the guanine ring of GDP. The G-5 region was composed of β8 strand and α6 helix, which spanned from residues P170 to A176, and was far from the GDP and Mg2+ molecules (Fig. 4b).
The GDP molecule interacted with residues D22, G24, K25, T26, T27, N138, D141, and A176 of EF-Tu, which were conserved in Mtb, E. coli, and T. thermophilus (Fig. 4b and Supplementary Fig. 2). Residues D22 and G24 were located in the first loop, which connected the β1 strand and α1 helix. Also, D141 was located in the loop connecting the β7 strand and α5 helix. Moreover, analyzing the electrostatic potential surface showed that GDP was bound in the pocket full of positive charges (Fig. 4c).
When superimposing the EF-Tu molecules in the EF-Tu•GDP and EF-Tu•EF-Ts structures, it showed similar features of overall folds with an RMSD value of 0.711 Å (Fig. 4d). A significant difference existed in Domains I and III, responsible for the interaction with EF-Ts. The EF-Tu in the recycling complex (EF-Tu•EF-Ts) had a much more compact shape than that in the inactive form (EF-Tu•GDP) (Fig. 4d). Significantly, the residues involved in the GDP-binding pocket changed positions. D22, G24, K25, T26, T27, A176, and D141 of EF-Tu shifted from 2.8 Å to 5.5 Å upon conformational change (Fig. 4e), which could explain how EF-Ts help EF-Tu to dissociate with GDP and reactivate EF-Tu during the elongation cycle of protein translation. The pocket was no longer suitable for binding with GDP because of the translocate of GDP-binding residues after EF-Ts binding, similar to the report in E. coli, in which the movement of residues K136 and N138 of EF-Tu made them away from the nucleotide-binding site and relaxed the interactions within the base (Fig. 4e)38.
Furthermore, we investigated the solution status of EF-Tu, EF-Ts, and EF-Tu•EF-Ts complex by the small-angle X-ray scattering (SAXS) method (Fig. 5a–i). All the proteins behaved well in solution. The maximum dimension (Dmax) from distance distribution function p(r) for the EF-Tu, EF-Ts, and EF-Tu•EF-Ts complex were 86 Å, 80 Å, and 106 Å, respectively (Fig. 5b, e, h). When superimposed the crystal structures with the envelopes generated from SAXS data, EF-Ts and EF-Tu•EF-Ts complex showed high similarities, whereas EF-Tu had discrepancies resulting from a highly flexible loop between Domain I and Domain II (Fig. 5c, f, i).
a The experimental scattering curve of the Mtb EF-Tu•EF-Ts complex. b The distance distribution function curve of the EF-Tu•EF-Ts complex. c The crystal structure of the EF-Tu•EF-Ts complex was fitted into the ab initio envelope obtained from SAXS. d The experimental scattering curve of Mtb EF-Tu. e The distance distribution function curve of Mtb EF-Tu. f The crystal structure of EF-Tu was fitted into the ab initio envelope obtained from SAXS. g The experimental scattering curve of Mtb EF-Ts. h The distance distribution function curve of Mtb EF-Ts. i The crystal structure of EF-Ts was fitted into the ab initio envelope obtained from SAXS. j Dynamic light scattering (DLS) analysis of the Mtb EF-Tu, EF-Ts, and EF-Tu•EF-Ts complex. Particle polydispersity was defined in the following terms: polydispersity (Pd), % polydispersity (% Pd).
The FDA-approved drug Osimertinib binds with Mtb EF-Tu and inhibits Mycobacterium growth
To identify the potential inhibitor of Mtb EF-Tu, we screened the FDA-approved drug library (total of 1971 drugs) with the nano-DSF method40. Compared to EF-Tu itself, sixteen drugs could dramatically affect the stability of the EF-Tu protein (Table 2). Significantly, Osimertinib, a medicine for treating non-small-cell lung carcinomas (NSCLC) with specific mutations41, significantly changed the unfolding transition midpoint from 52.5 to 60.4 °C (Fig. 6a, b). Next, 16 drugs were added to different bacterial strains to check the in vivo function separately. The results showed that Osimertinib did not change the growth of E. coli; however, it dramatically inhibited the growth of M. smegmatis and Mtb H37Ra strains at around 10 µM concentration (Fig. 6c–e). In contrast, other drugs did not show significant antibacterial activities, including hydroquinone and saractinib (Supplementary Fig. 4a, b). Also, Osimertinib significantly inhibited the growth of the M. Bovis BCG strain (Fig. 6f). The first-line anti-MTB drug isoniazid (also called INH) inhibits Mtb growth by disturbing the biosynthesis of mycolic acid. Consistently, isoniazid showed a strong ability to inhibit the growth of Mtb H37Ra and M. smegmatis, but did not change the growth of E. coli (Supplementary Fig. 4d–f). Thus, Osimertinib had a similar bacterial inhibition capacity with isoniazid in bacterial specificity. To further identify whether Osimertinib could directly bind with Mtb EF-Tu, we performed the microscale thermophoresis (MST) method with the purified protein. The result showed that Osimertinib bound to EF-Tu in vitro with medium intensity (Kd = 207 μM) (Fig. 6g), with the binding of GDP and EF-Tu as a positive control (Supplementary Fig. 4c).
a, b Thermal unfolding curves and unfolding transition midpoints of EF-Tu and EF-Tu/Osimertinib were detected by the Nano-DSF method. c–f Osimertinib did not affect E. coli growth, but significantly inhibited the growth of M. smegmatis, Mtb H37Ra, and M. bovis BCG strains. Different concentrations of Osimertinib were added to the bacterial strains, and cell densities were detected at different times (c–e). After treatment with varying concentrations of Osimertinib, the BCG strains were diluted 100 or 1000 times, and the numbers of colonies were calculated (f). g The MST result showed that Osimertinib is bound with EF-Tu in vitro. h The in silico modeling structure of EF-Tu and Osimertinib complex. EF-Tu was shown in a cartoon model. Osimertinib was shown in a stick model. The error bars represented the standard deviations (SD). *P < 0.05, **P < 0.01, ns no significance.
We next screened thousands of crystallization conditions for solving the Osimertinib-bound EF-Tu complex structure; however, no qualified diffraction datasets were obtained. Thus, we built the in silico modeling complex structure of EF-Tu and Osimertinib using the AlphaFold2 software. The structure showed that Osimertinib bound with the Domain I and II of EF-Tu, composing of flexible loops and different from the site of the GDP-binding pocket (Fig. 6h). Also, the electrostatic potential surface of the osimertinib-binding pocket was full of negative charges (Supplementary Fig. 5a). Furthermore, the binding pocket of osimertinib in the human EGFR/osimertinib complex structure (PDB ID: 6LUD) is composed of flexible loops (Supplementary Fig. 5b). Next, we analyzed the binding sites of four known inhibitors in EF-Tu. The crystal structure of antibiotic enacyloxin/EF-Tu complex (PDB ID: 2BVN) shows that enacyloxin locates on the cleft between Domain I and III of E. coli EF-Tu (Supplementary Fig. 5c), whereas antibiotic GE2270A locates on Domain II and does not interact with Domain I and III of E. coli EF-Tu (PDB ID: 3U6K) (Supplementary Fig. 5d). In addition, kirromycin shares a similar location with enacyloxin (Supplementary Fig. 5e), while pulvomycin locates in the middle hole, which interacts with three domains of T. ther EF-Tu (Supplementary Fig. 5f), indicating that Osimertinib docking site may overlap partially with GE2270A and pulvomycin. Thus, Osimertinib might inhibit Mtb growth by blocking the rearrangement of different domains of EF-Tu.
Discussion
Elongation factors play essential roles in synthesizing new proteins through translation in the ribosome. EF-Tu is one of the most critical proteins in bacteria, as it is the only protein delivering aminoacyl-tRNA to the ribosome for polypeptide synthesis23,24. Once EF-Tu exhibits dysfunction, the protein synthesis would be interrupted, and the growth of bacteria would be inhibited42. Unlike other bacteria, Mtb exhibits specific properties, including slow growth and latency, making it challenging to develop therapeutic targets1. Although the EF-Tu structures of E. coli and T. thermophilus have been solved for years, little is known about the Mtb EF-Tu. Our current work revealed the Mtb EF-Tu•EF-Ts complex structure (Fig. 1), and found that EF-Ts changed conformation upon binding with and reactivating EF-Tu (Figs. 2–4). Moreover, we identified the essential residues of EF-Ts, R13, N82, and H149, which affected the EF-Tu/EF-Ts complex formation using the SEC methods (Fig. 2). Although EF-Tu shared high sequence similarity and overall architecture, it bound to EF-Ts with different ratios between Mtb and E. coli. Most important, the structural difference existed in the C-terminal domain of E. coli EF-Ts, which stretched out and might block the interacting interface between EF-Ts and EF-Tu (Fig. 3).
Furthermore, our data revealed that the EF-Tu structure was retrieved back upon conformational change from the inactive form (EF-Tu•GDP) to the recycling form (EF-Tu/EF-Ts), explaining the recycling process of Mtb EF-Tu (Fig. 4). The GDP-binding pocket of EF-Tu was conserved across all GTPase proteins; however, the essential related residues dramatically shifted upon binding with EF-Ts, resulting in GDP dissociated from EF-Tu (Fig. 4b, e). This could clarify how EF-Tu as a G-protein was reactivated by EF-Ts during protein translation in Mtb.
EF-Tu is a promising target for exploring antibiotics23. Four kinds of antibiotics were designed to block the function of EF-Tu. Kirromycin binds with the 30S subunit of E. coli ribosome and interferes with the polypeptide synthesis43. Also, kirromycin significantly affects the nucleotide binding of unphosphorylated Mtb EF-Tu, but not the phosphorylated protein44. Pulvomycin can inhibit polypeptide synthesis in B. brevis and E. coli45. In addition, enacyloxin Iia is active against different Gram-positive and Gram-negative bacteria except for Mtb and M. Smegmatis46. Antibiotic GE2270A can inhibit Gram-positive bacteria47. However, none was used for curing tuberculosis48. In this study, Osimertinib was identified to inhibit different Mycobacterium strains by directly targeting EF-Tu (Fig. 6). As a medicine for NSCLC with specific mutations, Osimertinib could potentially be used to treat TB, despite further investigations needing to be performed.
The structure prediction software, like AlphaFold2, is very powerful for providing the initial model. In this case, the AlphaFold2-predicted EF-Ts structure was successfully used as a search model to solve the Mtb EF-Tu•EF-Ts complex structure. When superimposed the two EF-Ts structures (PDB ID: 7VMX and P9WNM1-F1), we found that the core and N-terminal domains shared the same conformation, and the RMSD value was 0.627 Å (Supplementary Fig. 1c). As it was challenging to obtain the EF-Tu/osimertinib complex structure via X-ray crystallography, we built the in silico model structure by autodocking. Compared with other structures of EF-Tu and its inhibitors, Osimertinib is more likely located on the cleft between Domain I and II (Fig. 6 and Supplementary Fig. 5), whereas the mechanism needs to be explored with more evidence. Together, the detailed Mtb EF-Tu•EF-Ts complex and EF-Tu•GDP structural information provided an exquisite image for understanding the polypeptide elongation process of protein synthesis and might have practical implications for designing new medicine to cure tuberculosis.
Methods
Protein expression and purification
The full-length EF-Tu (Uniprot ID: P9WNN1) and EF-Ts (Uniprot ID: P9WNM1) were subcloned from the Mtb genomic library into the pSMT3 vector with a His-SUMO tag. The wild-type and mutants of EF-Tu and EF-Ts plasmids were transformed into E. coli BL21 (DE3) or Rosetta (DE3) cells for protein expression. E. coli cells were cultured in LB medium with 25 µg/ml kanamycin and 34 µg/ml chloramphenicol at 37 °C. Isopropyl β-D-1-thiogalactopyranoside (IPTG, 0.2 mM) was added to cells to induce protein expression at 20 °C for 18 h when the OD600 reached 0.6–0.8. Then, the cells were harvested by centrifugation at 6000 rpm for 15 min at 4 °C. The cell pellets were resuspended in a lysis buffer (Tris-HCl pH 8.0, 100 mM NaCl, 10 mM imidazole pH 8.0, 5% glycerol, 4 mM MgCl2, and 2 mM β-mercaptoethanol) and disrupted using a high-pressure homogenizer (JNBIO, China). The cell debris was removed by centrifugation at 17,000 rpm for 60 min at 4 °C. Next, the supernatant was purified using a His TrapTM HP column (GE Healthcare) and was eluted in a buffer (50 mM Tris-HCl, 500 mM NaCl, 250 mM imidazole, 5% glycerol, pH 8.0). The His-sumo tag was removed by the ULP1 enzyme cleavage, followed by additional Ni-NTA affinity chromatography as previously described49. The protein was further purified by gel filtration using a Superdex200 16/600 column (GE Healthcare) in a gel-filtration buffer (20 mM Tris-HCl pH 7.4, 100 mM NaCl, 2 mM DTT). The EF-Tu•EF-Ts complex was obtained by premixing the EF-Tu and EF-Ts proteins with a 1:1 molar ratio on ice for 30 min, and then the mixture was applied to a gel-filtration chromatography (Superdex200 16/600 GL).
Mutagenesis
The EF-Ts mutants K24A, L21A, R13A, D77A, N82A, H149A, D154A, and EF-Tu-N358A were generated by whole-plasmid PCR in a 20-cycle reaction with steps at 98 °C for 10 s, 55 °C for 30 s, and 72 °C for 3 min per cycle. After digestion with the enzyme DpnI, the PCR products were transformed into E. coli Top10 cells. The positive constructs were determined by DNA sequencing.
Dynamic light scattering (DLS) measurement
The DLS data were collected on the DYNAMICS software from DynaPro NanoStar (Wyatt Technology), operating at a light source wavelength of 658 nm and a fixed scattering angle of 90°. The fresh proteins were diluted to 1 mg/mL with a buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 2 mM TCEP, and 5% glycerol at 25 °C.
Crystallization and data collection
The EF-Tu•GDP and EF-Tu•EF-Ts complexes were crystallized using the sitting-drop vapor-diffusion method by mixing 0.2 μL protein and 0.2 μL reservoir solution at 18 °C. EF-Tu and GDP were premixed with a molar ratio of 1:5 on ice for 30 min before screening the crystallization conditions. EF-Tu•GDP crystals were grown in a reservoir solution containing 26% (w/v) PEG 6000 and 100 mM HEPES pH 7.0 with a protein concentration of 10 mg/mL. For the EF-Tu•EF-Ts complex, the two proteins were premixed with a molar ratio of 1:1 on ice for 30 min before screening. EF-Tu•EF-Ts crystals were grown in a reservoir solution containing 8% PEG 6000, 2 mM ZnCl2, and 0.1 M Tris-HCl pH 8.0 with a protein concentration of 5 mg/mL. All of the crystals were briefly soaked in a cryoprotectant solution consisting of 25% (v/v) glycerol dissolved in their corresponding mother liquors before being flash-cooled directly in a liquid-nitrogen stream at 100 K. The X-ray diffraction data were collected at the BL17U1 and BL19U1 beamlines of the Shanghai Synchrotron Radiation Facility. The process of data integration, scaling, and merging was performed using the HKL3000 package and the XDS program.
Structure determination and refinement
The crystal structure of Mtb EF-Tu•GDP was determined by molecular replacement using the crystal structure of E. coli EF-Tu (PDB ID: 1EFC) as the search model. The EF-Tu•EF-Ts complex structure was determined by molecular replacement using EF-Tu•GDP and AlphaFold2-predicted EF-Ts structure (https://alphafold.ebi.ac.uk/entry/P9WNM1) as search models. Cycles of refinement and model building were carried out by using Phenix and COOT programs. The quality of the final model was evaluated with PROCHECK. All of the structures were displayed and analyzed using PyMOL program. The collected data and refinement statistics are summarized in Table 1.
Model building of EF-Tu and Osimertinib complex
The model building process was described previoulsy50. In brief, the Mtb EF-Tu structure (PDB: 7VMX) and the simplified molecular-input line-entry system (SMILES) file of osimertinib were initially placed in the models. Optimized binding poses of Mtb EF-Tu/osimertinib complex were predicted by iteratively employing molecular docking using AutoDock Vina51. The 3D structure model was generated from the predicted distance and orientation using constrained minimization. Next, five best models of the top 50 folded structures satisfying the restraints were selected according to Rosetta energy and GOAP energy51. Then, the best complex model was selected and depicted by Pymol.
Small angels X-ray scattering (SAXS)
The SAXS data of EF-Tu, EF-Ts, and EF-Tu•EF-Ts were collected at beamline BL19U2 of the Shanghai Synchrotron Radiation Facility, with a radiation wavelength of 1.03 Å. The protein samples were prepared at 0.1–10 mg/mL concentrations in the buffer (20 mM Tris-HCl pH 7.4, 100 mM NaCl). The samples were measured in triplicate, and the sample measurements were adjusted by subtracting the scattering from the buffer alone. Data were analyzed using the software package BioATSAS (https://www.embl-hamburg.de/biosaxs/). The scattering images were averaged and subtracted from the buffer-scattering images. Then, using the indirect Fourier transform method, the Rg was estimated. The distribution function p(r) was calculated from the parameter as Dmax. The SAXS envelopes of EF-Tu, EF-Ts, and EF-Tu•EF-Ts were built by GASBOR, as previously reported33.
Isothermal titration calorimetry (ITC)
ITC assays were carried out on the MicroCal ITC200 calorimeter at 25 °C. The titration buffer for the proteins was 10 mM HEPES pH 8.0, 50 mM NaCl, and 2 mM TCEP. The protein concentrations were 0.1–1 mM. The fitting curve with a single-binding-site model was performed by the ITC data analysis module provided by the manufacturer.
Nano differential scanning fluorimetry (Nano-DSF) experiments
Nano-DSF experiments were carried out on the Nano Temper Promethus NT48 instrument. The protein samples were prepared at 0.5 mg/mL concentration in the buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 2 mM TCEP, and 5% glycerol) after gel-filtration purification. Protein samples were transferred to capillary tubes after centrifugation (12,000 rpm, 20 °C, 10 min). The data were collected under the fluorescence absorption value within the range of 200–2000. The start temperature was 20 °C, and the highest temperature was 95 °C, with the heating rate being 1 °C/min to detect the Tm value of the protein. EF-Tu and Osimertinib were mixed in a molar ratio of 1:3 at room temperature for 20 min with the same data collection process for EF-Tu.
MicroScale thermophoresis (MST) experiments
The purified EF-Tu was dialyzed into the SEC buffer and labeled by the cyanine 5-NHS ester iodide (MCE, HY-135414B) according to the protocol. Osimertinib (Selleck, S7297) was dissolved in DMSO at 10 mM. The MST experiments were carried out on the Nano Temper Monolith NT.115 instrument. Labeled EF-Tu (200 nM) was mixed with the indicated concentrations of Osimertinib in the SEC buffer. The MST data were collected under 20% light-emitting diode power and 40% infrared laser power. The Kd value was determined by the Nano Temper analysis software (v.1.5.41).
Bacterial culture
Mtb H73Ra (ATCC 25177), M. Smegmatis (ATCC 700084), and M. Bovis BCG (ATCC 35733) strains were cultured in the Middlebrook 7H9 medium with an extra 10% oleic acid-albumin-dextrose-catalase-enriched Middlebrook (OADC). All the bacteria were cultured at 37 °C.
Antimicrobial activity assay
The Mtb H37Ra (ATCC 25177) and M. Smegmatis (ATCC 70084) strains were cultured to the logarithmic stage and transferred into a 96-well plate at a 10% inoculation rate, with 100 µL fresh Middlebrook 7H9 medium and 10% OADC per well. Strains were mixed with the indicated concentrations of Osimertinib in a 7H9 medium. The antimicrobial activity assay was carried out on the Spectra M5 plate reader (Molecular Devices) instrument. The absorption value of 600 nm (OD600) was collected every 30 min at 37 °C. M. Bovis BCG Danish strain (ATCC 35733) was grown in Middlebrook 7H9 broth and on Middlebrook 7H10 agar supplemented with 10% OADC at 37 °C. After three days of incubation of M. Bovis BCG with Osimertinib, the culture was diluted and cultured in 7H10 agar plates at 37 °C for 3–4 more weeks. Then, the CFU of bacteria was counted.
Statistics and reproducibility
Each experiment was performed at least three times (n = 3). All experiment data were analyzed using GraphPad Prism 8.0 (GraphPad Software Inc., USA) and were presented as the mean ± SD. Statistical analysis was performed using Student’s t test or two-way ANOVA. A value of P < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The coordinates and structural factors have been deposited in the Protein Data Bank with accession codes 7VMX (EF-Tu•EF-Ts complex) and 7VOK (EF-Tu•GDP complex). The SAXS data and models have been deposited in the SASBDB database with accession codes SASDQB5 (EF-Tu), SASDQC5 (EF-Ts), and SASDQD5 (EF-Tu/EF-Ts complex). All data generated or analyzed during this study are included in this published article (and its supplementary information files). The source data for all plots are shown in Supplementary Data 1. The uncropped and unedited gel images are included in Supplementary Data 2. All other data are available from the corresponding author upon reasonable request.
References
Yang, L. et al. Opportunities for overcoming tuberculosis: emerging targets and their inhibitors. Drug Discov. Today 27, 326–336 (2022).
Gegia, M. et al. Treatment of isoniazid-resistant tuberculosis with first-line drugs: a systematic review and meta-analysis. Lancet Infect. Dis. 17, 223–234 (2017).
Zimhony, O. et al. Pyrazinamide inhibits the eukaryotic-like fatty acid synthetase I (FASI) of Mycobacterium tuberculosis. Nat. Med. 6, 1043–1047 (2000).
Kashyap, A., Singh, P. K. & Silakari, O. Chemical classes targeting energy supplying GyrB domain of Mycobacterium tuberculosis. Tuberculosis 113, 43–54 (2018).
Singh, V. & Chibale, K. Strategies to combat multi-drug resistance in tuberculosis. Acc. Chem. Res. 54, 2361–2376 (2021).
Leach, K. L. et al. Linezolid, the first oxazolidinone antibacterial agent. Ann. N. Y Acad. Sci. 1222, 49–54 (2011).
de Wet, T. J., Warner, D. F. & Mizrahi, V. Harnessing biological insight to accelerate tuberculosis drug discovery. Acc. Chem. Res. 52, 2340–2348 (2019).
Schmeing, T. M. & Ramakrishnan, V. What recent ribosome structures have revealed about the mechanism of translation. Nature 461, 1234–1242 (2009).
Guo, H. Specialized ribosomes and the control of translation. Biochem Soc. Trans. 46, 855–869 (2018).
Voorhees, R. M. & Ramakrishnan, V. Structural basis of the translational elongation cycle. Annu Rev. Biochem. 82, 203–236 (2013).
Noel, J. K. & Whitford, P. C. How EF-Tu can contribute to efficient proofreading of aa-tRNA by the ribosome. Nat. Commun. 7, 13314 (2016).
Yamamoto, H. et al. EF-G and EF4: translocation and back-translocation on the bacterial ribosome. Nat. Rev. Microbiol. 12, 89–100 (2014).
Krab, I. M. & Parmeggiani, A. Mechanisms of EF-Tu, a pioneer GTPase. Prog. Nucleic Acid Res. Mol. Biol. 71, 513–551 (2002).
Scheffzek, K. & Ahmadian, M. R. GTPase activating proteins: structural and functional insights 18 years after discovery. Cell Mol. Life Sci. 62, 3014–3038 (2005).
Maracci, C. et al. Ribosome-induced tuning of GTP hydrolysis by a translational GTPase. Proc. Natl Acad. Sci. USA 111, 14418–14423 (2014).
Nissen, P. et al. The ternary complex of aminoacylated tRNA and EF-Tu-GTP. Recognition of a bond and a fold. Biochimie 78, 921–933 (1996).
Stark, H. et al. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol. 9, 849–854 (2002).
Kavaliauskas, D. et al. Structural dynamics of translation elongation factor Tu during aa-tRNA delivery to the ribosome. Nucleic Acids Res. 46, 8651–8661 (2018).
Morse, J. C. et al. Elongation factor-Tu can repetitively engage aminoacyl-tRNA within the ribosome during the proofreading stage of tRNA selection. Proc. Natl Acad. Sci. USA 117, 3610–3620 (2020).
Clark, B. F. & Nyborg, J. The ternary complex of EF-Tu and its role in protein biosynthesis. Curr. Opin. Struct. Biol. 7, 110–116 (1997).
Lathe, W. C. 3rd & Bork, P. Evolution of tuf genes: ancient duplication, differential loss and gene conversion. FEBS Lett. 502, 113–116 (2001).
Hughes, D. Both genes for EF-Tu in Salmonella typhimurium are individually dispensable for growth. J. Mol. Biol. 215, 41–51 (1990).
Parmeggiani, A. & Nissen, P. Elongation factor Tu-targeted antibiotics: four different structures, two mechanisms of action. FEBS Lett. 580, 4576–4581 (2006).
Arenz, S. & Wilson, D. N. Bacterial protein synthesis as a target for antibiotic inhibition. Cold Spring Harb. Perspect. Med. 6, a025361 (2016).
Fabbretti, A. et al. A derivative of the thiopeptide GE2270A highly selective against Propionibacterium acnes. Antimicrob. Agents Chemother. 59, 4560–4568 (2015).
Prezioso, S. M., Brown, N. E. & Goldberg, J. B. Elfamycins: inhibitors of elongation factor-Tu. Mol. Microbiol. 106, 22–34 (2017).
Kawashima, T. et al. The structure of the Escherichia coli EF-Tu.EF-Ts complex at 2.5 A resolution. Nature 379, 511–518 (1996).
Wang, Y. et al. Crystal structure of the EF-Tu.EF-Ts complex from Thermus thermophilus. Nat. Struct. Biol. 4, 650–656 (1997).
Kjeldgaard, M. & Nyborg, J. Refined structure of elongation factor EF-Tu from Escherichia coli. J. Mol. Biol. 223, 721–742 (1992).
Schmeing, T. M. et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–694 (2009).
Nissen, P. et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270, 1464–72. (1995).
Jeppesen, M. G. et al. Crystal structure of the bovine mitochondrial elongation factor Tu.Ts complex. J. Biol. Chem. 280, 5071–5081 (2005).
Kuang, S. et al. Structure insight of GSDMD reveals the basis of GSDMD autoinhibition in cell pyroptosis. Proc. Natl Acad. Sci. USA 114, 10642–10647 (2017).
Li, Z. et al. Structural insights into the complex of trigger factor chaperone and ribosomal protein S7 from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 512, 838–844 (2019).
Miller, D. L. & Weissbach, H. Studies on the purification and properties of factor Tu from E. coli. Arch. Biochem Biophys. 141, 26–37 (1970).
Cai, Y. C. et al. Interaction of mitochondrial elongation factor Tu with aminoacyl-tRNA and elongation factor Ts. J. Biol. Chem. 275, 20308–20314 (2000).
Palmer, S. O. et al. Cloning and characterization of EF-Tu and EF-Ts from Pseudomonas aeruginosa. Biomed. Res. Int. 2013, 585748 (2013).
Wieden, H. J. et al. Mechanism of elongation factor (EF)-Ts-catalyzed nucleotide exchange in EF-Tu. Contribution of contacts at the guanine base. J. Biol. Chem. 277, 6032–6036 (2002).
Schuette, J. C. et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 28, 755–765 (2009).
Ahmad, M. U. D. et al. Nano-differential scanning fluorimetry for screening in fragment-based lead discovery. J. Vis. Exp. 171, e62469 (2021).
Tan, C. S., Gilligan, D. & Pacey, S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 16, e447–e459 (2015).
Andersen, G. R., Nissen, P. & Nyborg, J. Elongation factors in protein biosynthesis. Trends Biochem Sci. 28, 434–441 (2003).
Wolf, H., Zahner, H. & Nierhaus, K. Kirromycin, an inhibitor of the 30 S ribosomal subunits function. FEBS Lett. 21, 347–350 (1972).
Sajid, A. et al. Interaction of Mycobacterium tuberculosis elongation factor Tu with GTP is regulated by phosphorylation. J. Bacteriol. 193, 5347–5358 (2011).
Assmann, D. & Wolf, H. Pulvomycin, an inhibitor of prokaryotic protein biosynthesis. Arch. Microbiol. 120, 297–299 (1979).
Watanabe, T., Izaki, K. & Takahashi, H. New polyenic antibiotics active against gram-positive and -negative bacteria. I. Isolation and purification of antibiotics produced by Gluconobacter sp. W-315. J. Antibiot. 35, 1141–1147 (1982).
Selva, E. et al. Antibiotic GE2270 a: a novel inhibitor of bacterial protein synthesis. I. Isolation and characterization. J. Antibiot. 44, 693–701 (1991).
Bhat, Z. S. et al. Drug targets exploited in Mycobacterium tuberculosis: pitfalls and promises on the horizon. Biomed. Pharmacother. 103, 1733–1747 (2018).
Chen, Z. et al. Structural basis of human helicase DDX21 in RNA binding, unwinding, and antiviral signal activation. Adv. Sci. 7, 2000532 (2020).
Han, Y. et al. Crystal structure of steroid reductase SRD5A reveals conserved steroid reduction mechanism. Nat. Commun. 12, 449 (2021).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput Chem. 31, 455–461 (2010).
Acknowledgements
We thank the staff of the Shanghai Synchrotron Radiation Facility beamlines BL17U1, BL18U, BL19U1, and BL19U2 for help with X-ray data and SAXS data collection. This work was supported by grants from the National Key Research and Development Project of China (2021YFC2301500, 2016YFA0500600), the National Natural Science Foundation of China (32161160323), and the Shanghai Committee of Science and Technology (20XD1400800).
Author information
Authors and Affiliations
Contributions
J.L. and G.Z. conceived and designed this study. B.Z., Y.G., W.G., Y.L., Z.L., Q.Q., X.L., H.S., and J.G. performed research and analyzed data; J.L. and G.Z. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Gene Chong.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhan, B., Gao, Y., Gao, W. et al. Structural insights of the elongation factor EF-Tu complexes in protein translation of Mycobacterium tuberculosis. Commun Biol 5, 1052 (2022). https://doi.org/10.1038/s42003-022-04019-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-022-04019-y
This article is cited by
-
Screening inhibitors against the Ef-Tu of Fusobacterium nucleatum: a docking, ADMET and PBPK assessment study
Molecular Diversity (2024)
-
Structural insights into RNase J that plays an essential role in Mycobacterium tuberculosis RNA metabolism
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.