Abstract
Color change during flower opening is common; however, little is understood on the biochemical and molecular basis related. Lilac (Syringa oblata), a well-known woody ornamental plant with obvious petal color changes, is an ideal model. Here, we presented chromosome-scale genome assembly for lilac, resolved the flavonoids metabolism, and identified key genes and potential regulatory networks related to petal color change. The genome assembly is 1.05 Gb anchored onto 23 chromosomes, with a BUSCO score of 96.6%. Whole-genome duplication (WGD) event shared within Oleaceae was revealed. Metabolome quantification identified delphinidin-3-O-rutinoside (Dp3Ru) and cyanidin-3-O-rutinoside (Cy3Ru) as the major pigments; gene co-expression networks indicated WRKY an essential regulation factor at the early flowering stage, ERF more important in the color transition period (from violet to light nearly white), while the MBW complex participated in the entire process. Our results provide a foundation for functional study and molecular breeding in lilac.
Similar content being viewed by others
Introduction
As one of the most important ornamental traits, floral color diversity has not only attracted much attention in the breeding and development of new varieties, but also play an important role in the evolution of floral traits and ecological effects1. Flower color change is a common phenomenon that accompanies the developmental period. The flower color of many plants is formed early and then fades during flower opening. For example, Malus hupehensis2 and Brunfelsia acuminata3 have red and purple flowers respectively in early flower opening stage, which gradually change to white with the opening. In the petals of Chrysanthemum morifolium4 and Cleome hassleriana5, the heavy pink coloration and anthocyanin content show the highest level before flower opening, and then the color gradually changed to light pink due to the continuous decrease in anthocyanin content. In addition, with the flower development the petal color of Paeonia 'Coral Sunset' and 'Pink Hawaiian Coral'6 fade from coral to light pink to pale yellow, presenting color group change. Petal color changes is often accompanied by both accumulation and loss of anthocyanins; therefore, a better understanding of the metabolic dynamics and molecular basis of petal color change is important for prolonging flower coloring and improving ornamental and economic value.
The accumulation of floral color has been studied more intensively and is mainly determined largely by distinct pigments, including flavonoids, carotenoids and/or alkaloids7. Among which, flavonoids as the third-largest group of natural products, are widely known for their important roles in plant fitness, health benefits and food quality8. Anthocyanin biosynthesis, as part of the flavonoids pathway, is essential for the appearance of these colors, which are expressed in plant tissues in a wide variety, from red to purple and even blue9. As we know, pigment accumulation is mainly determined by biosynthetic genes and their transcriptional regulation via transcription factors (TFs), which plays a major role in flower color development10,11. Currently, advances in genetic engineering make rare flower color development and directed breeding possible11,12.
Actually, there have been many studies on the biosynthesis and regulatory mechanisms of flower color, yet the widespread phenomenon of petal color change has been neglected. The genus Syringa (Oleaceae), known for the outstanding beautiful corollas, consists of more than 27 species and 2000 cultivars worldwide, of which 22 are distributed in China, including 18 endemic species13. As the representative species, Syringa oblata (lilac) is native to China with more than 1000 years of cultivation13, owing to its elegant colors, unique fragrance, beautiful corollas, large inflorescence, and early flowering. Moreover, as one of the rare trees with purple flowers, S. oblata is the most widely cultivated as ecological and ornamental tree, spanning subtropical, warm temperate, temperate, and the edge of the cold temperate zones14. Since the 20th century, there have been more than 200 new cultivars bred with S. oblata as excellent hybrid parents, including Syringa × hyacinthiflora and cultivars breeding by crossing between S. oblata and S. vulgaris. It is observed that lilac petals are darkest and red-purple in the bud stage, then gradually change to violet and finally fade close to white. Differences in anthocyanins lead to flower color changes and the production of various flower colors11; however, the biochemical and molecular basis for this change is still unclear. Lilac is considered as an excellent material to study the mechanism of petal color change because of its phenotypic advantages.
In lilac flowers, only two anthocyanins have been reported in the full blooming stage15, so a comprehensive assay and the metabolic dynamic analysis is lacking. Several studies have delved into the expression difference of some synthesis genes in the flavonoid biosynthetic pathway during flower developmental stage and cloned and verified the functions of some genes16, however, with the absence of genomic evidence, it is difficult to explore their regulatory mechanisms16. Therefore, the whole-genome sequence of S. oblata is needed to develop an in-depth understanding of the evolution and potential regulatory networks of petal color formation underlying biochemical process in successive flower development stages12.
Here, we present a high-quality chromosome-scale genome assembly of S. oblata, and it was determined by a combination of long-read sequencing and Hi-C scaffolding technologies. In total, 1.05 Gb and 93.75% of the assembled genome was anchored on 23 chromosomes with a scaffold N50 of 46.33 Mb. Furthermore, we predicted to contain 35,313 protein-coding genes and clarified the species WGD history. The metabolic dynamic results systematically revealed the composition and relative quantification of compounds related to flavonoid biosynthesis in lilac petals. Moreover, combining gene annotation, transcriptome and metabolome evaluations, we revealed a time-ordered co-expression network of color-changing petals, and identified the enzyme genes and potential regulatory factors controlling the key pigments associated with petal color change. This reference genome sequence, metabolic dynamic results and the generated gene regulatory network for petal color change are expected for further functional genomics investigations and molecular breeding.
Results
Genome sequencing, assembly, and quality assessment
Genome sequencing resulted in a total of 237.19 Gb of nanopore long-reads (Supplementary Fig. 1; Supplementary Table 1), with approximately 204-fold depth of 1.16 Gb genome, as estimated based on K-mer frequency analysis (Supplementary Fig. 2; Supplementary Table 2). Furthermore, short-reads PCR-free sequencing generated 45.65 Gb (39-fold) reads (Supplementary Table 3); from which 154.31 Gb (133-fold) Hi-C paired-end reads were produced (Supplementary Fig. 3; Supplementary Table 3). A high genome heterozygosity of 1.92% and a moderate repeat rate of 51.79% were assessed by 25-mer analysis (Supplementary Fig. 2). Here, we obtained an assembly ready for Hi-C scaffolding, which was 1.12 Gb in the length of contig with an N50 length of 3.96 Mb (Supplementary Table 4).
With the Hi-C scaffolding, the final assembly was generated by anchoring 443 contigs to 23 chromosome-scale super scaffolds with a total length of 1.05 Gb (Table 1; Supplementary Table 5; Supplementary Fig. 3), accounting for 93.75% (1.05/1.12Gb) of the assembly, with a scaffold N50 of 46.33 Mb (Supplementary Table 5). The number of pseudochromosomes matched the experimentally determined number of chromosome17 (2n = 46) (Fig. 1) and the pseudomolecules ranged in size from 34.29 to 69.21 Mb (Supplementary Table 6).
a Photo of S. oblata in Beijing, China (by Bo Ma, 2021). b Twenty-three pseudochromosomes were ordered by size. Chr1-Chr23 refers to the 23 assembled chromosomes. (i) Assembled chromosomes (Mb, orange); (ii) Gene density (green); (iii) Transposon density (blue); (iv) Tandem repeat density (purple); (v) GC content (yellow); Line in the center of the circle represent chromosome collinearity.
The short-reads genome mapping rate was 99.37%. Further, 1559 complete BUSCO (96.60%) were assembled, among which only 27 (1.70%) had fragmented matches, and 28 (1.70%) were entirely missing (Supplementary Table 7), indicating that the assembled genome had high accuracy, contiguity and completeness.
Genome annotation
A combination of three different strategies was used to perform the repeat annotation, including ab initio, homology-based, and transcriptome-based predictions. In total, we identified 572,494,098 bp repeat elements, accounting for 54.30% of the assembled genome (Table 1; Supplementary Table 8). This proportion is higher than that reported for other sequenced plants of Oleaceae, including Fraxinus excelsior18 35.90%, Osmanthus fragrans19 49.35%, and Olea europaea20 42.95%. These repeat elements contained 43.91% predominantly known transposable elements (TEs) (38.87% retrotransposons and 5.04% DNA transposons), 6.33% unclassified TEs, 3.71% tandem repeat, and 0.34% low complexity. Among all TEs, 37.79% consisted of LTR-RTs and most of them are LTR/Gypsy, occupying 23.37% of the genome, followed by the LTR/Copia (12.54%) (Supplementary Table 8).
A total of 35,313 protein-coding genes were predicted (Table 1), with average gene length of 4515 bp, of which 33,268 (94.21%) were assigned functional annotation (Supplementary Table 9), and 91.77% of the predicted genes had complete structure (Supplementary Table 10). The average length of transcripts and coding regions were 1650 bp and 1148 bp, respectively, with a mean of 6.41 exons (Supplementary Table 9). In total, 30,454 (86.24%), 13,431 (38.03%), 15,733 (44.55%), 33,242 (94.14%), 25,882 (73.29%), 29,765 (84.29%), and 29,833 (84.48%) gene models were annotated to Cluster of Orthologous Groups of proteins (COG)21, Gene Ontology (GO)22, Kyoto Encyclopedia of Genes and Genomes (KEGG)23, the NCBI Non-Redundant Protein Sequence Database (NR), Swiss-Prot, TAIR, and MSU Rice Genome Annotation Project Database24, respectively (Supplementary Table 9). Among which, in the GO annotation, 6681 genes were assigned to the biological process, 10,653 genes were assigned to the cellular compound, and 6958 genes were assigned to the molecular function categories. Additionally, we identified non-coding RNAs, including 231 miRNAs, 724 transfer RNAs (tRNA), 138 ribosomal RNAs, and 3690 small nuclear RNAs (snRNA) (Table 1; Supplementary Table 11).
We identified 3035 genes encoding TFs from 95 TF families in the lilac genome, accounting for 8.59% of protein-coding genes. The most members in the TF families were WD40 (308), AP2/ERF-ERF (205), MYB (180), bHLH (159), C2H2 (144), NAC (142), and WRKY (117).
Gene family analysis and phylogenetic tree construction
To investigate the evolution of the lilac genome, we performed a comparative genomic analysis with three Oleaceae plants (O. europaea, F. excelsior, and O. fragrans), three sequenced flowering plants (Prunus mume, Rhododendron delavayi, and Carica papaya), two representative plants of Solanaceae family (Solanum lycopersicum and S. tuberosum), two rosid plants (Arabidopsis thaliana and Vitis vinifera), and a monocot plant (Oryza sativa) (Supplementary Table 12). A total of 29,856 gene families encompassing 386,920 genes were identified among the 12 species, from which 14,829 gene families (33,058 genes) were found in S. oblata (Supplementary Table 13). The sequences of the 356 conserved single-copy orthologous genes identified were used to reconstruct the phylogenetic relationship with O. sativa serving as the outgroup (Fig. 2a). The revealed phylogeny was in agreement with previous studies20,25, which indicated that S. oblata had the closest relationship with Oleaceae (F. excelsior, O. fragrans, and O. europaea) and formed a clade with Solanaceae (S. lycopersicum and S. tuberosum) followed by R. delavayi, and was distant from the other species. In addition, lilac was separated from the Oleaceae common ancestor at ~ 43.20 (38.23–48.18) million years ago (Mya) (Fig. 2a).
a Phylogenetic tree, gene family expansions/contractions in lilac and 11 other species based on a concatenated alignment of 356 single-copy orthologs, and divergence times. b Venn diagram illustrating the shared and unique gene families among S. oblata and three Oleaceae plants (O. europaea, F. excelsior, and O. fragrans). c Ks distribution of S. oblata, F. excelsior, O. europaea, O. fragrans, S. oblata-F. excelsior, S. oblata-O. fragrans, S. oblata- S. lycopersicum and V. vinifera. d Chromosome collinearity analysis among lilac, O. europaea, O. fragrans and V. vinifera. Green lines represent four syntenic blocks in lilac match up to one region in V. vinifera.
We also identified unique and shared gene families among these species. A total of 11,952 gene families were shared with Oleaceae, and 2982 unique gene families were found in S. oblata genome compared with O. europaea (5590), F. excelsior (3174), and O. fragrans (3625) (Fig. 2b); 400 gene families (2255 genes) were found to be specific to the assembled S. oblata genome when compared with the other 11 genomes (Supplementary Table 13). Moreover, 990 gene families (3876 genes) were found in expansion, while 1599 gene families (3513 genes) were found in contraction in the S. oblata lineage (Fig. 2b). Functional enrichment analysis showed that expanded gene families were enriched in GO terms mostly involved in 'binding', 'transport', 'metabolic process', and 'signal transduction' (Supplementary Fig. 4). In contrast, KEGG enrichment analysis revealed that the expanded genes were mainly from pathway implicated in plant defense response, development, and monoterpenoid biosynthesis (Supplementary Fig. 5).
Gene synteny analysis and WGD events
Synonymous substitutions per site (Ks) age distribution, synteny analyses, and inferred dating times were used to unveil WGD events and to date the duplication events26. The divergence time of lilac and the other three Oleaceae species was approximately predicted at 43.20 Mya (Ks = 0.2) (Fig. 2a), simultaneously, they produced the same signature Ks peaks of WGDs at about 0.2519,20,27 (Fig. 2c), higher than 0.2, suggesting that the most recent WGD event, estimated at around 54 MYA (Ks = 0.25), occurred before the divergence of Oleaceae family (Fig. 2c, Supplementary Fig. 4). Similarly, the main Ks peak value of 0.25, was smaller than the Ks peak suggesting divergence between Oleaceae and Solanaceae (approximately 1.1) (Fig. 2c), indicating that the most recent WGD event occurred after the divergence of Oleaceae and Solanaceae. Moreover, the intra-genomic synteny analyses showed that 15,786 gene pairs distributed along 855 blocks in lilac (Supplementary Fig. 6; Supplementary Table 14), with synteny analyses of O. fragrans (Supplementary Fig. 7; Supplementary Table 14), which further supported this Oleaceae-specific WGD event. At the same time, the inter-genomic analyses between S. oblata and O. fragrans exhibited relatively conservative collinearity (Fig. 2d; Supplementary Fig. 8; Supplementary Table 14), supporting a close evolutionary relationship.
We also observed a weaker signal of an additional ancient WGD shared by Oleaceae lineage (Fig. 2c), which has been previously reported based on genomic data20. Subsequently, we detected a 4:1 syntenic relationship between S. oblata and V. vinifera (Fig. 2d), and built a gene homology dot plot (Supplementary Fig. 9; Supplementary Table 14), which provided additional evidence for the identified two WGD events19,20 after the Gamma paleo-hexaploidy in the stem lineage of core eudicots28,29. Moreover, the Ks analysis of four Oleaceae plants supported the WGD events (Fig. 2c), which were inferred from the lilac genome (as well as from O. europaea20, F. excelsior18 and O. fragrans19) and were specific to Oleaceae.
Reconstruction of flower pigments biosynthesis pathway in S. oblata
Our high-quality genome assembly and transcriptome data allowed reconstruction of the flower pigment biosynthesis pathway, specifically capturing the implicated enzymatic genes in this process. We unveiled 124 genes encoding functioning enzymes in the flavonol (from colorless to pale yellow) and colored anthocyanin (from pink-red to violet) biosynthesis pathway (Fig. 3); 14 important gene families in this pathway were identified, among which, 4CL (19), F3H (15), DFR (15), and F3oGT (24) gene families had a large number of members (Fig. 3). In addition, we found that members of the PAL, F3H, F3'H|F3'5'H, DFR, F3oGT and RT gene families were replicated, especially F3H and F3oGT, which might enhance flavonoids production. Gene expression profiles showed that during the flower opening period the CHS (So16G045740 and So19G016880), F3H (So13G050140 and So15G003400), and F3'H|F3'5'H (So05G012500 and So07G050530) increased firstly and then decreased, and F3H (So17G006510 and So22G037260) and F3'H|F3'5'H (So01G039330) gradually decreased, which was consistent with the pattern of anthocyanin contents (Figs. 3 and 4b; Supplementary Fig. 11).
The metabolic pathways of flavonol and anthocyanin. Gene expression profiles (log10(FPKM+1)) at different flower opening stages (from left to right were T1-T4), and the bar color change from blue to red indicates the expression from low to high. The left side of the heat map was the corresponding enzymes, and the right was the genes id within the genome. Genes identified in tandem clusters were marked in yellow color, and the top5 metabolites of petals at the four periods were marked with purple box. PAL phenylalanine ammonia-lyase, C4H cinnamate-4-hydroxylase, 4CL 4-coumarate CoA ligase 4, CHS chalcone synthase, CHI chalcone isomerase, F3H flavanone 3-hydroxylase, F3'H flavonoid 3'-hydroxylase, F3'5'H flavonoid 3'5'-hydroxylase, FLS flavonol synthase, DFR dihydroflavonol 4-reductase, ANS anthocyanidin synthase, F3oGT flavonoide-3-O-glucosyltransferase, RT rhamnosyltransferase.
Delphinidin-based anthocyanins which had the most contents in lilac petals usually were the major constituents of violet and blue flowers in nature, and F3'5'H was the key synthesis enzyme30. We noticed that the So07G050530 expression first increased and then decreased during the flower opening period, which showed the high levels at T2 and T3 (Fig. 3). Its expression profile was consistent with blue-violet anthocyanins petunidin-3-O-rutinoside (Pt3Ru) contents (Fig. 4b), suggesting that the expression of So07G050530 may be related to Pt3Ru synthesis.
Metabolic dynamic results revealed the petal color change in S. oblata
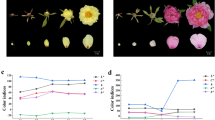
Before blooming, there was a rapid pigment accumulation process in the lilac flower, and gradually decreased during the flower opening16, with color change from dark to light purple (Fig. 4a). For further study the lilac's petal color change, four flower opening period were divided according to phenotypic characteristics (Fig. 4a, T1-T4). At the T1 stage (flower bud stage), the flower buds were red-purple and close to the calyx, no corolla tubes and lobes were showing or even barely exposed; at T2 (initial flower opening stage), corolla tubes were elongated, flower buds were enlarged, corolla lobes were about ready to open, and flower color was from red-purple to violet; at T3 (full flower opening stage), corolla lobes were stretched with almost 90° angle between the corolla lobes and tubes, and flower color was light violet; and at T4 (last flower opening stage), flowers color decayed and wilted, the corolla lobes were almost reversely curled, and the color of the flower became lighter than other three stages and tend to be in light violet nearly white (Fig. 4a).
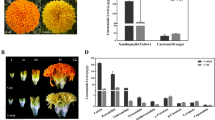
a Four flower opening stages of lilac from T1 to T4 (photographed by Bo Ma in 2021). b Relative contents of main anthocyanins including Dp3Ru, Cy3Ru, and Pt3Ru at the four flower opening stages. Data are presented as mean ±SE, n = 3 biologically independent samples (Supplementary Data 2); statistical significance was calculated by one-way ANOVA with multiple comparisons between adjacent stages, and was indicated by asterisks, in which n.s. (no significance), p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), and p < 0.0001 (****). c PCA distance biplot. The PCA biplot shows both the PC scores (PC1 and PC2 here) of samples (here: three petal samples for each of the four flower opening periods, as dots) and the loadings of variables (here: the relative content of metabolites in each petal sample, as vectors) in the 2-dimension space defined by the first two PCs. We can observe the difference between petal samples at different flower opening periods, the relative contribution of metabolite variables to this difference, and the relationship of the difference. d Predicted regulatory network and the connection among metabolites, structural genes of flavonoids (Fig. 3) and TFs. The purple dots on the top represent metabolites; the pink dots in the middle represent structural genes in the flavonoid metabolic pathway (Fig. 3); at the bottom, inside the blue circle, the green dots represent the MYB genes, the red dots represent the ERF genes, the orange dots represent the WD40 genes, the pink dots represent the bHLH genes, the blue dots represent the WRKY genes; and outside of the circle, blue dots represent the remaining TFs. L1 to L8 indicate the levels identified in the time-ordered gene co-expression network. The PCC value increases from small to large, the width of the edges changes from thinner to thicker in the networks. e the heatmaps of average normalized FPKMs for each flower opening periods at each level identified in the time-ordered gene co-expression network. The three stages of flower color change are determined based on the gene expression profile and metabolome, as early stage (T1, red-purple), transition stages (T2-T3, violet), and fading stage (T4, light violet nearly white).
A total of 118 flavonoid metabolites (Supplementary data 1) were detected, including 9 anthocyanins and 40 flavonols, and anthocyanins accumulation was the main factor determining flower color. The top 10 compounds accounted for 71.86% of the total content, including 2 anthocyanins, Dp3Ru (tended to be violet-blue31, 22.76%) and Cy3Ru (tended to be red-purple31, 14.33%) (Fig. 4b); 4 kaempferol-based flavonoids, kaempferol-3-O-rutinoside (8.33%), kaempferol-4'-O-glucoside (5.11%), kaempferol-3-O-glucoside (4.83%), and kaempferol-3-O-β-D-glucoside-7-α-L-rhamnoside (4.49%); 4 quercetin-based flavonoids, isorhamnetin-3-O-rutinoside (3.66%), isohyperoside (3.03%), quercetin-3-O-xylosyl(1→2)glucoside (2.73%), and quercetin-3-O-galactoside (2.57%) (Supplementary Fig. 10; Supplementary data 1). For the top 5 metabolites (Fig. 3), except for kaempferol-3-O-rutinoside, the contents of remaining 4 metabolites yielded the lowest values at T4 (Supplementary Fig. 10), consistent with flower color changes as seen with phenotypic observation (Fig. 4a). The latter three flavonoids (kaempferol-3-O-rutinoside, kaempferol-4-O-glucoside, and kaempferol-3-O-glucoside) determined the pure yellow flower color, and showed an overall trend of first increasing and then decreasing, all of which reached the highest contents at T2 (Supplementary Fig. 10).
Subsequently, we focused on the 9 anthocyanins, namely delphinidin-based (4 types, tended to be violet-blue), cyanidin-based (4 types, tended to be red-purple), and pelargonidin-based (1 type, tended to be orange-red) anthocyanins (Fig. 4b; Supplementary Fig. 11). The first two, Dp3Ru and Cy3Ru determined lilac flower color and showed the highest contents in T1; and at T2 and T3, the contents of Dp3Ru was still high, while Cy3Ru only showed one-third of content at T1 (Fig. 4b). In addition, the content change of pelargonidin-3-O-rutinoside (Pg3Ru) was consistent with Dp3Ru, which was much higher than the others, and cyanidin-3-O-glucoside (Cy3G) was consistent with Cy3Ru (Supplementary Fig. 11). Different from the monotonously decreased contents of first three pigments, the Pt3Ru, a delphinidin-based anthocyanin, first showed an increasing pattern and then decreased along with flower opening, with the highest value at T2 (Fig. 4b). Interestingly, the differential accumulation of pigments demonstrated that the cyanidin-based anthocyanins caused the color change of lilac from T1 to T3, while Dp3Ru did not differ significantly among the first three flower opening stages, causing color changes from T3 to T4 (Fig. 4b).
Furthermore, the PCA based on 20 differentially expressed major metabolites, including 8 anthocyanins (marker in red and purple in Fig. 4c; Supplementary Fig. 11) and 12 flavonoids with high contents (marker in yellow in Fig. 4c; Supplementary Fig. 12), showed that the first two principal components (PCs) cumulative contribution of the accounted for 84.6% of the variation (Fig. 4c). It is worth noting that PC1 (accounted for 67.5%) and can separate all four stages (Fig. 4c). Furthermore, the samples were arranged clearly in order of flower opening time (from left to right, Fig. 4c), with which anthocyanins, the higher contents at the early flower opening stage (Cy3Ru, Cy3G, Dp3Ru, Pg3Ru, and delphinidin (Dp)) were negatively correlated, while Pt3Ru and cyanidin (Cy) were positively correlated. As can be seen, the metabolic dynamics of these two pigments, Cy3Ru and Pt3Ru, were closely related to the color change of lilac flowers at the early and late blooming stages, respectively (Fig. 4b–c). Therefore, PCA clearly revealed the dynamic changes of the anthocyanin and other flavonoids contents with flower opening stages, which was an important basis for the dynamic integration of flower color phenotype and relevant gene expression.
Dynamic regulation of petal color change in S. oblata
The 12 samples of the four stages were clearly separated by metabolome and transcriptome PCA analysis (Supplementary Fig. 13). Between any two flower opening stages (T1 to T4), we found a total of 20,984 differentially expressed genes (DEGs) (2507 TFs and 18,477 structural genes) with an average Fragments Per Kilobase Million (FPKM) greater than 0.5 and 80 metabolites exhibiting significant differentiation. Then, a TF gene of MYB family, So16G008570, expressing in a high level at T1 and monotonically decreased until T4, was selected as the initial node to build the time-ordered gene co-expression network (TO-GCN). According to Pearson's correlation coefficient (PCC) values, eight time-ordered expression levels (denoted as L1 to L8, Fig. 4d) were obtained, which comprised 137,523 edges and 1427 nodes. Finally, 1347 genes including 1268 significantly differentially expressed TFs genes and 79 structural genes of anthocyanin and flavonol biosynthesis pathway, and 80 metabolites (including 8 anthocyanins) made up the TO-GCN (Fig. 4d). These 8 levels matched the expression peaks of DEGs at the four flower opening stages, as shown by the red squares (high expression levels) along the diagonal in the heat-map, which formed the basis for the inference of upstream and downstream genes/metabolites regulatory relationships (Fig. 4e).
The analysis revealed that most network nodes appeared at T1 (corresponding to time-ordered levels L1-L3), including 31 enzymatic genes in the anthocyanin and flavonol biosynthesis, 612 TFs, and 41 metabolites. There were 265 genes (including 247 TFs), 10 metabolites at T2 (corresponding to time-ordered level L4), and 287 genes (including 262 TFs), 15 metabolites at T3 (corresponding to time-ordered levels L5-L6). The least genes, only 5 structural genes appeared at T4 (corresponding to time-ordered levels L7-L8), when there were 147 TFs and 14 metabolites. The distribution of structural genes in time-ordered levels showed that the early biosynthetic genes (EBGs: PAL, C4H, 4CL, CHS, CHI, F3H, F3'H)8 of flavonoid biosynthesis pathway were mainly clustered in the latter levels (Fig. 4d).
For better description, we further distinguished the early flower opening stage (T1, when flowers were red-purple), the transition stage (T2 and T3, violet), and the fading stage (T4, nearly white). At T1, we found that among the 583 TFs that may directly regulate structural genes of the flavonoid pathway, the bHLH family contained the most members (59), followed by the WRKYs family (53) (Fig. 5a). In addition, in the potential regulatory sub-network (Fig. 5b) the structural genes were regulated mostly by MYB, bHLH, and WD40, including F3oGT (So01G017180; So01G079250; So01G079260; So01G079270; So18G022420; So18G022460; So19G037390; So07G011940; So08G012630), RT (So13G042380), F3H (So22G037260), and F3'H|F3'5'H (So01G039330). To analyze the change of lilac flower color, the sub-networks of differential expressed anthocyanins were performed (Fig. 5c; Supplementary Figs. 14–15). We found that Cy3Ru (Fig. 5c) and Cy3G (Supplementary Fig. 14) were two main high-content cyanidin-based pigments that caused red color fade, among the five analyzed anthocyanins distributed at T1 (Fig. 5c; Supplementary Figs. 14–15). The regulatory network of F3'H|F3'5'H (So01G039330) was constructed (Fig. 5c), which encoded a key enzyme for the synthesis of DHQ and was directly related to the Cy3Ru. We initially speculated the potential regulatory networks in which WRKY played an important role together with MYB, bHLH, WD40 and ERF (Fig. 5c–d). Meanwhile, composition of the 5'UTR upstream cis-acting elements of the F3'H|F3'5'H gene and the expression levels of related genes supported the networks (Supplementary Fig. 19).
a Sub-network for flavonoids metabolism. The network consists of metabolites (purple), structural genes (pink) and TFs from the outside to the inside. b Sub-network for flavonoids metabolism containing only the three families of TFs MYB (green), bHLH (pink), and WD40 (orange) at the early flowering stage. The structural genes were linked to bHLH, MYB and WD40 in red, blue, and orange lines, respectively. c Resolved one of the metabolisms of Cy3Ru and regulation for F3'H|F3'5'H gene involved. And 38 TFs (sixteen bHLH, five MYB, four WD40, nine WRKY and four ERF) identified as its direct regulators. d TFs (from Fig. 5c) heat-maps of gene expression profiles (log10(FPKM+1)) at the four flower opening stages and DNA binding sites of F3'H|F3'5'H gene (from Fig. 5c) in 5'UTR upstream 2Kb. The bar color change from blue to red indicates the expression from low to high. The PCC value increases from small to large, the width of the edges changes from thinner to thicker in the networks.
Among the 425 potential direct regulatory factors at the transition stage (T2 and T3), the WD40 family contained most members (65), which was nearly two-thirds more than that at T1, followed by MYB and ERF families (Fig. 6a). Similarly, the potential regulatory network MYB, bHLH, and WD40 families and their targets (Supplementary Fig. 16), revealed a different situation from T1. At the transition stage, the structural genes with the most edges were 4CL (So16G047200), C4H (So15G050380), CHS (So16G045740), CHI (So22G046560), F3'H|F3'5'H (So07G050530), DFR (So02G010260), ANS (So03G007060), and F3oGT (So09G055690; So01G036790) (Supplementary Fig. 16), most of which belonged to EBGs. Three anthocyanins including cyanidin (Cy), cyanidin-3-O-(2''-O-glucosyl)glucoside, and Pt3Ru were distributed at the transition stage (Fig. 6a). And, one structural gene (4CL, So08G002910) associated with Cy, and seven flavonols and anthocyanins enzyme genes (PAL, So03G064720; 4CL, So13G051200; CHS, So19G016880; FLS, So13G055200; F3H, So15G003400; F3'H|F3'5'H, So05G012500; F3oGT, So18G014040) potentially regulated the Pt3Ru synthesis, which were with high expressions (PCC > 0.9 between metabolites and enzyme genes) (Fig. 6a; Supplementary Fig. 19).
a Sub-network for flavonoids metabolism. The network consists of metabolites (purple), structural genes (pink) and TFs from the outside to the inside. b DNA binding sites of the one direct regulatory gene and two potential upstream regulatory genes in 5'UTR upstream 2Kb. c Resolved one of the metabolisms of Pt3Ru and hierarchical regulation for CHS gene involved. From right to left, the network consists of metabolite (Pt3Ru in L5), structural gene (CHS in L4), TFs that play a direct regulatory role (L4), and upstream regulatory TFs (L3 and L2). The expression profiles at the four flower opening stages were displayed as heat-maps (log10(FPKM+1)). The bar color change from blue to red indicates the expression from low to high. The PCC value increases from small to large, the width of the edge changes from thinner to thicker in the networks.
Pt3Ru was the most important pigment between the transition and the fading stages (Fig. 4b). CHS was the first key enzyme in the synthesis and metabolism pathway of anthocyanins/flavonols, whose upstream regulation mechanism was unclear yet. Here, an important sub-network (Fig. 6b) inferred from TO-GCNs, showed that CHS gene (So19G016880) with significantly high expression levels (Supplementary Fig. 19), was directly related to Pt3Ru, simultaneously, and was the potential first-order to third-order upstream regulators. In the co-expression network that we have resolved, CHS gene (high expression levels in T2 and T3) might be regulated by ERF So15G039730 (high expression level in T2) as an upstream third-level regulator, GATA So01G024070 (high expression level in T2) as an intermediate secondary regulator, and WD40 So01G077230 (high expression level in T2 and T3) as the direct regulatory factor. The consistency of qRT-PCR data (Supplementary Fig. 19) and the result of DNA binding site prediction may initially confirm our inference (Fig. 6c). At the same time, two bHLH family members, So17G021180 and So09G026360, which exhibited significantly high and differential expressions during the four flower opening stages and were consistent with the expression level trend of WD40 So01G077230 (Supplementary Fig. 19), maybe potentially interacted with the WD40 So01G077230 accompanied by high PCC with 0.998 and 0.981. In addition, we also found that ERFs acted as both the direct regulators (So11G003650 and So18G001910) and the upstream regulator (So15G039730), all with significantly high expressions (Supplementary Fig. 19). Furthermore, F3'H|F3'5'H So05G012500 and So07G050530, which contributes to the red-based cyanidin and the blue-based delphinidin pigment, were analyzed for potential regulatory networks (Supplementary Figs. 17–18). Genes, ERF So11G003650 and ERF So15G039730, were also identified as the important direct and upstream regulators in these networks.
Discussion
In many horticultural plants, it is common for the flower color to fade or deepen with the flower opening. Although the research on anthocyanin accumulation has been very detailed, the metabolic dynamics and molecular regulation mechanisms of petal color change are still poorly understood. Not only does lilac flowers have the obvious color change, but the elegant purple color is an important ornamental trait and widely recognized. Here, we reported on a chromosome-scale genome assembly of S. oblata in the genus Syringa of family Oleaceae. Compared with the published genomes of Oleaceae plants (F. excelsior, O. fragrans, O. europaea, and F. suspensa), the present S. oblata genome assembly had the highest heterozygosity (1.92% vs 1.45% of O. fragrans, the highest known)19, and the genome size (1.05 Gb) was less than O. europaea with 1.127–1.48 Gb20 and much larger than O. fragrans with 733.2625–740.71 Mb19, F. excelsior with 866.8 Mb18, and F. suspensa with 737.47Mb32. This assembled genome is expected to be a valuable resource for genome-wide studies for genetic dissection of key traits, molecular breeding, and evolutionary studies in lilac.
Comparative analyses revealed that lilac experienced and shared two rounds of WGDs20 along with F. excelsior, O. fragrans, and O. europaea within the Oleaceae family. The more recent WGD event is obviously shared with four Oleaceae plants, suggesting an Oleaceae-specific whole-genome duplication, which was consistent with previous reports for other Oleaceae species18,19,20,27. However, the older WGD event was barely visible may be due to the accelerated deletion of the oldest syntenic pairs after the recent WGD, a classically observed phenomenon33 additionally supported by collinearity evidence. This lilac genomic information is expected to further clarify the evolution of Oleaceae plants10.
Here, we performed the metabolome assay of time-ordered lilac flower samples to grasp the key flavonoid metabolites determining flower color change accurately and comprehensively. Metabolome results showed that Dp3Ru and Cy3Ru were the two major metabolites with the highest contents among the identified 118 flavonoid metabolites. Throughout the flower opening period, these two key anthocyanins showed distinctive and specific pattern of accumulation, distribution and fading. At the early stage of flower opening (T1), these two anthocyanins presented the highest relative contents, and the content of Cy3Ru was slightly higher than Dp3Ru (53.76% vs 46.24%); at the transition stage (T2-T3), the relative content of Cy3Ru was only two-thirds of T1, and on the contrary, the latter Dp3Ru was the main one (T2, 72.44% and 27.56%; T3, 70.25% and 29.75%); at the fading stage (T4), the lowest relative content was just one-sixth of T1, and Dp3Ru still was the main component with a proportion of 70.98%. Moreover, delphinidin-based anthocyanins were the major constituents of violet and blue flowers in nature, which has always been one of the most interesting topics in ornamental plant research, such as the world-famous cut flowers, roses, chrysanthemums, and lilies; such high contents of delphinidin-based anthocyanins (Dp, Dp3Ru, Pt3Ru, Dp3Ru7G) in lilac petals provide ideas for future research.
It has been revealed that pelargonidin, cyanidin and delphinidin were attached to the B ring of the molecule with one, two and three hydroxyl groups, respectively, the increase of which would shift the color from redder to bluer9. Our analysis revealed that anthocyanin content was closely related to the lilac petals color, which shifted from reddish-purple to violet and then to light violet nearly white along with the flower opening period. This may be closely related to self-association or/and an intramolecular co-pigmentation34. In addition, glycosylation, including mono-, di-, and tri-saccharides, generally improved the stability and water solubility of the pigment35; with more stable diglycosides than their corresponding monoglycosides36; at the same time, location, number, and size of glycosidic substitutions played important roles on the anthocyanins color expression37. These also explained why the content of rutinoside (diglycoside) in the lilac petals was much higher than those of glucoside (monoglycoside) and pigments without sugar groups.
In particular, it is relatively rare to find anthocyanin rutinosides in ornamental plants, but more reports often focus on the fruits of plants with higher nutritional value, such as black currant38, Solanum betaceum39, and Liriope platyphylla40. Black currant is rich in anthocyanins and the content of anthocyanidin 3-O-rutinosides is significantly higher than that of anthocyanidin 3-O-glucosides; among which, Cy3Ru and Dp3Ru account for about 77% of the total anthocyanin content, and Cy3G and Dp3G account for about 21%. Anthocyanins extracted from blackcurrant fruit have been reported to be important in the treatment of cancer41, cardiovascular disease and free radical scavenging, and are found intact in the blood when absorbed by humans as anthocyanin rutinosides38. In the petals of lilac, Cy3Ru and Dp3Ru account for about 98% of the total anthocyanin content, which may be of great value in future medical research and derivative products.
Based on the gene annotation and expression profile, we reconstructed the entire anthocyanin and flavonol metabolic pathways and found there were multiple-copy genes for key enzymes playing major roles. The followed analyses of the time-ordered gene co-expression networks, combining transcriptomic and metabonomic data, unraveled important TF families and members which may play key roles in anthocyanins regulation in lilac petals. MBW-dependent (MYB-bHLH-WDR transcriptional complexes)8,42,43 regulators were evidenced to control anthocyanin biosynthesis and meolism in our study, and WD40 seem to be more significant at the transition stage. It is worth noting that WRKY and ERF may play important roles at the early and transition periods, respectively. Several recent studies revetabaled the novel mechanisms by which WRKY TFs together with the MBW complex regulated the anthocyanin pigment pathway, and they even proposed the 'MBWW quartet' model42. Also, PyERF3 was found to interact with PyMYB114 and PybHLH3 to co-regulate fruits anthocyanin biosynthesis in Asian red-skinned pears43. Therefore, we speculated that WRKY and ERF families, which were known to play crucial roles in plant tolerance against abiotic and biotic stresses, were potential regulators in flavonoid biosynthesis pathway42,43, flower color change and patterning12. In addition, as the ethylene response factor, ERF was identified as the important direct as well as upstream regulators in our flower color change regulatory network, so we speculate that ethylene may affect petal color change, which may provide ideas for our future work.
In conclusion, the high-quality chromosome-level genome assembly presented here is expected to provide a reference for the further understanding of Oleaceae evolution and diversification. Furthermore, our transcriptome and metabolome investigations also provided an in-depth insight into lilac dynamic flower coloration. Genomic data from this study will be an essential resource in many traits for further functional study with genome editing and also for molecular marker-assisted breeding to promote genetic studies and novel cultivars breeding for Syringa.
Methods
Plant materials
A 10-year-old S. oblata individual, with red-purple inflorescence, strong fragrance, and stable flower traits was chosen from the nursery of Beijing University of Agriculture (China) for genome sequencing and de novo genome assembly (Fig. 1). Fresh leaves were collected for the genome sequencing library preparation, and flowers at four blooming stages (Fig. 4a) were collected with three replicates to determine metabolites (flavonoids), and to perform transcriptome sequencing in support of genome annotation and gene expression analysis. Plant materials were immediately frozen in liquid nitrogen and stored at −80 °C for subsequent genomic DNA and RNA extractions.
Library construction and sequencing
Three kinds of libraries were constructed, including short-reads RNA-free sequencing with DNBSEQTM, long-read Nanopore, and Hi-C, following manufacturers' instructions.
Petals' RNA was isolated using QIAGEN RNeasy plant mini kit (QIAGEN, Hilden, Germany) for short-read sequencing with DNBSEQTM. One library with insert sizes of 300–400 bp was constructed, and then a BGISEQ Platform was used to sequence the reads, resulting in 45.65Gb clean data, providing ~39.35× genome coverage.
High-quality and intact genomic DNA was extracted from fresh leaves using DNAsecure Plant Kit (Tiangen Biotech, Co. Ltd, Beijing, China). For long-reads sequencing, three Nanopore libraries with 20 kb insert size were generated and sequenced on the GridION X5 sequencer platform (Oxford Nanopore Technologies, UK) at the Beijing Genomics Institute (Wuhan, China). Hi-C library was sequenced on a BGISEQ platform.
Genome size and heterozygosity estimation
Size and heterozygosity of the lilac genome was estimated by K-mer analysis. Firstly, Jellyfish (v2.1.4) was used to quickly count the depth distribution of K-mer = 17–31, and the peak depth was clearly observed from the distribution data; then the frequency spectrum of K-mer was fitted by GenomeScope44 (v1.0.0) to predict the genome characteristics.
De novo genome assembly and quality assessment
De novo genome assembly employed the following steps: first, we used the NECAT (https://github.com/xiaochuanle/NECAT) to assemble the nanopore long-reads into contigs; subsequently corrected three times using Racon45 software; followed by mapping short-reads to the reference genome using BWA-MEM46 (v0.7.17), finally, further polished with the second round of pilon using clean DNBSEQTM reads.
The Hi-C data were used for scaffolding the preliminary assemblies and to improve assembly continuity. The main Hi-C scaffolding workflow was as follows: clean Hi-C data was obtained by filtering Hi-C data from MboI digestion by Hic-pro software; then, clean Hi-C data were mapped to the corrected contigs genome by Juicer47 (v1.5.6); 3D-DNA pipeline48 was used for chromosome-level scaffolding by clustering, ordering, and orienting the previous assemblies based on genomic proximity information between Hi-C read pairs; finally, with the help of manual adjustment, the HIC cross-linking signal was redrawn by HiCPlotter49 software and the adjacent anchored scaffolds were connected forming 23 superscaffolds corresponding to 23 pseudochromosomes.
Given that the S. oblata genome had higher heterozygosity, Purge_dup50 was used to remove heterozygous and redundant sequences from the corrected genome to obtain corrected contigs. We evaluated the completeness of the assembled genome through BUSCO v5.2.2 analysis using gene content from the embryophyta_odb1051, and mapping short-reads to the genome for quality assessment.
Genome annotation
First, repetitive elements were discovered by a combined strategy based on homologous sequence alignment (homolog) and de novo in the lilac genome. For homolog-based prediction, the RepBase52 (v.21.12) library was applied to search against the lilac genome by RepeatMasker (v.4.0.7) (http://www.repeatmasker.org) and RepeatProteinMasker (v.4.0.7) with default parameters. For de novo-based approach prediction, a de novo repetitive element library was established with Piler53, RepeatScout54 and RepeatModeler, then predicted by RepeatMasker software. Furthermore, to identify the Tandem Repeats and LTR-RT, Tandem Repeats Finder (TRF)55 (v4.09) and LTR_FINDER56 (v1.06) were employed, respectively.
Gene structure prediction was based on a combination of evidence-based and de novo predictions. We used the Maker57 software to carry out evidence-based assembly by evidence files, which included downloaded transcriptome data and mRNA/protein sequences of O. europaea subsp. europaea, F. excelsior, and O. fragrans. Transcriptome reads were sequenced on the BGISEQ-MGI2000RS platform and assemblies were generated (Supplementary Table 3) with Trinity (v.2.1.1) for genome annotation. De novo prediction was carried out using Augustus58. After repetitive sequence masking, the gene sets predicted by various methods were integrated into a non-redundant and more complete gene set. Finally, we used BUSCO (BUSCO, RRID:SCR_ 015008) to evaluate the quality of the gene set.
For non-coding RNAs (ncRNAs) annotation, the tRNAs were predicted using tRNAscan-SE59 (v1.3.1) software. For highly conserved rRNAs, we chose related species' rRNA sequence as references, and predicted rRNAs sequences using BLASTN60. Through sequence information searches against the Rfam (https://rfam.org) database using INFERNAL61 software, miRNAs and snRNAs were predicted.
With the help of eggNOG-mapper (http://eggnog-mapper.embl.de), gene functional annotation was performed using KEGG23, GO22 and COG21 databases by comparing the eggNOG5.0 database. The advantages of eggNOG-mapper are: (i) ability to distinguish between paralogous and orthologous genes, and achieve more accurate gene function annotation and (ii) faster database searches through HMMER (http://www.hmmer.org) (v.3.0) or DIAMOND. In addition, functional annotation of protein-coding genes was carried out by performing BLASTP60 (E-value ≤ 1e−5) searches against SWISS-PROT62, NCBI NR protein databases, MSU Rice Genome Annotation database15 and Arabidopsis protein databases.
Gene family, phylogenetic analysis, and divergence time estimation
OrthoFinder63 (v2.3.3) software was used to cluster paralogous and orthologous groups from genomes of other 11 species. CAFE (Computational Analysis of gene Family Evolution)64 (v4.2.1) was used to analyze the expansion and contraction of gene family among these species. GO classification and KEGG pathways using the R package clusterProfiler65. After concatenating single-copy genes to form a super matrix following alignment by MUSCLE66 (v3.8.31), the conservative regions from the generated matrix were extracted, and the gap information in the sequence was deleted to form a new matrix with Gblocks_0.91b67. Then RAxML (v8)68 was used to construct the phylogenetic tree, and the FigTree program (v1.4.3) (http://tree.bio.ed.ac.uk/software/figtree/) was used for root determination. Codeml and MCMCtree programs, within the PAML package69 (v4.5) software, were used to estimate the molecular clock (replacement rate) and the differentiation time between species. Finally, combined with the divergence time of known species from published literature or TimeTree database (http://www.timetree.org/) as the calibration (S. lycopersicum-S. tuberosum 5–9 Mya, A. thaliana-C. papaya 63–82 Mya, O. europaea-O. fragrans 7–45 Mya, F. excelsior-O. fragrans 33–47 Mya, A. thaliana-O. sativa 115–308 Mya), the lilac divergence time was calculated and estimated to be between 38–48 Mya.
Genome collinearity/synteny and whole-genome duplication (WGD) analyses
For intergenomic comparison, we performed all-against-all BLASTP60 with E-value ≤ 1e−5 (v2.2.26) to align protein sequences, and the WGDI70 and JCVI71 (v0.8.12) package was used to determine high-confidence collinear blocks, paralogous and orthologous, in these species. Then, the Ks (the number of synonymous substitutions per synonymous site) values of the homologs within syntenic blocks were calculated by PAML to identify WGDs. WGD time was estimated with F. excelsior-S. oblata divergence time (mean: 43.20 Mya) as an age constraint. Ks peaks of F. excelsior-S. oblata syntenic orthologs allowed calculating γ = Ks/(2 × (divergence time))33. The same γ value was applied to calculate the time of WGD events in lilac.
Identification of flavonoid biosynthesis genes, transcription factors (TFs) and transcription factor binding site (TFBS)
Proteins in the flavonoid biosynthesis pathway including PAL (phenylalanine ammonia-lyase), C4H (cinnamate-4-hydroxylase), 4CL (4-coumarate CoA ligase), CHS (chalcone synthase), CHI (chalcone isomerase), F3H (flavanone 3-hydroxylase), F3'H (flavonoid 3'-hydroxylase), F3'5'H (flavonoid 3'5'-hydroxylase), FLS (flavonol synthase), DFR (dihydroflavonol 4-reductase), ANS (anthocyanidin synthase), F3oGT (flavonoide-3-O-glucosyltransferase), and RT (rhamnosyltransferase) were searched in the lilac genome annotation. Then, we added Hidden Markov Model (HMM) profiles of PAL (PF00221.20), C4H (PF00067.23), 4CL (PF00501.29, PF13193.7), CHS (PF02797.16, PF00195.20, PF00108.24), CHI (PF02431.16, PF16035.6), F3H (PF03171.21, PF14226.7), F3'H|F3'5'H (PF00067.23) (the F3'H|F3'5'H genes of other plants such as V. vinifera, Petunia hybrida and A. thaliana were also downloaded to construct phylogenetic tree to determine), FLS (PF03171.21, PF14226.7), DFR (PF01370.22, PF16363.6), ANS (PF03171.21, PF14226.7), F3oGT (PF00201.19), and RT (PF00201.19) from Pfam database (http://pfam.xfam.org/), to search against protein sequences databases employing the HMMER (v.3.0) in the S. oblata genome.
We used iTAK software72 to identify TFs in the genome of the 12 assessed species. In addition, PlantRegMap was used to identify the WD40 family by homology to A. thaliana. The 2 kb sequences upstream of the genes were used to identify TFBSs present in the promoters of genes using PlantRegMap with threshold p-value ≤ 1e−4 (http://plantregmap.gao-lab.org/binding_site_prediction.php).
Metabolome and transcriptome analyses
For the metabolome, after freeze-drying, crushing, weighting, dissolving, centrifugating, absorbing and filtrating, the samples were measured by Ultra Performance Liquid Chromatography (UPLC), and Tandem mass spectrometry (MS/MS).
Qualitative and quantitative analyses of the raw data obtained via UPLC-MS/MS were performed using Analyst v1.6.3 software. To determine the repeatability of metabolite extraction and detection, one quality control sample (QC) was inserted per ten samples, and then the overlapped total ion current chromatogram was analyzed. Principal Component Analysis (PCA), Fold Change (FC), and Orthogonal Partial Least Squares-Discriminant Analysis (OPLS-DA) were used to screen out differential metabolites (Supplementary Table 15), with FC > 2 or FC < 0.5, and variable importance in project (VIP based on OPLS-DA) greater 1. Then, we selected all eight anthocyanins and 12 additional flavonoids with high content in the differential metabolites for the PCA distance biplot.
Petals at four flower opening stages were collected with three replicates to perform transcriptome sequencing. For transcriptome analysis, clean reads were 304.3 million and Q20 and GC rate were 95% and 36.2%, respectively (Supplementary Table 3). DEGs (Supplementary Table 16) among the four flower blooming stages were identified using DESeq2, with adjusted p-value (FDR)<0.05 and |log2FC| ≥ 1.
Regulation network construction based on TO-GCNs
We used a recently developed method of reconstructing TO-GCNs73 for joint analysis of transcriptome and metabolome data. Finally, 80 differential metabolites and 20,984 DEGs (2507 TFs and 18,477 structural genes) were incorporated. The PCC cutoff values were 0.90 between structural genes and TFs, and 0.73 between metabolites and genes. TO-GCNs network was visualized using Cytoscape74.
Quantitative real-time PCR analysis
The key genes analyzed were verified by quantitative real-time PCR with the expression level of lilac actin gene as an internal reference. QRT‐PCR was performed using Green qPCR SuperMix (TransStart) on a CFX Connect Detection System (Bio-Rad). Using the 2-ΔΔCt method to calculate relative expression levels, and each analysis included three replicates. The specific primers for qRT‐PCR were listed in Supplementary Table 17.
Statistics and reproducibility
In Fig. 4b, Supplemental Figs. 10–12, and Supplemental Fig. 19, we used n = 3 biologically independent samples. Statistical analyses of metabolites relative contents and qRT-PCR were performed with GraphPad Prism 8. All the values were given as the mean ± SE and were analyzed by one-way ANOVA with multiple comparisons between adjacent stages (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
References
Essenberg, C. J. Intraspecific relationships between floral signals and rewards with implications for plant fitness. AoB Plants 13, plab006 (2021).
Han, M. L. et al. Analysis of flavonoids and anthocyanin biosynthesis‐related genes expression reveals the mechanism of petal color fading of Malus hupehensis (Rosaceae). Braz. J. Bot. 43, 81–89 (2020).
Li, M. Study on the molecular mechanism of petal fading of Brunfelsia acuminata and the technology of adjusting light and temperature. (Ph.D. Thesis) (2020).
Wang, Y. G. et al. An R2R3-MYB transcription factor CmMYB21 represses anthocyanin biosynthesis in color fading petals of chrysanthemum. Sci. Horticulturae 293, 110674 (2022).
Nozzolillo, C. et al. Novel aspects of the flowers and floral pigmentation of two Cleome species (Cleomaceae), C. hassleriana and C. serrulate. Biochemical Syst. Ecol. 38, 361–369 (2010).
Guo, L. P. et al. Transcriptome and chemical analysis reveal putative genes involved in flower color change in Paeonia 'Coral Sunset'. Plant Physiol. Biochem. 138, 130–139 (2019).
Pu, X. et al. The honeysuckle genome provides insight into the molecular mechanism of carotenoid metabolism underlying dynamic flower coloration. N. Phytol. 227, 930–943 (2020).
Xu, W., Dubos, C. & Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185 (2015).
Yoshida, K., Oyama, K. & Kondo, T. Chemistry of flavonoids in color development. Recent Adv. Polyphen. Res. 3, 99–129 (2012).
Wessinger, C. A. & Rausher, M. D. Lessons from flower colour evolution on targets of selection. J. Exp. Bot. 63, 5741–5749 (2012).
Zhao, D. & Tao, J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 6, 261 (2015).
Yang, F. S. et al. Chromosome-level genome assembly of a parent species of widely cultivated azaleas. Nat. Commun. 11, 5269 (2020).
Ming, J., Gu, W. C., Liu, C., Liu, K. & Wang, L. N. Advances in germ plasm resources of Syringa Linn. research. World Forestry Res. 20, 20–26 (2007).
Yao, J. M., Chen, L., Cui, H. X. & Yao, T. Responses of photosynthesis and heat-tolerance in seedling of four lilacs to high-temperature. Grassl. Turf 30, 50–55+58 (2010).
Zhang, J. Studies on petal anthocyanins compositions and coloration mechanism of four oramental plants. (Thesis) (2011).
Zheng, J. et al. Transcriptome analysis of Syringa oblata Lindl. inflorescence identifies genes associated with pigment biosynthesis and scent metabolism. Plos One 10, e0142542 (2015).
Taylor, H. Cyto-taxonomy and phylogeny of the Oleaceae. Brittonia 5, 337–367 (1945).
Sollars, E. S. et al. Genome sequence and genetic diversity of European ash trees. Nature 541, 212–216 (2017).
Yang, X. et al. The chromosome-level quality genome provides insights into the evolution of the biosynthesis genes for aroma compounds of Osmanthus fragrans. Hortic. Res. 5, 72 (2018).
Unver, T. et al. Genome of wild olive and the evolution of oil biosynthesis. Proc. Natl Acad. Sci. USA 114, E9413–E9422 (2017).
Koonin, E. V. et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 5, 28 (2004).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kawahara, Y. et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4 (2013).
Chen, H. et al. Whole-genome resequencing of Osmanthus fragrans provides insights into flower color evolution. Hortic. Res. 8, 98 (2021).
Cheng, S. P. et al. Haplotype-resolved genome assembly and allele-specific gene expression in cultivated ginger. Hortic. Res. 8, 188 (2021).
Rao, G. et al. De novo assembly of a new Olea europaea genome accession using nanopore sequencing. Hortic. Res. 8, 64 (2021).
Jaillon, O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007).
Jiao, Y. et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 13, R3 (2012).
Katsumoto, Y. et al. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 48, 1589–1600 (2007).
Lou, Q., Wang, L., Liu, H. & Liu, Y. Anthocyanin profiles in flowers of grape hyacinth. Molecules 22, 688 (2017).
Li, L. F., Cushman, S. A., He, Y. X. & Li, Y. Genome sequencing and population genomics modeling provide insights into the local adaptation of weeping forsythia. Horticulture Res. 7, 130 (2020).
Badouin, H. et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546, 148–152 (2017).
Yoshida, K., Mori, M. & Kondo, T. Blue flower color development by anthocyanins: from chemical structure to cell physiology. Nat. Prod. Rep. 26, 884–915 (2009).
He, J. & Giusti, M. M. Anthocyanins: natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 1, 163–187 (2010).
Sinopoli, A., Calogero, G. & Bartolotta, A. Computational aspects of anthocyanidins and anthocyanins: A review. Food Chem. 297, 124898 (2019).
Sigurdson, G. T., Robbins, R. J., Collins, T. M. & Giusti, M. M. Impact of location, type, and number of glycosidic substitutions on the color expression of o-dihydroxylated anthocyanidins. Food Chem. 268(DEC.1), 416–423 (2018).
Slimestad, R. & Solheim, H. Anthocyanins from black currants (Ribes nigrum L.). J. Agriculture Food Chem. 50, 3228–3231 (2002).
Osorio, C. et al. Chemical characterisation of anthocyanins in tamarillo (Solanum betaceum Cav.) and Andes berry (Rubus glaucus Benth.) fruits. Food Chem. 132, 1915–1921 (2012).
Lee, J. H. & Choung, M. G. Identification and characterisation of anthocyanins in the antioxidant activity-containing fraction of Liriope platyphylla fruits. Food Chem. 127, 1686–1693 (2011).
León-González, A. J. et al. Delphinidin-3-O-glucoside and delphinidin-3-O-rutinoside mediate the redox-sensitive caspase 3-related pro-apoptotic effect of blackcurrant juice on leukaemia Jurkat cells. J. Funct. Foods 17, 847–856 (2015).
Lloyd, A. et al. Advances in the MYB-bHLH-WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58, 1431–1441 (2017).
Yao, G. et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 92, 437–451 (2017).
Vurture, G. W. et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33, 2202–2204 (2017).
Vaser, R., Sovic, I., Nagarajan, N. & Sikic, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746 (2017).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: Genomics, 2013.
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95 (2017).
Akdemir, K. C. & Chin, L. HiCPlotter integrates genomic data with interaction matrices. Genome Biol. 16, 198 (2015).
Guan, D. et al. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics 36, 2896–2898 (2020).
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A. & Zdobnov, E. M. BUSCO Update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evolution 38, 4647–4654 (2021).
Jurka, J. et al. Repbase update, a database of eukaryotic repetitive elements. Cytogenetic Genome Res. 110, 462–467 (2005).
Edgar, R. C. & Myers, E. W. PILER: identification and classification of genomic repeats. Bioinformatics 21, I152–I158 (2005).
Price, A. L., Jones, N. C. & Pevzner, P. A. De novo identification of repeat families in large genomes. Bioinformatics 21, I351–I358 (2005).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–W268 (2007).
Holt, C. & Yandell, M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 491 (2011).
Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–W439 (2006).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic acids Res. 25, 955–964 (1997).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Nawrocki, E. P. & Eddy, S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013).
Boeckmann, B. et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31, 365–370 (2003).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
De Bie, T., Cristianini, N., Demuth, J. P. & Hahn, M. W. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22, 1269–1271 (2006).
Yu, G. C., Wang, L. G., Han, Y. Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics-a J. Integr. Biol. 16, 284–287 (2012).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evolution 17, 540–552 (2000).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Yang, Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Computer Appl. Biosci.: CABIOS 13, 555–556 (1997).
Sun, P. C. et al. WGDI: A user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. BioRxiv (2021).
Tang, H. B., Krishnakumar, V. & Li, J. P. jcvi: JCVI utility libraries. Zenodo (2015).
Zheng, Y. et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 9, 1667–1670 (2016).
Chang, Y. M. et al. Comparative transcriptomics method to infer gene coexpression networks and its applications to maize and rice leaf transcriptomes. Proc. Natl Acad. Sci. USA 116, 3091–3099 (2019).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
This study was supported by the National Key R&D Program of China (2018YFD1000406), the Joint Project of Beijing Municipal Education Commission and Beijing Municipal Natural Science Foundation (KZ201810020028). We thank Dr. Qiang Gao for assistance with the genome assembly.
Author information
Authors and Affiliations
Contributions
P.S.L., J.W., Z.H.H., and J.F.M. conceived and designed the study; B.M., T.L.S., J.W., Y.Y.Y., W.B.W., and W.B.X. prepared the materials, conducted the experiments, analyzed the data and prepared the results; B.M. wrote the manuscript; J.F.M., P.S.L., J.W., Z.H.H., Y.Z., S.C.S., Y.C.Y., I.P., and Y.A.E. edited and improved the manuscript; all authors approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Nan Ma, Jiang Zhi Huang and Jing Wang for their contribution to the peer review of this work. Primary Handling Editors: Diane Saunders and George Inglis. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ma, B., Wu, J., Shi, TL. et al. Lilac (Syringa oblata) genome provides insights into its evolution and molecular mechanism of petal color change. Commun Biol 5, 686 (2022). https://doi.org/10.1038/s42003-022-03646-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-022-03646-9
This article is cited by
-
High quality genomes produced from single MinION flow cells clarify polyploid and demographic histories of critically endangered Fraxinus (ash) species
Communications Biology (2024)
-
Comparative transcriptome analysis of dioecious floral development in Trachycarpus fortunei using Illumina and PacBio SMRT sequencing
BMC Plant Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.