Abstract
The ABC multidrug exporter MsbA mediates the translocation of lipopolysaccharides and phospholipids across the plasma membrane in Gram-negative bacteria. Although MsbA is structurally well characterised, the energetic requirements of lipid transport remain unknown. Here, we report that, similar to the transport of small-molecule antibiotics and cytotoxic agents, the flopping of physiologically relevant long-acyl-chain 1,2-dioleoyl (C18)-phosphatidylethanolamine in proteoliposomes requires the simultaneous input of ATP binding and hydrolysis and the chemical proton gradient as sources of metabolic energy. In contrast, the flopping of the large hexa-acylated (C12-C14) Lipid-A anchor of lipopolysaccharides is only ATP dependent. This study demonstrates that the energetics of lipid transport by MsbA is lipid dependent. As our mutational analyses indicate lipid and drug transport via the central binding chamber in MsbA, the lipid availability in the membrane can affect the drug transport activity and vice versa.
Similar content being viewed by others
Introduction
The passive movement of phospholipids between the leaflets in a membrane bilayer is extremely slow, in the order of hours to days, as shown in model membranes1,2. This is, at least in part, due to the energetically unfavourable movement of the hydrophilic headgroup across the hydrophobic core of the bilayer. For this reason, the cell requires membrane-embedded enzymes that facilitate the transbilayer movement of lipids within seconds. Examples of mammalian phospholipid transporters include (i) the calcium-activated nhTMEM16 scramblase that catalyses the shuffling of phospholipids between the inner and outer leaflets of the plasma membrane independent of metabolic energy3, (ii) the type IV P-type ATPase flippases that catalyse the inward translocation of phospholipids from the outer leaflet to the inner leaflet of the plasma membrane4, and (iii) the liver ATP-binding cassette (ABC) floppase ABCB45 for which in vivo data indicate a role in the outward translocation of phosphatidylcholine (PC) and its biliary secretion from hepatocytes into the canaliculi6. Phospholipid transport is equally important in microorganisms. The dimeric ABC exporter MsbA is an essential lipid transporter in Gram-negative bacteria. In vivo observations point to a role of MsbA in the translocation of phospholipids and core Lipid-A from their point of synthesis at the inner leaflet of the plasma membrane to the outer leaflet7,8. In further steps at the outer leaflet of the plasma membrane, a polysaccharide moiety (O-antigen) is ligated to core Lipid-A to form full-length lipopolysaccharides (LPS). The Lipid-A (endotoxin) domain of LPS is a unique, glucosamine-based phospholipid that serves as the hydrophobic anchor of LPS and is the bioactive component of the molecule that is associated with Gram-negative septic shock. After it is formed, LPS is transported across the periplasm into the outer leaflet of the outer membrane by the LPS translocase LptB2FGCADE9. But exactly how MsbA mediates the transport of naturally abundant lipids and how this reaction is related to MsbA’s ability to transport amphiphilic drugs10,11,12,13,14,15 has not been investigated in detail. Here we study the energetics of lipid transport by purified MsbA in proteoliposomes and compare the lipid and drug transport activities in a mutagenesis approach.
Results
Energetics of lipid floppase activity
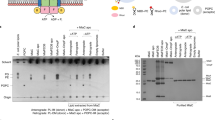
We conveniently express Escherichia coli MsbA in Lactococcus lactis, a Gram-positive bacterium that lacks Lipid-A and LPS as well as an E. coli-like periplasm and outer membrane10,11,15. We use these cells as a source of MsbA for lipid transport studies in proteoliposomes (Fig. 1). As (i) a previous study detected the transport of phospholipids and Lipid-A by MsbA in a 32Pi and 14C-acetate labelling approach in E. coli cells16, (ii) PE is the predominant phospholipid in E. coli making up 70–80% of the total phospholipid pool17,18, and (iii) advanced studies by native mass spectrometry detected the binding of PE to MsbA19, we established a transport assay for physiologically relevant long-acyl-chain (2 × C18) PE in proteoliposomes in which MsbA was incorporated in an inside-out fashion (Fig. 1a). Lipids labelled with fluorescent nitrobenzoxadiazole (NBD) have frequently been used in transport studies with ABC transporters20,21 and P-type ATPases22. However, as MsbA mediates the efflux of fluorescent dyes10,11,13,15, this ABC transporter might interact with NBD-lipid as a drug analogue rather than a lipid. Moreover, NBD-labelled lipids are usually detected using fluorescence quenchers such as Co2+ and dithionite, the intrinsic membrane permeability of which reduces the ability to discriminate between labelled lipids in the outer and inner leaflet of the membrane. In our assay, 1 in 17 PE molecules in the proteoliposomes contained a non-invasive biotin moiety that was covalently linked to the ethanolamine head group of the lipid (Fig. 1b). We detected PE flopping by inside-out oriented MsbA as a relocation of the lipid from the outer to the inner leaflet of the membrane. The reduction in the amount of external biotin-PE over time was quantified from the emission of membrane-impermeable fluorescence-tagged avidin when a quencher in complex with the avidin is displaced by the biotin moiety of the PE (Fig. 1a). The potential sources of metabolic energy that can drive substrate transport by MsbA, ATP and the transmembrane chemical H+ gradient (∆pH), were applied using well-established methods15. Briefly, proteoliposomes prepared in buffer pH 6.8 were diluted in buffer pH 8.0, imposing a difference between the interior pH and external pH by a pH jump (pHin 6.8/pHout 8.0) that was stabilised by the dissociation of NH4+ in the lumen of proteoliposomes and the outward diffusion of NH3. Using the fluorescent pH indicator BCECF trapped in the proteoliposomal lumen, measurements of the interior pH over time demonstrate that this pH difference was sustained for the 20 min duration of the biotin-lipid floppase assays (Supplementary Fig. 1). Where required, ATP was included in the external buffer. Remarkably, a significant PE floppase activity for MsbA-WT was observed only in the simultaneous presence of 5 mM Mg-ATP and the ∆pH (pHin 6.8/pHout 8.0) (Fig. 1c). The increased biotin-PE signal obtained with the addition of Triton-X100 to disrupt these proteoliposomes at the end of the floppase assay (Fig. 1d), demonstrates that PE flopping is based on the translocation of lipid to the inner leaflet of the membrane. This conclusion is strengthened by the finding that free biotin is neither transported by MsbA, nor liberated from biotin-PE by spontaneous hydrolysis during the floppase assay (Supplementary Fig. 2). No significant PE floppase activity was observed in MsbA-WT-containing proteoliposomes in the presence of ATP without the ΔpH (pHin 6.8/pHout 6.8 and pHin 8.0/pHout 8.0) or the ΔpH (pHin 6.8/pHout 8.0) only (Fig. 1c), indicating the importance of the simultaneous presence of both forms of metabolic energy. Furthermore, PE floppase activity was not detected in empty liposomes (Fig. 1e) or proteoliposomes containing MsbA-ΔK382 that lacks the catalytic Walker A lysine residue and has strongly reduced ATPase activity11 (Fig. 1f). A previous cryo-electron microscopic (cryo-EM) study identified the quinoline compound G907 as a specific MsbA inhibitor that binds to and stabilises the inward-facing MsbA dimer, thereby preventing the transition to the outward-facing conformation23. We synthesised G907 (Supplementary Methods) and tested its efficacy in our biotin-PE transport assay. At a 100 nM concentration (5x the reported in-vitro IC5023), G907 completely inhibited active biotin-PE transport in MsbA-WT-containing proteoliposomes (Fig. 1c), thus confirming that this activity is mediated by MsbA. These findings indicate that inside-out oriented MsbA transports biotin-PE from the outer leaflet to the inner leaflet of the proteoliposomal membrane. This floppase activity is based on apparent H+/biotin-PE antiport and is facilitated by ATP binding and hydrolysis in the presence of a ΔpH (interior acidic in the proteoliposomes). The lipid transport assays were repeated using the relatively large E. coli hexa-acylated (C12-C14) Lipid-A that is biotin-labelled on the disaccharide core (Fig. 2a)24. Different from biotin-PE, significant biotin-Lipid-A transport was observed in the proteoliposomes with ATP only; this activity was not affected by the imposed ΔpH (interior acidic) (Fig. 2b and Supplementary Fig. 1). Therefore, the transport of Lipid-A and PE has different energetic requirements.
a Schematic showing proteoliposomes containing headgroup-labelled biotin-PE (headgroup depicted as red circle with two C18 acyl chains in grey) in the inner and outer leaflet of the membrane, and the MsbA homodimer inserted in an inside-out fashion. Step 1, the PE transport reaction (red arrow) by MsbA is initiated by the addition of Mg-ATP in the external buffer and the imposition of a ΔpH (interior acidic) across the membrane. ATP binding and hydrolysis (brown arrow) at the nucleotide-binding domain dimer (in orange and slate blue) and simultaneous proton conduction (blue arrow) by the membrane domains (MD) (in light orange and light blue) provide metabolic energy for the transport of PE from the outer leaflet of the membrane to the inner leaflet. Step 2, following transport activity, the remaining amount of biotin-PE in the outer leaflet is quantified from the fluorescence emission (yellow lightning bolts) of fluorescence-tagged avidin (blue concave rectangles) when a bound quencher (green circles) is displaced by the binding of the biotin moiety. b Structural formula of the 1,2-dioleoyl (C18) biotin-PE in which the orange bracket highlights the phosphodiester moiety. c, d Biotin-PE transport assays with MsbA-WT before (c) or after (d) the addition of 1% (v/v) detergent Triton X-100 at the end of the transport reaction. Note the change in the fluorescence scale in d. Data refer to different provisions of metabolic energy: (i) Control (no metabolic energy, pHin 6.8/pHout 6.8) (set at 100%), (ii) imposed ΔpH (pHin 6.8/pHout 8.0), (iii) ATP (pHin 6.8/pHout 6.8), (iv) imposed ΔpH (pHin 6.8/pHout 8.0) plus ATP, or (v) ATP 8/8 (pHin 8.0/pHout 8.0). Control 2 in d refers to the Control after Triton X-100 addition. The MsbA inhibitor G907 was used in c. e Biotin-PE transport assays with liposomes without MsbA proteins. f–i Biotin-PE transport assays with ATPase-deficient MsbA-ΔK382 (f) and substrate binding chamber mutants MsbA-TripRA (R78A R148A R296A) (g), MsbA-K299A (h) and MsbA-DED (D41N E149Q D252N) (i). Data represent observations in three experiments (n = 3) with independently prepared batches of proteoliposomes. Lines and error bars in scatter dot plots refer to mean ± s.e.m. (one-way analysis of variance; ****P < 0.0001). Asterisks above the square brackets refer to comparisons with the control without metabolic energy.
a Structural formula of biotin-Lipid-A containing four primary hydroxymyristate (C14) acyl chains and one secondary myristate and laurate (C12) acyl chain. The orange brackets highlight the two phosphate groups in the glucosamine moieties of the lipid. b Biotin-Lipid-A assays with MsbA-WT and substrate binding chamber mutant MsbA-TripRA (R78A R148A R296A). Data refer to different provisions of metabolic energy: (i) Control (no metabolic energy, pHin 6.8/pHout 6.8), (ii) ATP (pHin 6.8/pHout 6.8) or (iii) imposed ΔpH (pHin 6.8/pHout 8.0) plus ATP. Data represent observations in three experiments (n = 3) with independently prepared batches of proteoliposomes. Lines and error bars in the scatter dot plot refer to mean ± s.e.m. (one-way analysis of variance; ****P < 0.0001). Asterisks above the horizontal line refer to comparisons with the control without metabolic energy.
Similar pathways for lipids and drugs
Recent cryo-EM and crystal structures of the MsbA-Lipid-A complex suggest that R78 (transmembrane helix (TMH) 2), R148 (TMH3), Q256 (TMH5), R296, K299 (TMH6) and interspersed carboxyl residues D41 (TMH1), E149 (TMH3) and D252 (TMH5) form a ring of polar interactions with the 1 and 4′ phosphate group and the glucosamine, ester and amide groups connecting the acyl chains in Lipid-A8,23,25 (Fig. 3). Similar acidic and basic residues coordinate Lipid-A binding in the cryo-EM structure of the LPS translocase LptB2FGC26. We tested the effect of mutations of the ‘ring’ residues in MsbA on lipid and drug transport. First, we focussed on the arginine residues in the ring and found a deficiency in active Lipid-A and PE transport in proteoliposomes for a triple mutant MsbA containing R78A R148A R296A substitutions (referred to as MsbA-TripRA) (Figs. 1g and 2b); the K299A mutation did not inhibit PE transport (Fig. 1h). Moreover, we tested the impact of the arginine substitutions on MsbA-mediated lipid transport in E. coli WD2 cells. These cells contain a genome-encoded MsbA mutant A270T that misfolds at non-permissive temperature (44 °C), causing a strong deficiency in cell growth. This phenotype could be rescued by the expression of plasmid-encoded MsbA-WT or single arginine substitution mutants but not by the (i) double arginine mutants R78A R148A, R78A R296A, R148A R296A, (ii) triple arginine mutant MsbA-TripRA, (iii) ATPase-deficient MsbA-ΔK382 or (iv) truncated MsbA-MD protein lacking the nucleotide-binding domain (Fig. 4). All mutants were expressed at a comparable level as MsbA-WT in the plasma membrane of E. coli WD2 cells (Supplementary Figs. 3 and 4). These in vivo experiments point to a requirement for two out of the three arginine side chains and functional nucleotide-binding domains in lipid transport by MsbA.
a Ribbon diagram of inward-facing MsbA with bound Lipid-A (PDB-ID: 5TV4) shows the location of arginine residues R78, R148, R296, K299 and carboxyl residues D41, E149 and D252 (referred to as the ‘ring’ residues) in the substrate binding chamber near the centre of phospholipid bilayer. View in the plane of the plasma membrane with the two half-transporters in light blue and light orange. b Close-up view from the external face of the membrane shows how the arginine and carboxyl side chains (in blue and red stick representation) in the MD of each half-transporter form a ring of hydrophilic interactions with the 1 and 4′ phosphate group (in orange spheres) and the glucosamine, ester and amide groups connecting the acyl chains in Lipid-A (in green stick representation). The colours of the residue numbers refer to the half-transporters in which the residues are located. The biotin tag in the disaccharide core of our biotin-Lipid-A (Fig. 2a) does not interfere with the interactions of Lipid-A with the ring residues. c Ribbon diagram of outward-facing MsbA (PDB-ID: 3B60) from which Lipid-A has dissociated, shown with the same arginine and carboxyl residues as in a. d Close-up view from the external face of the membrane shows that R78-D41 and R296-E149 form salt-bridges within each half-transporter (indicated by circles with distance in Å). Figure was generated in PyMol v2.4.1.
E. coli WD2 expresses a genome-encoded temperature-sensitive MsbA protein containing an alanine to threonine substitution at position 270 in TMH516. At non-permissive temperature (44 °C), this MsbA mutant rapidly inactivates. Due to a deficiency in the MsbA-mediated transport of phospholipids and Lipid-A to the outer membrane, the biogenesis of the outer membrane is impaired, prohibiting cell growth and division7,41. In contrast, E. coli WD2 cells exhibit normal growth at 30 °C. To test whether the expression of plasmid-encoded MsbA proteins can rescue the E. coli WD2 cells at 44 °C, the cells were transformed with pBAD24-based plasmids containing wild-type or mutant msbA genes. Medium without cells (Medium) (blue) and cells with empty plasmid (Control) (red) served as controls. a Expression of MsbA-WT (green), single substrate binding chamber mutants R78A (magenta), R148A (orange) or R296A (black), ATPase-deficient MsbA-ΔK38211 (light brown) or drug transport-active MsbA-MD lacking the nucleotide-binding domain15 (dark blue). b Expression of MsbA-WT (green), double mutants R78A R148A (magenta), R148A R296A (orange) or R78A R296A (black), MsbA-TripRA (R78A R148A R296A) (light brown) or triple substrate-binding chamber mutant MsbA-DED (D41N E149Q D252N) (dark blue). Histograms show OD600 levels near 375 min; bar colours match those in the traces. The results demonstrate that ATP hydrolysis at the nucleotide-binding domains and arginine and carboxyl side chains in the central binding cavity are important for in vivo lipid transport by MsbA. All mutants were equally well expressed as MsbA-WT in the plasma membrane (Supplementary Figs. 3 and 4). Data points represent observations in three experiments (n = 3) with independently prepared batches of cells. Values in histograms show mean ± s.e.m. (one-way analysis of variance; **P < 0.01; ***P < 0.001; ****P < 0.0001). In the data for 44 °C, the asterisks above bars refer to comparison with the non-expressing control, whereas the asterisks above the square brackets refer to comparisons with MsbA-WT.
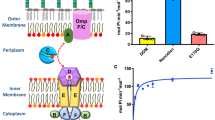
In addition to lipids, MsbA transports small molecules like ethidium, erythromycin and chloramphenicol10,11,15. The efflux of ethidium can conveniently be monitored in L. lactis cells from the decrease in fluorescence emission when the dye dissociates from intracellular nucleic acids27. In contrast to the observations for Lipid-A and PE, the R78A R148A double mutant and TripRA mutant showed a significantly enhanced efflux of ethidium compared to MsbA-WT (Fig. 5a). Analysis of protein expression levels and the rate of ethidium transport as a function of the dye concentration identified an increased ethidium binding affinity of MsbA as the underlying mechanism for the enhanced efflux activity (Fig. 5b and Supplementary Figs. 5 and 6). Hence, the arginine side chains facilitate the binding of Lipid-A and PE in the chamber but inhibit the binding of ethidium.
a ATP-depleted cells were preloaded with 2 µM ethidium after which ethidium efflux was initiated by the addition of glucose (+Glc) as a source of metabolic energy11,15. Active efflux by MsbA-WT was abolished in the presence of 2.5 µM of the MsbA inhibitor G907 (0.7x the reported in-vivo IC50 in cells23). MsbA-mediated ethidium efflux was inhibited by the triple DED mutations in the substrate binding chamber and was enhanced by K299A and sequential R-to-A replacements in the chamber. Fluorescence traces are the mean of three independent experiments. Values in histogram show significance of fluorescence levels near 600 s. All mutants were equally well expressed as MsbA-WT in the plasma membrane (Supplementary Figs. 5 and 6). b Effect of R-to-A mutations on the apparent affinity (Km) of MsbA for ethidium in the transport reaction. Histogram data represent observations in three experiments (n = 3) with independently prepared batches of cells and are expressed as mean ± s.e.m. (one-way analysis of variance; *P <0.05; **P < 0.01; ****P < 0.0001). Asterisks above the square brackets refer to comparisons with the MsbA-WT, whereas asterisks above the horizontal line refer to comparisons with the non-expressing control.
Second, we focussed on carboxyl residues D41, E149 and D252 that, in inward-facing MsbA, are interspersed between R78, R148 and R296 and located in the vicinity of the polar headgroup of bound Lipid-A (Fig. 3). In our lipid transport assays, we observed that the D41N, E149Q and D252N triple mutation (referred to as MsbA-DED) is tolerated in biotin-PE transport in proteoliposomes (Fig. 1i). However, MsbA-DED showed considerably reduced lipid transport in E. coli WD2 cells at non-permissive temperature (Fig. 4b) suggesting that the carboxylates are important for Lipid-A transport. MsbA-DED is also impaired in ethidium efflux in cells (Fig. 5a), which agrees with a previous study where the DED mutation was found to strongly inhibit ATP-dependent ethidium+/H+ antiport by MsbA15. When taken together, these data indicate that ethidium, biotin-PE and biotin-Lipid-A are transported via the central substrate binding chamber in which the basic residues R78, R148 and R296 and acidic residues D41, E149 and D252 promote interactions of MsbA with Lipid-A. The basic residues are important for the interactions with PE but inhibit the interactions with cationic ethidium. Instead, ethidium transport is facilitated by the presence of the acidic residues.
Discussion
During cell growth and division of Gram-negative bacteria, phospholipids and core Lipid-A are synthesised in the inner leaflet of the plasma membrane and exported (flopped) by MsbA to the outer leaflet, from where they are transported by the LPS translocase LptB2FGCADE9,26 to the expanding outer membrane. Structural studies suggest that MsbA is a Lipid-A floppase but no direct biochemical measurements of the flopping of Lipid-A with purified MsbA have been reported to date. Consequently, the energetic requirements of the MsbA-mediated lipid transport reactions have not been studied in detail. These measurements are technically challenging because there is no net change in the amount of lipid in the membrane; the lipid reorients from one side to the other.
In our Lipid-A and PE transport assays, we measured the metabolic energy-dependent floppase activity of purified MsbA in proteoliposomes, prepared with E. coli lipids and egg yolk PC, as a relocation of biotinylated lipid from the external membrane surface of proteoliposomes to the internal membrane surface. The biotin-lipid remaining in the outer leaflet of the membrane is detected by the binding of membrane-impermeable avidin (Fig. 1a). Control experiments show that free biotin is neither transported by MsbA, nor liberated from the biotinylated lipids in the time course of the assays (Supplementary Fig. 2). In further control experiments, the lipid floppase activity (i) does not occur in empty liposomes but requires functional MsbA protein, (ii) is inhibited by the MsbA inhibitor G907, and (iii) is affected by the ring residues in the substrate binding chamber that are known to coordinate Lipid-A binding (Figs. 1–5). The location of the biotin tag in the disaccharide core of our biotin-Lipid-A (Fig. 2a) ensures that the biotin does not interfere with the interactions of Lipid-A with the ring residues (Fig. 3a, b). Given the biotin-labelling of 1 in 17 PE molecules in our proteoliposomal membranes, the excess of unlabelled PE over biotin-Lipid-A, and the chemical differences between the biotinylated lipids and the native lipids they mimic, the measured PE and Lipid-A transport activities most likely underestimate the in-situ activity of MsbA as a lipid floppase.
The findings in this study represent important advancements in our understanding of lipid transport by MsbA. First, the process of Lipid-A transport from the plasma membrane to the outer membrane in E. coli requires two major transport systems, MsbA and LptB2FGCADE, the activity of which could be intrinsically connected. As summarised in the schematic in Fig. 6a–c, our results demonstrate that MsbA can mediate the transport of Lipid-A and PE in the absence of auxiliary proteins. Therefore, MsbA functions as an independent lipid floppase in the plasma membrane that supports LptB2FGCADE, and possibly also the MlaFEDB phospholipid translocase28,29,30, in anterograde lipid trafficking from the outer leaflet of the plasma membrane to the outer membrane. The amount of LPS, and its underacylated forms31, versus phospholipid that is ultimately transported to the outer membrane might, therefore, be regulated at the level of MsbA. Second, we show that the activity of ATP-dependent biotin-PE transport in proteoliposomes is strongly stimulated by the imposition of a ΔpH (interior acidic) (Figs. 1 and 6c). The repetition of these measurements with long-acyl-chain (2 × C18) NBD-PE also demonstrates the importance of the ΔpH + ATP in the MsbA-mediated transport of this lipid (Supplementary Fig. 7), and extends previous observations on the low ATP-dependent transport activity of MsbA for long-acyl-chain NBD-PE in proteoliposomes32. As the efficient transport of ethidium, chloramphenicol and erythromycin by MsbA requires the input of ATP and ΔpH (Fig. 6b), in which the ATPase reaction is stimulated by the ΔpH15, the energetics of PE transport is most similar to that of small-molecule transport. On the other hand, the Lipid-A transport reaction is ATP-dependent without a noticeable contribution of the ΔpH (Figs. 2 and 6a). The requirement for ATP binding and hydrolysis in Lipid-A and PE transport is in agreement with the phospholipid and Lipid-A-stimulated ATPase activity of purified MsbA33 and the dimerisation of the nucleotide-binding domains in the MsbA dimer upon Lipid-A binding13. We conclude that the role of the ΔpH in ATP-dependent lipid transport by MsbA is independent of the biotin modification but is linked to the structural properties of the transported lipid. Third, our data suggest that lipids and drugs share similar pathways in MsbA. Both types of substrates are transported via the central substrate binding chamber of MsbA where they interact with the same ring of side chains in the substrate binding chamber. R78, R148, R296, K299, and interspersed D41, E149 and D252 are known to form hydrophilic interactions with the two anionic phospho-glucosamine moieties in Lipid-A in inward-facing MsbA and to rotate away in the outward-facing state, thus enabling electrostatic interactions between R78-D41 and R296-E149 and facilitating lipid release (Fig. 3). These basic and acidic residues are important for Lipid-A transport in our experiments (Figs. 2, 4 and 6a). The arginine residues in the ring also support the transport of the smaller PE ligand that contains a single phosphodiester moiety but no glucosamine groups (Figs. 1 and 6c). In agreement with the PE data, basic residues contribute to the coordination of the phosphodiester moiety of phospholipid substrates in the crystallised lipoxygenase enzyme LOX34 and periplasmic phospholipid transport protein Ttg2D35 from Pseudomonas aeruginosa. Reported molecular dynamics simulations for the phospholipid scramblase nhTMEM16 from Nectria haematococca also point to a role of basic residues in the coordination of phospholipid binding36. Although the R78, R148, R296 residues in the ring are essential for PE transport, they are inhibitory for ethidium transport. In particular, the ethidium efflux rate and apparent binding affinity increased with the removal of basic residues in the MsbA-R78A R148A and MsbA-TripRA mutants (Fig. 5). The replacement of K299 in the ring by A (MsbA-K299A) showed a similar enhancement of ethidium transport (Fig. 5a) but did not affect the transport of PE (Fig. 1h). The binding sites for PE and ethidium are, therefore, partially overlapping. As our biotin-PE contains C18 acyl chains, whereas structural evidence suggests that the interior binding chamber can accommodate Lipid-A with C12 and C14 acyl chains23, the possibility exists that, similar to small molecules, only the headgroup of biotin-PE is accommodated in the central binding chamber. The acyl chains would remain in the phospholipid bilayer. Related mechanisms were proposed for phospholipid flipping by eukaryotic type IV P-type ATPases4,37 and the transport of lipid-linked oligosaccharide by the ABC floppase PglK from Campylobacter jejuni38. However, with two acyl chains in PE versus 6 acyl chains in Lipid-A, the acyl chain region in PE is much less densely packed and, due to the rotational freedom around the sp3 hybridised carbon atoms, it might have sufficient conformational flexibility to be accommodated in the chamber.
a In Lipid-A flopping, the arginine side chains of R78, R148 and R296 (indicated by blue B) and interspersed acidic side chains of D41, E149 and D252 (indicated by red C) in the central binding chamber (indicated by half blue ovals) coordinate a multitude of polar interactions with the two glucosamine and phosphate moieties of Lipid-A (in cyan blue). The sequence of reaction steps 1–4, underlying the alternating access of the binding chamber, is dependent on ATP binding and hydrolysis in our experiments and is based on structures for inward-open apo MsbA (PDB-ID: 3B5W), inward-facing G907-Lipid-A-bound MsbA (PDB-ID: 6BPL) and outward-closed ADP-vanadate-bound MsbA (PDB-ID: 5TTP) from E. coli and outward-facing AMP-PNP-bound MsbA from Salmonella typhimurium (PDB-ID: 3B60). Out and in refer to the outside and inside of the plasma membrane. b Ethidium transport is ATP binding and hydrolysis and ΔpH (interior alkaline)-dependent. Deprotonation of the C residues in inward-facing conformation in Step 1′ is followed by ethidium (Et+) binding near these residues. The B residues are not essential in this reaction and are omitted in the schematic. Step 2′, Transition to outward-facing state. Step 3′, Ethidium release and protonation of C residues. Step 4′, ΔpH-dependent transition to inward-facing state. c In PE flopping, Steps 1”–4” represent analogous ATP and ΔpH-dependent reactions as shown for ethidium, in which the B residues coordinate the binding of the phosphodiester group in PE. As the C residues are not essential in our PE transport measurements, proton coupling will involve protonatable groups (indicated by [nH+]) other than D41, E149 and D252.
In the transport of ethidium by MsbA, the carboxyl residues in the ring are essential for transport activity (Fig. 5). We suggest that, following the binding of cationic ethidium near the ring carboxylates in the inward-facing state, proton binding to these carboxylates in the outward-facing conformation aids in the dissociation of the bound ethidium towards the extracellular environment and the reorientation of MsbA to the inward-facing state (Fig. 6b). The transport cycle is completed by the deprotonation of the carboxyl residues in the cellular interior and binding of ethidium. In this way, the ATP-dependent ethidium transport reaction in cells is facilitated by the imposition of the ΔpH (interior alkaline) across the plasma membrane. Proton coupling in ATP-dependent biotin-PE transport will involve protonatable groups other than D41, E149 and D252.
In conclusion, our findings demonstrate MsbA-mediated Lipid-A and PE transport with different energy requirements and provide important details in the structural mechanisms proposed for MsbA based on X-ray crystallography and cryo-EM techniques. MsbA transports lipids and amphiphilic drugs out of cells by shared translocation pathways. The lipid availability in the membrane can, therefore, affect the drug transport activity and vice versa. This conclusion will also be relevant for other multidrug transporters that can transport drugs and lipids as alternative substrates.
Materials and methods
Compound synthesis
The synthesis of biotin-labelled E. coli Lipid-A from the glutaryl glucose linker-containing Lipid-A derivative and biotin-dPEG4TM-hydrazide (Invitrogen) is described by Fujimoto et al.24. The selective MsbA inhibitor G907 was synthesised by the procedure reported by Ho et al.23 and is described in full in the Supplementary Methods.
Bacterial strains, mutagenesis and protein expression
The drug-hypersensitive L. lactis NZ9000 ΔlmrA ΔlmrCD39, lacking expression of the endogenous multidrug transporters LmrA and LmrCD, was used for the expression of His6-tagged MsbA proteins from nisin-inducible pNZ8048 plasmids in the nisin controlled expression (NICE) system (NIZO Food Research, The Netherlands)40. The expression of MsbA proteins from arabinose-inducible pBAD24 plasmid (ATCC product 87399) in E. coli WD2 cells is described below under ‘Cell growth experiments’.
For site-directed mutagenesis of MsbA, the pGEM-5Zf(+) (Promega)-derived cloning vector pGEM-MsbA containing coding regions for an amino-terminal His6-tag and thrombin cleavage site followed by the wild-type E. coli msbA gene with silenced internal NcoI sites (at position 595 and 1375)10,11, was propagated in E. coli XL1-Blue cells (New England Biolabs) grown in Luria-Bertani Broth medium supplemented with 100 µg/mL ampicillin. Mutant msbA genes were generated in pGEM-MsbA by round-the-horn mutagenesis using the following primers: R78A, forward primer 5′-GCCGGTATCACCAGCTATGTCTCCAGC-3′, reverse primer 5′-TAAAATCATCAGCCCGATCACC-3′; R148A, forward primer 5′-GCAGAAGGTGCGTCGATCATCG-3′, reverse primer 5′-CACAACAGTAATCAGTGCGCC-3′; R296A, forward primer 5′-GCCCCGCTGAAATCGCTGACC-3′, reverse primer 5′-CATCAGTGCAATCATTGAAGAGAAAAC-3′; K299A, forward primer 5′-GCTTCGCTGACCAACGTTAACGCC-3′, reverse primer 5′-CAGCGGACGCATCAGTGCAATCA-3′. The mutant msbA genes were sequenced to ensure that only the intended changes had been introduced. Similar to wild-type msbA and previously described mutants10,11,15, the mutant genes were subcloned as NcoI/XbaI fragments into linearised pNZ8048 and pBAD24 plasmid DNA.
Overnight cultures of the lactococcal cells containing pNZ plasmid encoding (i) wild-type MsbA (referred to as MsbA-WT)10, (ii) MsbA-MD lacking the nucleotide-binding domain15, (iii) the single MsbA mutants: ΔK38211, K299A, R78A, R148A, or R296A, (iv) the double mutants: R78A R148A, R78A R296A, or R148A R296A, or (v) the triple mutants: R78A R148A R296A (referred to as MsbA-TripRA) or D41N E149Q D252N (referred to as MsbA-DED)15 were diluted 1:25 (v/v) in M17 medium (Formedium) supplemented with 25 mM glucose and 5 µg/mL chloramphenicol. Cells were allowed to grow at 30 °C to an OD660 of 0.55–0.60, after which 10 pg mL−1 nisin-A11 was added to induce protein expression for 1 h.
Preparation of inside-out membrane vesicles
Lactococcal cells from 2 L culture were harvested at 4 °C by centrifugation (13,000 × g, 10 min). The cell pellet was washed once (100 mM KPi, pH 7.0) and resuspended in 25 mL of the same buffer. Chicken egg white lysozyme (3 mg mL−1, Sigma-Aldrich) was added together with half a tablet of Complete-Protease Inhibitor Cocktail (Sigma-Aldrich) and the mixture was incubated for 30 min at 30 °C. To lyse the cells, the mixture was passaged twice through a cell disrupter (Basic Z 0.75-kW Benchtop Cell Disrupter, Constant Systems) at 20,000 p.s.i. Subsequently, 10 μg mL−1 DNase and 10 mM MgSO4 were added, and the resultant mixture was incubated for 30 min at 30 °C to digest DNA and RNA. In all, 15 mM K-EDTA (pH 7.0) was then added, and the mixture was centrifuged for 40 min at 13,000 × g and 4 °C. The supernatant containing the membrane vesicles was transferred to a clean tube and centrifuged for 1 h at 125,000 × g and 4 °C. The membrane vesicle pellet was resuspended in 50 mM KPi buffer (pH 7.0) containing 10% (v/v) glycerol to a total membrane protein concentration of 60 mg mL−1. The membrane vesicle suspension was stored in aliquots in liquid nitrogen. The presence of MsbA proteins in membrane vesicles was assessed on western blot probed with a 1:1000 (v/v) dilution of primary mouse anti-polyhistidine tag antibody (Sigma-Aldrich, cat. no.: H1029) followed by a 1:5000 (v/v) dilution of horseradish peroxidase-conjugated secondary goat anti-mouse antibody (Sigma-Aldrich, cat. no.: A4416). The histidine-tagged MsbA signals were detected by chemiluminescence using SuperSignal™ West Pico PLUS Chemiluminescent Substrate (ThermoFisher Scientific) and Odyssey Fc Imaging System (Li-Cor Biosciences).
Purification of His-tagged MsbA proteins
Histidine-tagged MsbA proteins were purified by Ni2+-nitrilotriacetic acid (NTA) affinity chromatography. In a typical experiment, 200 µg nickel-Ni2+-NTA resin with a binding capacity of more than 15 mg mL−1 and bead size between 45 and 165 µm, was pre-equilibrated by washing 5 times with 5 resin volumes of ultrapure water and twice with 5 resin volumes of wash Buffer A (50 mM KPi (pH 8.0), 0.1 M NaCl, 10% (v/v) glycerol, 0.05% (w/v) n-dodecyl-β-D-maltoside (DDM) and 20 mM imidazole (pH 8.0)). After every wash, the resin was collected by centrifugation (175 × g, 1 min, 4 °C). To solubilise the target protein, 600 µL membrane vesicles (36 mg total protein) were thawed and added to 4 mL of solubilisation buffer (50 mM KPi (pH 8.0), 10% (v/v) glycerol, 0.1 M NaCl and 1% (w/v) DDM). The mixture was shaken gently at 4 °C for 4 h. The mixture was added to 200 μg washed Ni2+-NTA resin and the suspension was mildly rotated overnight at 4 °C. The resultant resin with bound protein was transferred to a 1.5 mL bed-volume disposable Biospin chromatography column (Bio-Rad), and was washed with 20 resin volumes of wash Buffer A and 30 resin volumes of Wash Buffer B (50 mM KPi (pH 7.0), 0.1 M NaCl, 10% (v/v) glycerol, 0.05% (w/v) DDM and 20 mM imidazole (pH 8.0)). Subsequently, the His-tagged protein was eluted with 500 μL of elution buffer (50 mM KPi (pH 7.0), 0.1 M NaCl, 5% (v/v) glycerol, 0.05% (w/v) DDM and 150 mM imidazole (pH 8.0)). The purified protein was kept on ice and used for further experiments immediately.
Preparation of proteoliposomes containing biotinylated lipids
Purified MsbA proteins were reconstituted in proteoliposomes in an inside-out fashion15. For this purpose, 8 mg of liposomes were prepared from (i) acetone-ether-washed total lipid extract from E. coli, (ii) egg yolk phosphatidylcholine, and (iii) 18:1 Biotinyl Cap PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl), referred to as biotin-PE) (all purchased from Avanti Polar Lipids Inc.) mixed at a ratio of 3:1:0.12 (w/w/w) in chloroform. For the preparation of biotin-Lipid-A containing proteoliposomes, the biotin-PE was replaced by 0.5% (w/w) biotin-Lipid-A24 (stored at 0.5 mg mL−1 in chloroform:methanol 9:1 (v/v)). The solvent was evaporated from the lipid mixture by flushing with N2 gas after which the lipid mixture was hydrated in Buffer 1 (20 mM KPi, 100 mM NH4SCN, 50 mM K2SO4, pH 6.8) at a concentration of 4 mg mL−1. Using a LiposoFast-Basic extruder (Avestin), lipids were extruded 11 times through a 400 nm polycarbonate filter to form unilamellar liposomes of homogenous size. The extruded liposomes were destabilised by adding Triton X-100 until the maximum OD540 was just passed. For reconstitution, 160 µg of purified protein was mixed with destabilised liposomes in a ratio of 1:50 (w/w) to a final concentration of 80 µg mL−1. The mixture was left shaking gently at RT for 30 min. To remove detergent, the proteoliposomes were successively incubated with polystyrene bio-beads (Bio-Bead SM-2, Bio-Rad): 80 mg mL−1 at room temperature for 2 h, 8 mg mL−1 at 4 °C for 2 h, and finally 8 mg mL−1 at 4 °C for 18 h. Before use, the bio-beads were pre-washed 3 times with methanol, once with ethanol, four times with ultrapure water and once with Buffer 1. Ready-made proteoliposomes were harvested by centrifugation at 165,000 × g and 4 °C for 20 min and resuspended in 200–300 μL Buffer 1. Samples were kept on ice and immediately used in further experiments.
Biotin–lipid floppase assays
MsbA-containing proteoliposomes and empty liposomes containing 3% (w/w) biotin-PE or 0.5% (w/w) biotin-Lipid-A were diluted 6-fold in Buffer 1 in which they were prepared (no gradient control) or Buffer 2 (20 mM KPi, 100 mM KSCN, pH 8.0) to impose a ΔpH (interior acidic). All buffers were pre-warmed at 30 °C and supplemented with 5 mM MgSO4 and, where indicated, 5 mM Mg-ATP. Following dilution, the proteoliposomes were incubated for 20 min at 30 °C. Subsequently, 9 to 15 samples of 10 μL were each mixed with 90 µL working solution of Pierce™ Fluorescence Biotin Quantitation Kit. The mixture was incubated at RT for 10 min, and the fluorescence intensity was measured in a CLARIOstar plate reader (BMG Labtech) with excitation and emission wavelength of 494 nm and 520 nm, respectively. For biotin-Lipid-A analyses, the Fp(e)/mean Fp(c) ratio was divided by the Fl(e)/mean Fl(c) for each data point × 100%, where p corresponds to MsbA-containing proteoliposomes, l corresponds to empty liposomes (without MsbA protein), e corresponds to the treatment with ATP or ΔpH+ATP and c corresponds to the control treatment without metabolic energy. For biotin-PE analyses, Fl(e) = Fl(c) in the ΔpH, ATP and ΔpH+ATP treatments. Therefore, we directly compared Fp(e)/mean Fp(c) ratios for the different treatments, and the mean Fp(c) was set at 100%.
Internal pH measurements in proteoliposomes
Proteoliposomes were prepared by the same protocol as described under ‘Preparation of proteoliposomes containing biotinylated lipids’. However, 100 μM 2′,7′-bis-(2-carboxyethyl)−5-(and-6)-carboxyfluorescein (BCECF) was added to Buffer 1 in which the lipids were hydrated before extrusion. The BCECF-loaded proteoliposomes were 100-fold diluted in 2 mL of prewarmed Buffer 1 or Buffer 2, with a composition as described under ‘Biotin–lipid floppase assays’, in a 3.5 mL glass cuvette. BCECF fluorescence was monitored in a LS 55B Luminescence Spectrometer (PerkinElmer) at excitation and emission wavelengths of 502 and 525 nm, and with slit widths of 5 nm each.
Cell growth experiments
E. coli WD2 cells expressing a chromosome-encoded temperature-sensitive MsbA A270T mutant7,41 were used to study Lipid-A transport by pBAD24 plasmid-encoded MsbA proteins. The cells were grown aerobically overnight at 30 °C in LB broth supplemented with 12.5 µg mL−1 tetracycline and 100 µg mL−1 ampicillin. The cell cultures were first diluted in antibiotic-containing LB medium and grown to an OD600 of 0.6–0.7. The cell cultures were then diluted 500-fold in fresh antibiotic-containing LB broth containing 0.02% (w/v) of the inducer arabinose, of which 300 µL aliquots were dispensed in the wells on a 96-well plate. To assess cell growth, the OD600 of the cell suspensions was monitored at 44 or 30 °C for 6.5 h in a CLARIOstar plate reader in absorbance mode at 600 nm.
Ethidium transport
To measure ethidium efflux, 50 mL cultures of L. lactis cells with or without expressed MsbA proteins were harvested by centrifugation at 4 °C (6500 × g, 10 min) and washed once with wash buffer (50 mM KPi, 5 mM MgSO4, pH 7.0). Cells were de-energised in the presence of 0.5 mM 2,4-dinitrophenol at 30 °C for 40 min, and washed 3 times with the wash buffer, and resuspended to an OD660 of 5. For each measurement, 200 μL of cell suspension was diluted in 1800 μL of pre-warmed wash buffer at 30 °C in a 3.5 mL glass cuvette. Ethidium bromide (2 μM) was added to the mixture and the fluorescence level was monitored in a LS 55B Luminescence Spectrometer (PerkinElmer) at excitation and emission wavelengths of 500 and 580 nm with slit widths of 10 and 5 nm, respectively. Fluorescence traces were collected in BioLight Studio FL v1.04.02. Ethidium efflux was initiated by the addition of 25 mM glucose after which the ethidium fluorescence intensity was monitored for a further 10 min. The apparent affinity of MsbA proteins for ethidium in the transport reaction was determined as described10,11, by measuring the ethidium concentration dependence of the rate of MsbA-facilitated ethidium uptake into de-energised cells. The rates in MsbA-expressing cells were corrected for passive ethidium influx in de-energised control cells without MsbA expression. For the determination of kinetic parameters (Km and Vmax) the data were fitted to the Michaelis–Menten equation in Graphpad Prism v 9.2.0.
NBD-PE floppase assay
MsbA-containing proteoliposomes and empty liposomes were prepared as described under ‘Preparation of proteoliposomes containing biotinylated lipids’ using a lipid mixture composed of (i) acetone-ether-washed total lipid extract from E. coli, (ii) 18:1 phosphatidylglycerol (1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)) and (iii) 18:1 headgroup-labelled NBD-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) at a ratio of 3: 1: 0.012 (w/w/w). In the floppase assay, the (proteo)liposomes were diluted 100-fold in 2 mL prewarmed Buffer 1 or Buffer 2, with a composition as described under ‘Biotin–lipid floppase assays’, in a 3.5 mL glass cuvette. Following dilution, the proteoliposomes were incubated for 5 min at 30 °C. Subsequently, 4 mM sodium dithionite was added to quench the NBD fluorescence in the outer leaflet of the membrane. The remaining NBD fluorescence in the inner leaflet was measured in a LS 55B Luminescence Spectrometer (PerkinElmer) at excitation and emission wavelengths of 464 and 536 nm with slit widths of 10 and 5 nm, respectively. Fluorescence traces were collected in BioLight Studio FL v1.04.02. For analysis, the fluorescence intensity was normalised to F/Fmax and, following the addition of the dithionite, the (F/Fmax)p/(F/Fmax)l ratios at plateau for the different treatments (ATP, ΔpH, and ΔpH+ATP) were compared with the control without metabolic energy (set at 100%), where p corresponds to MsbA-containing proteoliposomes, l corresponds to liposomes without MsbA protein, and Fmax and F correspond to the observed fluorescence plateau before and after dithionite addition.
Statistics and reproducibility
All experiments were performed at least three times using three independent batches of cells and proteoliposomes. Significance of data was tested by one-way analysis of variance and Tukey’s multiple group comparisons. Asterisks shown in histograms and scatter dot plots either refer to comparisons with control (above the bar) or comparisons of specific pairs (horizontal brackets): *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data that support the findings of this study have been deposited in the University of Cambridge research repository Apollo with link https://doi.org/10.17863/CAM.75215 (ref. 42) or are available from the corresponding author upon reasonable request. The source data for the figures and supplementary figures are included in Supplementary Data 1. The DNA sequences of pNZ-MsbA, pGEM-MsbA and pBAD-MsbA plasmid and the wild-type and mutant msbA genes, as well as the amino acid sequences of the MsbA proteins, are included in Supplementary Data 1. Requests for unique materials should be addressed to the corresponding author. The pNZ plasmids utilise the nisin controlled expression (NICE) system, which was obtained from NIZO Food Research, The Netherlands.
References
Kornberg, R. D. & McConnell, H. M. Inside-outside transitions of phospholipids in vesicle membranes. Biochemistry 10, 1111–1120 (1971).
Nakano, M. et al. Flip-flop of phospholipids in vesicles: kinetic analysis with time-resolved small-angle neutron scattering. J. Phys. Chem. B 113, 6745–6748 (2009).
Bethel, N. P. & Grabe, M. Atomistic insight into lipid translocation by a TMEM16 scramblase. Proc. Natl Acad. Sci. USA 113, 14049–14054 (2016).
Vestergaard, A. L. et al. Critical roles of isoleucine-364 and adjacent residues in a hydrophobic gate control of phospholipid transport by the mammalian P4-ATPase ATP8A2. Proc. Natl Acad. Sci. USA 111, E1334–E1343 (2014).
Olsen, J. A., Alam, A., Kowal, J., Stieger, B. & Locher, K. P. Structure of the human lipid exporter ABCB4 in a lipid environment. Nat. Struct. Mol. Biol. 27, 62–70 (2020).
Smit, J. J. et al. Homozygous disruption of the murine mdr2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell 75, 451–462 (1993).
Doerrler, W. T., Gibbons, H. S. & Raetz, C. R. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 279, 45102–45109 (2004).
Mi, W. et al. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 (2017).
Owens, T. W. et al. Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 567, 550–553 (2019).
Woebking, B. et al. Drug-lipid A interactions on the Escherichia coli ABC transporter MsbA. J. Bacteriol. 187, 6363–6369 (2005).
Woebking, B. et al. Functional role of transmembrane helix 6 in drug binding and transport by the ABC transporter MsbA. Biochemistry 47, 10904–10914 (2008).
Doshi, R., Woebking, B. & van Veen, H. W. Dissection of the conformational cycle of the multidrug/lipidA ABC exporter MsbA. Proteins 78, 2867–2872 (2010).
Doshi, R. & van Veen, H. W. Substrate binding stabilizes a pre-translocation intermediate in the ATP-binding cassette transport protein MsbA. J. Biol. Chem. 288, 21638–21647 (2013).
Doshi, R. et al. Molecular disruption of the power stroke in the ATP-binding cassette transport protein MsbA. J. Biol. Chem. 288, 6801–6813 (2013).
Singh, H. et al. ATP-dependent substrate transport by the ABC transporter MsbA is proton-coupled. Nat. Commun. 7, 12387 (2016).
Doerrler, W. T., Reedy, M. C. & Raetz, C. R. An Escherichia coli mutant defective in lipid export. J. Biol. Chem. 276, 11461–11464 (2001).
Raetz, C. R. & Dowhan, W. Biosynthesis and function of phospholipids in Escherichia coli. J. Biol. Chem. 265, 1235–1238 (1990).
Bogdanov, M. et al. Phospholipid distribution in the cytoplasmic membrane of Gram-negative bacteria is highly asymmetric, dynamic, and cell shape-dependent. Sci. Adv. 6, eaaz6333 (2020).
Gupta, K. et al. Identifying key membrane protein lipid interactions using mass spectrometry. Nat. Protoc. 13, 1106–1120 (2018).
Aye, I. L., Singh, A. T. & Keelan, J. A. Transport of lipids by ABC proteins: interactions and implications for cellular toxicity, viability and function. Chem. Biol. Interact. 180, 327–339 (2009).
Neumann, J., Rose-Sperling, D. & Hellmich, U. A. Diverse relations between ABC transporters and lipids: an overview. Biochim. Biophys. Acta Biomembr. 1859, 605–618 (2017).
Shin, H. W. & Takatsu, H. Substrates of P4-ATPases: beyond aminophospholipids (phosphatidylserine and phosphatidylethanolamine). FASEB J. 33, 3087–3096 (2019).
Ho, H. et al. Structural basis for dual-mode inhibition of the ABC transporter MsbA. Nature 557, 196–201 (2018).
Fujimoto, Y., Kimura, E., Murata, S., Kusumoto, S. & Fukase, K. Synthesis and bioactivity of fluorescence- and biotin-labeled lipid A analogues for investigation of recognition mechanism in innate immunity. Tetrahedron Lett. 47, 539–543 (2006).
Padayatti, P. S. et al. Structural insights into the lipid A transport pathway in MsbA. Structure 27, 1114–1123 e1113 (2019).
Li, Y., Orlando, B. J. & Liao, M. Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature 567, 486–490 (2019).
Raturi, S. et al. Engineered MATE multidrug transporters reveal two functionally distinct ion-coupling pathways in NorM from Vibrio cholerae. Commun. Biol. 4, 558 (2021).
Chi, X. et al. Structural mechanism of phospholipids translocation by MlaFEDB complex. Cell Res. 30, 1127–1135 (2020).
Coudray, N. et al. Structure of bacterial phospholipid transporter MlaFEDB with substrate bound. Elife 9, e62518 (2020).
Tang, X. et al. Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB. Nat. Struct. Mol. Biol. 28, 81–91 (2021).
Douglass, M. V., Cleon, F. & Trent, M. S. Cardiolipin aids in lipopolysaccharide transport to the gram-negative outer membrane. Proc. Natl Acad. Sci. USA 118, e2018329118 (2021).
Eckford, P. D. & Sharom, F. J. The reconstituted Escherichia coli MsbA protein displays lipid flippase activity. Biochem. J. 429, 195–203 (2010).
Doerrler, W. T. & Raetz, C. R. ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 277, 36697–36705 (2002).
Garreta, A. et al. Structure and interaction with phospholipids of a prokaryotic lipoxygenase from Pseudomonas aeruginosa. FASEB J. 27, 4811–4821 (2013).
Yero, D. et al. The Pseudomonas aeruginosa substrate-binding protein Ttg2D functions as a general glycerophospholipid transporter across the periplasm. Commun. Biol. 4, 448 (2021).
Jiang, T., Yu, K., Hartzell, H. C. & Tajkhorshid, E. Lipids and ions traverse the membrane by the same physical pathway in the nhTMEM16 scramblase. Elife 6, e28671 (2017).
Andersen, J. P. et al. P4-ATPases as phospholipid flippases-structure, function, and enigmas. Front. Physiol. 7, 275 (2016).
Perez, C. et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524, 433–438 (2015).
Venter, H., Velamakanni, S., Balakrishnan, L. & van Veen, H. W. On the energy-dependence of Hoechst 33342 transport by the ABC transporter LmrA. Biochem. Pharmacol. 75, 866–874 (2008).
de Ruyter, P. G., Kuipers, O. P. & de Vos, W. M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62, 3662–3667 (1996).
Reuter, G. et al. The ATP binding cassette multidrug transporter LmrA and lipid transporter MsbA have overlapping substrate specificities. J. Biol. Chem. 278, 35193–35198 (2003).
Guo, D. et al. Energetics of lipid transport by the ABC transporter MsbA is lipid dependent. University of Cambridge Research Repository Apollo, https://doi.org/10.17863/CAM.75215 (2021).
Acknowledgements
This research was funded by Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/R00224X/1 (to H.W.v.V). D.G. and Y.T. were funded by China Scholarship Council – Cambridge Trust PhD Scholarships. C.G. was funded by a BBSRC Doctoral Training Partnership (DTP) Targeted PhD studentship (project 2114197). T.N. received a student grant from Christ’s College Cambridge.
Author information
Authors and Affiliations
Contributions
D.G., H.S., C.G., Y.T., T.N. and H.W.v.V. planned and performed cloning, protein expression and reconstitution, lipid floppase and drug resistance and drug transport experiments. A.S. and K.F. planned and performed the synthesis of biotin-Lipid-A. S.M.R. and D.R.S. planned and performed the synthesis of MsbA inhibitor G907. H.W.v.V. and D.G. wrote the manuscript with input from the other authors. Supplementary Information on G907 synthesis was written by S.M.R. and D.R.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review information
Communications Biology thanks Qinghai Zhang and the other anonymous reviewers for their contribution to the peer review of this work. Primary handling editor: Eve Rogers.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guo, D., Singh, H., Shimoyama, A. et al. Energetics of lipid transport by the ABC transporter MsbA is lipid dependent. Commun Biol 4, 1379 (2021). https://doi.org/10.1038/s42003-021-02902-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-021-02902-8
This article is cited by
-
Cerastecins inhibit membrane lipooligosaccharide transport in drug-resistant Acinetobacter baumannii
Nature Microbiology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.