Abstract
Patients with Alzheimer’s disease (AD) often have osteoporosis or osteopenia. However, their direct link and relationship remain largely unclear. Previous studies have detected osteoporotic deficits in young adult Tg2576 and TgAPPsweOCN mice, which express APPswe (Swedish mutant) ubiquitously and selectively in osteoblast (OB)-lineage cells. This raises the question, whether osteoblastic APPswe contributes to AD development. Here, we provide evidence that TgAPPsweOCN mice also exhibit AD-relevant brain pathologies and behavior phenotypes. Some brain pathologies include age-dependent and regional-selective increases in glial activation and pro-inflammatory cytokines, which are accompanied by behavioral phenotypes such as anxiety, depression, and altered learning and memory. Further cellular studies suggest that APPswe, but not APPwt or APPlon (London mutant), in OB-lineage cells induces endoplasmic reticulum-stress driven senescence, driving systemic and cortex inflammation as well as behavioral changes in 6-month-old TgAPPsweOCN mice. These results therefore reveal an unrecognized function of osteoblastic APPswe to brain axis in AD development.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is the most common form of dementia. It is pathologically characterized by cortical and cerebrovascular β-amyloid (Aβ) plaques, phosphor-tau containing neurofibrillary tangles, reactive glial cell (astrocyte and microglial cell)-associated chronic brain inflammation, and neuron-loss1,2. Interestingly, in addition to brain pathologies, patients with AD, both early and late onset, often have osteopenia or osteoporosis3,4,5,6,7,8,9,10, a condition characterized by the loss of bone-mass or bone mineral density (BMD) with micro-architectural deterioration of bone tissue, and a higher rate of hip fracture. However, little is known regarding the underlying mechanisms of AD association with bone loss.
A growing list of genetic risk genes has been identified in patients with early onset and late onset AD. Intriguingly, many of the AD risk genes, such as TREM2 (triggering receptors expressed on myeloid cells-2) and PYK2, are highly expressed in immune cells and bone cells, and encode proteins that regulate not only neuron synaptic functions, but also immune responses and bone homeostasis11,12,13,14,15. APOE, another AD risk gene, is also identified as a risk factor for osteoporosis16,17,18. Among the various risk genes for AD development, we chose Swedish mutant APP (APPswe) to address the question regarding AD association with bone loss for the following reasons. The Swedish mutations in the APP gene are initially identified in patients with early-onset (EO)AD, which promote the generation of Aβ by favoring its proteolytic cleavage performed by β- and γ-secretases19,20,21. Much research has focused on the impacts of Aβ on the brain, even though APP or APPswe is known to be expressed not only in the brain, but also in periphery tissues, including osteoblast (OB)-lineage cells22,23. Although APPswe is only detected in a small fraction of AD patients, it is commonly used to generate AD animal models, such as Tg2576 and 5XFAD24,25. APPswe in these animal models (in particularly Tg2576) is expressed ubiquitously, in both the brain and periphery tissues, including OB cells22,23. While investigating the phenotypes of these APPswe–based animal models have provided valuable insights into Aβ brain pathology and impairments in mouse cognitive functions, the function of APPswe in peripheral tissues, such as OBs, remains poorly understood. Previous examinations of bone structures in Tg2576 mice have identified early-onset osteoporotic deficits, months before any brain-pathologic defect that was detected22,23. Knocking out App (in APP−/− mice), or selective expressing APPswe in osteocalcin (OCN) promoter driven Cre (OCN-Cre)+ OB-lineage cells (in TgAPPsweOCN mice) recapitulates the osteoporotic defects in Tg2576 mice23,26. These observations raise an interesting question, could problems in the bone cells conversely contribute to AD pathology in the brain?
Here, we provide evidence that TgAPPsweOCN mice express APPswe largely in the OB-lineage cells, with little to weak expression in the dorsal dentate gyrus (dDG) of the hippocampus. These mice develop age-dependent [starting at 6-month-old (MO)] and brain-region selective pathologies, and exhibit anxiety- and depression-like behaviors, as well as altered cognitive functions. While these mice at 6-MO showed brain-pathy (including glial activations and elevated pro-inflammatory cytokines) largely in the cortex, these mice at 12-MO showed brain-pathy mainly detected in the hippocampus. Further mechanistic studies demonstrate that APPswe, but not APPwt or APPlon (London mutant), in OB-lineage cells increases endoplasmic reticulum (ER)-stress, senescence, and SASPs (senescence associated secretory phenotypes). Inhibition of ER-stress abolishes APPswe-induced senescence, and suppression of senescence diminishes brain and behavioral phenotypes in 6-MO TgAPPsweOCN mice. Taken together, these observations suggest that APPswe in OB-lineage cells contributes to the brain-region selective inflammation and glial activation and induces anxiety- and depression-like behaviors in age-dependent manner, which are largely due to elevated OB-senescence, SASPs, and systemic inflammation. These results thus uncover a link between APPswe in the OB-lineage cells and AD development.
Results
Selective APPswe expression in OB-lineage cells in TgAPP swe OCN mice
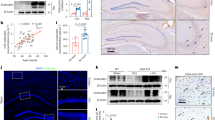
To investigate osteoblastic APPswe’s function in AD development, we took advantage of TgAPPsweOCN mice, in which human APPswe expression in LSL-hAPPswe mice depends on the removal of LSL by the OCN-Cre (Fig. 1a)23. Although OCN-Cre mice express Cre primarily in mature/adult OB-lineage cells27,28, our recent study showed Cre activity in neurons of dDG hippocampus, olfactory bulb, and cerebellum29. Thus, it is important to verify APPswe’s expression in bone cells and brain tissues of TgAPPsweOCN mice. Notice that the hAPPswe protein was detected in the OB-lineage BMSCs (bone marrow stromal cells), but not in the hippocampus or cortex of the TgAPPsweOCN mice (6-MO) (Fig. 1b, c). We then asked if this is due to hAPPswe’s cleavage (to produce Aβ40 or Aβ42) in the brain tissues. ELISA measuring human Aβ40 and Aβ42 levels showed little-to-no Aβ increase in the hippocampus, cortex, or serum samples (Fig. 1d, e); but slight increases of both Aβ40 and Aβ42 in the OB-lineage cells, of TgAPPsweOCN mice (6-MO), as well as in the brain tissues and serum samples of 6-MO Tg2576 mice (Fig. 1d, e). These results eliminate the possibility of β- and γ-cleavages of hAPPswe in the brain of 6-MO TgAPPsweOCN mice, suggesting little hAPPswe expression in the mutant brain at this age. We further tested this view by RT-PCR analysis of hAPPswe’s transcripts in the mutant mice. Using specific primers for human APP, a weak hAPPswe expression (~1.5 fold over control) was detected in the TgAPPsweOCN brain regions (e.g., hippocampus, olfactory bulb, and cerebellum) where OCN-Cre is expressed29, but not in the OCN-Cre negative cortex (Fig. 1f). Notice that the hAPPswe’s transcripts were much more abundant in the BMSCs (~70 fold over control) than in the brain (Fig. 1f), implying a much weaker Cre activity in neurons than in OB-lineage cells of the OCN-Cre mice. This viewpoint is consistent with the RT-PCR findings that Cre is expressed largely in the OB-lineage cells (~128 fold over control), weakly (~18 fold over control) in the hippocampus, and undetectable in the cortex of OCN-Cre mice (Fig. 1g). Taken together, these results suggest that the hAPPswe is highly expressed in OCN-Cre+ OB-lineage cells, but little to weakly expressed in the OCN-Cre+ dDG, olfactory bulb, and cerebellum neurons, of TgAPPsweOCN mice.
a Illustration of the transgene and generation of the conditional transgenic mice selectively expressing human APPswe in an OCN-Cre dependent manner. b, c Western blot analysis of human APP (hAPP) protein levels in BMSCs, hippocampus, and cortex of 6-MO control (LSL-APPswe) and TgAPPsweOCN mice. b Representative blots; and c quantification. d, e ELISA analyses of human Aβ40(d) and Aβ42(e) levels in serum, BMSCs (50 μg in total protein), and brain homogenates including hippocampus and cortex (300 μg total protein) from 6-MO control, TgAPPsweOCN, and Tg2576 mice. f RT-PCR analysis of hAPP gene expression in BMSCs, olfactory bulb, cerebellum, hippocampus, and cortex of 6-MO control and TgAPPsweOCN mice. g RT-PCR analysis of Cre expression in BMSCs, hippocampus, and cortex of 6-MO control (LSL-APPswe) and TgAPPsweOCN mice. All data were presented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 (n = 3 mice). Mann–Whitney U test was used in c and g, and one-way ANOVA followed by Tukey post hoc test was used in d–f.
Age-dependent and brain region-selective elevations in reactive astrocytes, microglial cells, and inflammatory cytokines, and an impairment in DG neurogenesis in TgAPP swe OCN mice
We then addressed whether TgAPPsweOCN mice exhibit any brain pathology that is similar to those of APPswe-based AD animal models (e.g., Tg2576)24,25,32,33,34, by performing the following studies.
First, we measured both Aβ40 and Aβ42 levels in the bone cells and brain tissues of TgAPPsweOCN mice at ages of not only 6-MO, but also 12-MO. Although little Aβ40 or Aβ42 levels were detected in 6-MO TgAPPsweOCN cortex and hippocampus (Fig. 1d, e), Aβ42, but not Aβ40, was slightly elevated in 12-MO TgAPPsweOCN hippocampus, but not cortex nor serum samples (Supplementary Fig. 1a, b). Additionally, little to no Aβ plaque was detected in 12-MO TgAPPsweOCN bone and brain sections, in contrast from brain sections from 5XFAD mice (4.5 MO) (Supplementary Fig. 1c–e). These findings support the view for a weak hAPPswe/Aβ42 expression in 12-MO TgAPPsweOCN hippocampal DG neurons.
Second, we examined neuronal distribution patterns and densities in the cortex and hippocampus of TgAPPsweOCN mice (at age of ~7-MO) by conducting co-immunostaining analysis using antibodies against NeuN (a marker for all neurons) and Ctip2 (a marker for Layer V–VI neurons in the cortex and neurons in CA1-2 and DG). Little change in the NeuN+ and Ctip2+ neuron distribution patterns and densities was detected in TgAPPsweOCN brains (Supplementary Fig. 2).
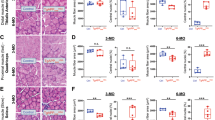
Third, we assessed the morphologies and densities of glial cells, including Olig2+ oligodendrocytes, S100β+ ependymal cells, GFAP+ astrocytes, and IBA1+ microglial cells, in the brain sections of control (LSL-APPswe) and TgAPPsweOCN mice. The Olig2+ oligodendrocytes and S100β+ ependymal cells appeared to be unchanged in the TgAPPsweOCN cortex or brain (Supplementary Fig. 3). Intriguingly, both GFAP+ astrocytes and IBA1+ microglial cells were increased in 6-MO TgAPPsweOCN cortex, particularly in layers I–III, but not in hippocampus (Fig. 2a–d), suggesting a brain region-selective activation of these glial cells. This view was further verified through a Western blot analysis, which showed increased GFAP and IBA1 protein levels in 6-MO TgAPPsweOCN cortex, but not in hippocampus (Fig. 2e, f). Because glial cell activation is often associated with increased inflammation30,31, we examined expressions of inflammation associated cytokines (e.g., Il1b, Il6, Il10, and Tnfa), growth factors (e.g., Tgfb1 and Csf2), and proteinase (e.g., Mmp3) in both the cortex and hippocampus of control and TgAPPsweOCN mice (at 6-MO) using RT-PCR analysis. The transcripts of Il1b, Il10, Tnfa, and Mmp3 were all increased in TgAPPsweOCN cortex, but not in hippocampus (Fig. 2g, h), supporting the view of cortex as a vulnerable brain region in 6-MO TgAPPsweOCN mice.
a Representative images of co-immunostaining with IBA1 (green), GFAP (magenta), and DAPI (blue) of hippocampal sections from 6-MO control (LSL-APPswe) and TgAPPsweOCN mice. Scale bars: 200 µm (upper) and 20 µm (lower). b Quantification of data in a. c Representative images of co-immunostaining with IBA1 (green), GFAP (magenta), and DAPI (blue) of cortex sections from 6-MO control (LSL-APPswe) and TgAPPsweOCN mice. Scale bars: 100 µm (upper) and 20 µm (lower). d Quantification of data in c. e Representative Western blots using antibodies against hAPP, GFAP, and IBA1 in homogenates of cortex and hippocampus of control and TgAPPsweOCN mice. GAPDH was used as a loading control. f Quantification of the data in e. g–h Real-time PCR (RT-PCR) analysis of indicated gene expressions in 6-MO control (LSL-APPswe) and TgAPPsweOCN cortex (g) and hippocampus (h). All quantification data were presented as mean ± SD (n = 3–4). *p < 0.05, **p < 0.01, ***p < 0.001. Student’s t test was used in b, d, and f–h.
Fourth, we found that the glial activation and inflammatory phenotypes in TgAPPsweOCN mice were not only brain-region selective, but also age-dependent. Whereas the cortex displayed the glial activation/inflammation in 6-MO TgAPPsweOCN mice, these phenotypes were not detected in 3-MO TgAPPsweOCN (Supplementary Fig. 4), but evidently more obvious in 12-MO TgAPPsweOCN hippocampus than the cortex (Supplementary Fig. 5), suggesting age-dependent changes in the brain-region selectivity of the glial activation/inflammation phenotypes.
Finally, we examined adult neurogenesis in hippocampal DG (dentate gyrus), which is also impaired in AD animal models32. EdU was injected into the mice ~12 h before sacrifice to label proliferative neural stem cells (NSCs). Hippocampal sections were co-immunostained EdU with antibodies against DCX (doublecortin) (a marker for newborn neurons derived from NSCs). While TgAPPsweOCN mice at 3-MO showed no difference in EdU+ and DCX+ cell densities compared to the controls, TgAPPsweOCN mice at 6-MO displayed significant reductions in EdU+ and DCX+ cell densities at both dorsal and ventral DG (Supplementary Fig. 6), demonstrating an age-dependent impairment in the hippocampal DG neurogenesis of TgAPPsweOCN mice, similar to that described in AD animal models32.
In aggregate, TgAPPsweOCN mice (starting at 6-MO) exhibit partial AD relevant brain pathologies, which include increased reactive astrocytes, microglial cells, and inflammatory cytokines in the cortex (at 6-MO)/hippocampus (at 12-MO), impaired DG neurogenesis, and elevated Aβ42 in 12-MO hippocampus.
Age-dependent anxiety- and depression-like behaviors in TgAPP swe OCN mice
Glial activation, brain inflammation, and decreased DG neurogenesis are often associated with depression- and/or anxiety-like behaviors33,34,35,36,37,38. We thus subjected TgAPPsweOCN and control mice to an open field test (OFT) for evaluation of TgAPPsweOCN mice’s anxiety and locomotor activity. TgAPPsweOCN mice at 6- and 12-MO, but not 3-MO, showed reduced center duration time but comparable total distance traveled to the controls (Fig. 3a, b and Supplementary Fig. 7a, b), suggesting a reduced exploratory, but not locomotor, activity, in the mutant mice, and implicating anxiety and/or depression. We further examined their behaviors using elevated plus maze test (EPMT) and light/dark transition test (LDT), both tests widely used to assess anxiety-related behavior in mouse models39,40. Indeed, TgAPPsweOCN mice, again at 6- and 12-MO, but not 3-MO, showed decreased open arm duration time and entries by EPMT (Fig. 3c, d and Supplementary Fig. 7c, d), and reduced time in light box room in the LDT (Fig. 3e and Supplementary Fig. 7e), supporting the view for anxiety-like behaviors. We then assessed their depression-like behaviors using tail suspension test (TST), force swimming test (FST), and sucrose preference test (SPT). TgAPPsweOCN mice (6- and 12-MO, but not 3-MO) appeared to be depressed, exhibiting increased immobility times in both TST (Fig. 3f and Supplementary Fig. 7f) and FST (Fig. 3g and Supplementary Fig. 7g) and reduced sucrose preference (Fig. 3h and Supplementary Fig. 7h). Together, these results suggest that TgAPPsweOCN mice experience age-dependent (starting at 6-MO) anxiety- and depression-like behaviors.
a, b OFT: Representative tracing images (a), and quantifications of total distance and center duration time (b) were shown. c, d EPMT: Representative tracing images (c), and quantifications of open arm duration time and entries (d) were shown. e LDT: Quantifications of the time spent in the light room and the number of transitions into the light room. f TST, g FST, and h SPT. In all these behavior tests, 6-MO and 12-MO control (LSL-APPswe) and TgAPPsweOCN mice (males) were examined. All quantification data were shown as mean ± SD (n = 10 mice). *p < 0.05, **p < 0.01, ***p < 0.001, Student’s t test.
Since hAPPswe is weakly expressed in dDG neurons in TgAPPsweOCN hippocampus (Fig. 1f), we wondered whether such a weak dDG expression of APPswe could induce similar behavior phenotypes to that in TgAPPsweOCN mice. The AAV-CamkII-Cre (Cre under the control of CamkII promotor for excitatory neuron expression) and AAV-CamkII-GFP (as control) were specifically injected into the dDGs of both sides of the hippocampus in LSL-APPswe mice (at age of 4-MO); and mice at 6-MO were subjected to the behavior tests (Supplementary Fig. 8a). While dDG neurons in LSL-APPswe mice were successfully infected with the viruses (indicated by the GFP, hAPPswe expression, and Aβ42 increase) (Supplementary Fig. 8b–g), little to no differences in behavior tests using EMPT, LDT, TST, FST, and SPT were detected between Cre and GFP virus injected mice (Supplementary Fig. 8h–l), unlike the TgAPPsweOCN mice. These results thus implicate that the anxiety- or depression-like behaviors in 6-MO TgAPPsweOCN mice are in large due to APPswe’s expression in OB-lineage cells, but not dDG neurons.
Age-dependent alterations in spatial learning and memory in TgAPP swe OCN mice
Although anxiety- and depression-like behaviors are present in AD animal models (e.g., Tg2576 and 5XFAD)41,42,43 and AD patients44,45,46, a key AD relevant functional deficit is the age-dependent cognition decline47,48. Therefore, we subjected TgAPPsweOCN and control (LSL-APPswe) mice to the Morris water maze (MWM) test (to access mouse spatial learning and memory function)49, and the novel object recognition (NOR) test (to evaluate mouse recognition memory)50,51. Interestingly, age-dependent changes in both MWM and NOR tests were detected in TgAPPsweOCN mice. No obvious difference in MWM or NOR task performance was observed between TgAPPsweOCN and control mice at 3-MO (Fig. 4a–c). Un-expectedly, at 6-MO, TgAPPsweOCN mice exhibited faster learning and better long-term memory in MWM (Fig. 4d, e), but no obvious difference in NOR task performance (Fig. 4f), compared to the age-matched controls, suggesting an improvement in spatial learning and memory in 6-MO TgAPPsweOCN mice. Interestingly, at ~12-MO, impairments in both MWM and NOR tasks were detected in TgAPPsweOCN mice (Fig. 4g–i). These results are intriguing, demonstrating age-dependent changes in spatial and novel object learning and memory of TgAPPsweOCN mice, in line with their age-dependent changes in the brain-region selectivity of the glial activation/inflammation.
a–c 3-MO control (LSL-APPswe) and TgAPPsweOCN male mice were subject to Morris water maze (MWM) (a, b) and Novel Object Recognition (NOR) (c) tests. d–f 6-MO control (LSL-APPswe) and TgAPPsweOCN male mice were subject to MWM (d, e) and NOR (f) tests. g–i 12-MO control (LSL-APPswe) and TgAPPsweOCN male mice were subject to MWM (g, h) and NOR (i) tests. In MWM tests, the latencies to reach the hidden platform during the training period were showed in a, d, and g; and the representative tracing images and quantification of time spent in target quadrant, platform crossing time, and swim speed were shown in b, e, and h. In NOR tests, the time spent with novel object per total time with both objects as the novel object preference was quantified, shown in c, f, and i. All values were presented as mean ± SD (n = 10 mice). *p < 0.05, one-way ANOVA followed by Tukey post hoc test was used in a, d, and g, and Student’s t test was used in b, c, e, f, h, and i.
Increased senescence and SASPs in APPswe + OB-lineage cells
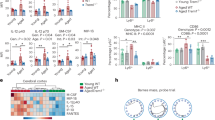
To investigate if and how APPswe in OCN-Cre+ OB-lineage cells gives rise to the brain and behavior phenotypes, we purified OCN-Cre+ BMSCs (marked by tdTomato+, believed to be OB progenitors28) from both 6-MO control (OCN-Cre; Ai9) and TgAPPsweOCN; Ai9 mice using fluorescence-activated cell sorting (FACS), and then subjected them to RNA-seq analysis (Fig. 5a). 917 up- and 1825 down-regulated genes were identified in APPswe+ OB progenitors (Fig. 5b). Among these genes, 154 up- and 269 down-regulated genes encode secreted proteins (Fig. 5b). Interestingly, GO analysis showed that most up-regulated genes are involved in inflammatory response, cytokine production, and cytokine/chemokine-mediated signaling pathways; and most down-regulated genes are implicated in cell cycle, cell proliferation, and bone mineralization (Fig. 5c). Further heat map analysis illustrated the up- and down-regulated genes for bone mass regulators, cytokines and chemokines, AD risk genes, and growth factors critical for neurogenesis (Fig. 5d). Some of these up/down regulated genes were verified by RT-PCR analyses (Fig. 5e). Notice that transcription factors (e.g., Sp7, Nfatc1, Satb2, Spp1, Sparc, and Jun) for OB-lineage cell proliferation, differentiation, or mineralization, and bone-mass regulators (e.g., Bmp2, Ihh, Lrp4, and Ctnnb1) were down regulated in APPswe+ OB progenitors (Fig. 5d, e), in line with our previous report23; and some of the AD-risk genes including Trem2, ApoE, Cd33, Sorl1, Vps35, Ptk2b, and Psen1/2 were altered in APPswe+ OB progenitors, and growth factors including brain-derived neurotrophic factor (Bdnf) and insulin-like growth factor 1 (Igf1) were decreased in APPswe+ OB progenitors (Fig. 5d, e).
a Schematic of purification and RNA-seq of Tdtomato+ (Td+) OB progenitors from control (OCN-Cre; Ai9) and TgAPPsweOCN; Ai9 mice. b–d Volcano plots (b), GO analysis of up/down-regulated genes (c), and heat map (d) of differentially expressed genes identified by RNA-seq. e RT-PCR analysis of AD risk gene App, Vps35, Trem2, Apoe, Ptk2b, and Sorl1; bone-mass regulator Sp7, Nfatc1, Col1a1, Spp1, Sparc, Bmp2, Lrp4, and Ctnnb1; cytokine Il1b, Il6, and Il10; chemokine Ccl5 and Cxcl1, growth factor Tgfb1 gene expression in purified Td+ OB progenitors from 6-MO control (OCN-Cre; Ai9) and TgAPPsweOCN; Ai9 mice. All values were presented as mean ± SD (n = 3 mice). *p < 0.05, **p < 0.01, and ***p < 0.001, by Mann–Whitney U test.
Interestingly, the increased cytokines and chemokines exhibits features of SASPs52,53. We thus asked if APPswe+ OB-progenitors undergo senescence. Indeed, senescence marker proteins, such as SA-β-gal (senescence associated β-galactosidase), P16Ink4a, P21, and P5354,55, were all elevated in APPswe+ OB-progenitors (derived from 3- and 6-MO mice) (Fig. 6), suggesting that APPswe induces OB-senescence. We also detected reductions in tdTomato+ (Td+) or OCN-Cre+ OB progenitors (Supplementary Fig. 9a, b) and in EdU+ proliferative cells in OB-progenitor cultures from TgAPPsweOCN; Ai9 mice (Supplementary Fig. 9c–e), indicating a growth arrest of these cells, another feature of cellular senescence in APPswe+ OB-progenitors.
a SA-β-gal staining of 3-MO and 6-MO BMSCs from control (LSL-APPswe) and TgAPPsweOCN mice. Scale bar, 20 µm. b Quantification of SA-β-gal+ cell densities (mean ± SD; n = 3 independent experiments). **p < 0.01, ***p < 0.001. c Western blot analysis of indicated protein expression in BMSCs from mice with indicated genotypes (at 6-MO). GAPDH was used as a loading control. d Quantification analyses of the data in c, *p < 0.05, ***p < 0.001. mean ± SD n = 3. Mann–Whitney U test.
Finally, we wondered whether other tissues in TgAPPsweOCN mice develop senescence-like phenotypes. The mRNAs from various tissues [including cortex, hippocampus, TA (Tibialis anterior) muscles, kidneys, and livers] of 6-MO control and TgAPPsweOCN mice were subjected to RT-PCR analyses with P16Ink4a and P53 transcripts––both markers of senescence. Interestingly, both P16Ink4a and P53 were increased in the cortex and TA muscles, but not kidney or liver, of 6-MO TgAPPsweOCN mice (Supplementary Fig. 10a–e). These results suggest brain-region and tissue selective senescence-like phenotypes in TgAPPsweOCN mice.
Diminished behavior phenotypes and brain pathology in TgAPP swe OCN mice treated with senescence inhibitor
To determine if the increased senescence and SASPs in TgAPPsweOCN mice contribute to the brain and behavior deficits, we treated TgAPPsweOCN mice with Dasatinib (D) + Quercetin (Q), or Veh control (10% PEG 400), because the combination of D + Q is a well examined effective senolytic drug in animal studies56,57. We first treated cultured OB progenitors (BMSCs) from TgAPPsweOCN mice with D + Q. As expected, the senescence markers (SA-β-gal, P53, and P16Ink4a) and the SASP-like factors (e.g., Il1b, Il6, Cxcl1, Ccl5, and Tgfb1) were all decreased in D + Q treated APPswe+ OB-progenitors (Supplementary Fig. 11), verifying D + Q’s inhibitory effects on OB-senescence. We then administered D + Q to TgAPPsweOCN mice as illustrated in Fig. 7a. Remarkably, nearly all the behavior phenotypes, including depression (by TST, FST, and SPT), anxiety (by OFT and EPMT), and improved spatial learning and memory (by MWM) in TgAPPsweOCN mice (at 6-MO) were all diminished by D + Q treatments (Fig. 7b–i), providing evidence for senescence as a potential pathological mechanism for these behavior changes. Notice that D + Q treatments had little effect on the anxiety-like behavior assessed by LDT (Fig. 7d), implicating additional mechanism(s) underlying this event.
a Schematic diagram of experimental design. 6-MO control (LSL-APPswe, n = 10 males) and TgAPPsweOCN mice were treated with Veh (10%PEG 400) (n = 10 males) or DQ (D 5 mg/kg, Q 50 mg/kg, dissolved in 10% PEG 400, once per two weeks) (n = 9 males), starting at age of 3-MO, and then subjected to indicated behavior tests at 6-MO. b OFT: Representative tracing images and quantifications of the total distance and the center duration time were shown. n.s. not significant, *p < 0.05. c EPMT: Representative tracing images and quantifications of the open arm duration time and entries were shown. *p < 0.05, **p < 0.01, ***p < 0.001. d LDT: Quantifications of the time spent in the light room and the number of transitions into the light room were shown. n.s. not significant, *p < 0.05. e TST, f FST, and g SPT were shown. *p < 0.05, **p < 0.01. h–i MWM: the latency to reach the hidden platform during the training period (h), and representative tracing image and quantification of the time spent in the target quadrant, platform crossing time and swim speed (i) were shown. *p < 0.05, **p < 0.01. One-way ANOVA followed by Tukey post hoc test. All data were presented as mean ± SD.
Moreover, the GFAP+ reactive astrocytes, IBA1+ cells, and SASP-like factors (e.g., Il1b, Tnfa, but not Il10 or Mmp3) in TgAPPsweOCN cortex were attenuated (Supplementary Fig. 12a–c), and the impaired hippocampal DG neurogenesis in TgAPPsweOCN mice was restored (Supplementary Fig. 12d, e) by D + Q treatments. In aggregates, these results suggest that APPswe-induced senescence and SASPs are likely to prompt cortical brain inflammation and glial activation, which may underlie the behavioral phenotypes in 6-MO TgAPPsweOCN mice.
Systemic inflammation in TgAPP swe OCN mice likely due to APPswe-induced OB-senescence and SASPs
To further understand how APPswe-induced OB-senescence and SASPs contribute to the brain pathology and behavior changes in TgAPPsweOCN mice, we speculate that APPswe induced OB-senescence and SASPs contribute to systemic inflammation, which promotes brain inflammation and behavior changes. To test this speculation, we addressed the following questions.
First, are the increased SASPs (such as cytokines and chemokines) in APPswe+ OB progenitors released and traveled through the circulation system of TgAPPsweOCN mice to induce the systemic inflammation? Using multiplexed antibody-based arrays to screen for altered serum/plasma proteins in TgAPPsweOCN mice (~7-MO) compared to their litter-mate control mice (LSL-APPswe), increases in chemokines (CCL2, 5, 9, 11, 22, CXCL1, 2, 10, 13, 16), cytokines (IL1β, 2, 3, 4, 5, 6, 7, 9, 10, 15, 16, 17, 20, 23, 25, 31), and cytokine modulators (HMGB1, MTOR, CDH5) in the serum samples of TgAPPsweOCN mice (Fig. 8a–c), indicating a systemic inflammation. The increases in serum IL1β and IL6 levels in TgAPPsweOCN mice were verified by ELISA analyses (Fig. 8d). Notice that 8 up-regulated proteins (IL1β, 6, 10; CCL2, 5; CXCL1, 10 and MMP3) and 2 down-regulated proteins (VEGFB and PF4) were identified not only by the serum antibody array assay, but also by the RNA-seq analysis of OB progenitors (Fig. 8e), suggesting that many of the serum cytokines come from the APPswe-induced OB-derived SASPs.
a Representative images of serum L-Series label-multiplex antibody arrays of ~7-MO control and TgAPPsweOCN mice. b Volcano plots analysis of a. c Heat map of data in a. n = 4, significant difference was set at p < 0.05. d Elisa assays of serum IL1β and IL6 levels in ~7-MO control and TgAPPsweOCN mice. The data were presented as mean ± SD (n = 4 mice). *p < 0.05 by Student’s t test. e Comparison between this antibody array with secreted factors by RNA-seq of purified Tdtomato+ BMSCs. f Comparison of the changes (upregulated secreted proteins in Tg2576 over control mice) to those detected in TgAPPsweOCN mice.
Second, is APPswe in OCN-Cre+ cells a key contributor to the systemic inflammation? Although APPswe is largely expressed in OB-lineage cells of TgAPPsweOCN mice (Fig. 1), we cannot rule out the potential contribution of APPswe’s weak expression in the hippocampal dDG to systemic inflammation. To this end, we examined the serum inflammatory cytokines and chemokines in mice (LSL-APPswe) injected with AAV-CaMKII-Cre or AAV-GFP into their dDGs; and the Cre-injected mice exhibited similar levels of APPswe/Aβ42 in the hippocampus compared to 12-MO TgAPPsweOcn mice (Supplementary Fig. 8g). Using a small-scale antibody array containing antibodies against multiple SASP-like pro-inflammatory cytokines and chemokines (Supplementary Fig. 13a), little to no change was detected between the serum samples from the Cre and GFP injected mice (Supplementary Fig. 13a, b). These results thus eliminate the possibility of dDG APPswe/Aβ42 contribution to the systematic inflammation, supporting APPswe in OCN-Cre+ OB-lineage cells as a major contributor of systemic inflammation. We also measured serum inflammatory factors in Tg2576 mice, a well-studied AD animal model that expresses APPswe ubiquitously24, using multiplexed antibody-based arrays, and compared the changes (upregulated secreted proteins in Tg2576 over control mice) with TgAPPsweOCN mice. Among 49 upregulated secreted proteins in Tg2576 mice, 31 (~63%) were increased in TgAPPsweOCN mice (Fig. 8f), providing additional support for the view.
Third, is the systemic inflammation results from the APPswe induced-senescence and SASPs? Measuring serum SASP-like cytokines and chemokines in TgAPPsweOCN mice treated with and without D + Q, as illustrated in Fig. 7a, demonstrate that many cytokines (IL1β, 2, 23, 27) and chemokines (CCL2, 11 and CXCL1, 2) were increased in serum samples of TgAPPsweOCN mice treated with Veh, but decreased in the mice with D + Q treatments (Supplementary Fig. 13c, d). Together, these results suggest that the systemic inflammation in TgAPPsweOCN mice is likely in large due to the APPswe-induced OB-senescence and SASPs.
Induction of ER stress-driven OB-senescence by expression of APPswe, but not APPwt or APPlon
To understand how APPswe in OB-lineage cells induces senescence, we re-analyzed the RNA-seq data (APPswe+ vs control OB progenitors) and found that, in addition to the increases in mRNAs of senescence genes, the transcripts of ER stress genes (e.g., Grp78, Atf6, and Hsp90) were elevated in APPswe+ OB-progenitor cells (Fig. 9a, b). The increase in ER stress proteins (e.g, GRP78 and ATF6) were further verified by Western blot (Fig. 9c, d). To investigate the relationship between APPswe-induced ER stress and senescence, we treated APPswe+ OB-progenitors with 4-PBA (4-Phenylbutyric acid), an inhibitor of ER stress58. 4-PBA treatments abolished the increases of the senescence marker proteins P16Ink4a, P53, and SA-β-gal (Fig. 9e–h), suggesting that APPswe likely increases OB-senescence by inducing ER stress.
a Heat map of differentially expressed ER stress or anti-stress related genes identified by RNA-seq in control (OCN-Cre; Ai9) and TgAPPsweOCN; Ai9 Td+ OB-progenitors (detail analysis was described in Methods). b RT-PCR analysis of ER stress-related genes Grp78, Atf6, Hsp90b1, Eif2ak3, Ern1, Hsp90aa1, and Hspa2 and anti-stress related gene Sirt3 gene expression in purified Td+ BMSCs from 6-MO control (OCN-Cre; Ai9) and TgAPPsweOCN; Ai9 mice, *p < 0.05, **p < 0.01, ***p < 0.001, mean ± SD, n = 3, Mann–Whitney U test. c Western blot analysis of indicated protein expression in BMSCs from mice with indicated genotypes (at 6-MO). GAPDH was used as a loading control. d Quantification of data in c, *p < 0.05, **p < 0.01. mean ± SD, n = 4, Student’s t test. e Western blot analysis of indicated protein expression in BMSCs from 6-MO control and TgAPPsweOCN with or without 0.25 mM 4-PBA (4-Phenylbutyric acid) treatment. f Quantification analyses of the data in e, *p < 0.05, n = 3. g SA-β-gal staining of 6-MO control and TgAPPsweOCN BMSCs with vehicle (Veh)(PBS) and 4-PBA treatment, respectively, scale bar, 20 µm. h Quantification of SA-β-gal+ cell densities in g (mean ± SD; n = 5, **p < 0.01, ***p < 0.001). Two-way analysis of variance test was used in f and h.
Notice that ER stress can be induced by the overexpression of membranous proteins59. It thus is necessary to determine if the increased ER stress in APPswe+ cells results from its over expression. To this end, MC3T3 cells (an OB cell line) expressing APPwt-YFP (wild type), APPswe-YFP, and APPlon-YFP were examined. MC3T3 cells expressing APPswe-YFP, but not APPwt-YFP or APPlon-YFP, showed an obvious increase in GRP78 (an ER stress sensor) (Supplementary Fig. 14a, b), indicating a more dramatic effect on ER stress by APPswe-YFP and demonstrating its specificity. Additionally, a more prominent co-localization of GRP78 with APPswe-YFP than those with APPwt-YFP or APPlon-YFP was observed (Supplementary Fig. 14a, c). Moreover, APPswe-YFP had an increased co-localization with EEA1, an early endosome marker, but decreased co-location with GM130, a marker for Trans-Golgi, compared with those of APPwt-YFP or APPlon-YFP (Supplementary Fig. 14d–g). These results demonstrate APPswe’s distinctive cellular features in its increase of GRP78 and its subcellular localizations. Finally, the senescence marker, SA-β-gal, was selectively increased in MC3T3 cells expressing APPswe-YFP, but not APPwt nor APPlon (Supplementary Fig. 14h–i), providing additional support for the specificality of the detrimental effects by APPswe, but not by the overexpression of APPwt or APPlon.
Discussion
Patients with AD often have osteopenia or osteoporosis3,4,5,6,7,8,9,10. The lower bone mineral density is often reported in the earliest clinical stages of AD patients (both men and women) and associated with their brain atrophy and memory decline8. However, it remains unclear if the AD patients carrying the Swedish mutations have osteoporosis-like deficit. Here, using TgAPPsweOCN mouse model that selectively expresses APPswe largely in the OB-lineage cells, we found that APPswe in OB-lineage cells induces senescence and SASPs, which appear to be a key contributor of systemic inflammation, and thus promote anxiety- and depression-like behaviors in TgAPPsweOCN mice. Our studies also suggest that the senescence may be insufficient to induce the cognitive decline detected in 12-MO TgAPPsweOCN mice, which may be associated with a weak expression of APPswe/Aβ42 in the dDG neurons of the hippocampus. These observations, summarized in Fig. 10a, lead to a working hypothesis depicted in Fig. 10b, and opens a box of additional questions for future investigations.
A prerequisite to a better understanding of the mechanisms of TgAPPsweOCN mice’ brain/behavior phenotypes is to reveal where exactly the APPswe is expressed. TgAPPsweOCN mice are generated by crossing OCN-Cre with the LSL-hAPPswe mice, and thus the APPswe expression is controlled not only by the CAG promoter in LSL-hAPPswe mice (for its mRNA expression), and but also by the OCN-Cre dependent removal of LSL (for hAPPswe protein expression)23. Although OCN-Cre mice express Cre largely in OB-lineage cells60, our recent study demonstrates the Cre activity in neurons at the dDG, olfactory bulb, and cerebellum of the brain in OCN-Cre mice29. Our further studies in this paper lead us to conclude that hAPPswe or OCN-Cre is largely expressed in the OB-lineage cells, but weakly expressed in the dDG neurons, in 12-MO TgAPPsweOCN mice (Fig. 1b–g). We thus believe that the cortical brain and behavior phenotypes in 6-MO TgAPPsweOCN mice are likely induced by the APPswe in OB-lineage cells. However, it is possible that the weak APPswe/Aβ42 expression in aged (12-MO) dDG hippocampal neurons contributes to the inflammation phenotypes in the mutant hippocampus and the cognitive decline (Fig. 10b).
How does APPswe in OB-lineage cells induce brain pathology? Several lines of evidence support the hypothesis that APPswe-induced OB-senescence and SASPs may underlie its effects on the brain, particularly the cortex, via systemic inflammation (Fig. 10b). Many SASP-like proteins were induced in cultured APPswe+ OB progenitors and increased in serum samples of TgAPPsweOCN mice (Figs. 5 and 8). Cultured APPswe+ OB progenitors and MC3T3 cells showed increased senescence cells (Fig. 6 and Supplementary Fig. 14h–i)52,61. While the OB-senescence phenotypes were temporally associated with APPswe-induced bone-deficits23, they occurred earlier than brain deficits, in TgAPPsweOCN mice (Fig. 6). The inhibition of senescence in TgAPPsweOCN mice diminished nearly all the brain and behavior phenotypes (Fig. 7 and Supplementary Fig. 12). In line with this hypothesis are the multiple literature reports that demonstrate cellular senescence as tightly linked to skeleton and brain aging and various degenerative diseases, including AD62,63,64,65,66,67, and the use of senolytic drugs to attenuate the disease process has been shown in several animal models of AD68,69.
In terms of the systemic inflammation, while it can be induced by deficits in multiple organs, our results suggest that APPswe -induced senescence and SASPs in OB-lineage cells appear to be a key contributor to this event. Many (31 over 49, ~63%) upregulated SASP-like factors detected in serum samples of TgAPPsweOCN mice were also detected in Tg2576 mice (Fig. 8f). Although APPswe is weakly expressed in the dDG neurons of TgAPPsweOCN mice (Fig. 1), examining the serum inflammatory cytokines and chemokines in mice (LSL-APPswe) injected with AAV-CaMKII-Cre or AAV-GFP into their dDGs showed an increase in APPswe/Aβ42 in the hippocampus of Cre injected mice (Supplementary Fig. 8g), but a comparable level of serum cytokines and chemokines between Cre and GFP injected mice (Supplementary Fig. 13a, b). Treatments with senescence inhibitors (D + Q) abolished nearly all the increased inflammatory cytokines in the serum samples of TgAPPsweOcn mice (Supplementary Fig. 13c, d). These results thus eliminate the possible contribution of the APPswe/Aβ42 at the dDG to systematic inflammation, and support the view.
How does APPswe in OB-lineage cells induce senescence and SASPs? We believe that APPswe-induced ER stress may underlie this process for the following reasons. First, expressing APPwt, APPswe, or APPlon in osteoblastic cell line, MC3T3 cells, results in an increased of β-gal+ SnCs specifically in APPswe+, but not APPwt+ or APPlon+, cells (Supplementary Fig. 14h–i), although APP or Aβ levels were increased in all three types of cells. These results not only suggest the specificity of APPswe in the induction of the senescence, but also implicate Aβ’s insufficiency or independency to this event. Second, APPswe, compared to APPwt or APPlon, exhibited distinctive features in its subcellular localizations and its induction of ER-stress, in addition to senescence (Supplementary Fig. 14a–g), revealing an association between the selective induction of the ER stress and senescence by APPswe, but not APPwt or APPlon, in line with the view that APPswe is processed by β-secretase or BACE1 in Golgi-derived vesicles, and APPwt is cleaved in the endosomes70. Third, both RNA-seq and Western blot analyses showed that APPswe+ OB progenitors have increased expressions of not only senescence associated genes, but also ER stress genes (e.g., Grp78, Atf6, and Hsp90) (Fig. 9a–d); and treatment of APPswe+ OB progenitors with an ER stress inhibitor 4-PBA abolished the increase of senescence marker proteins P16Ink4a, P53, and β-gal+ SnCs (Fig. 9e–h), supporting the view for ER stress as an driver of senescence. Notice that Hashimoto et al. report an absence of ER stress responses in AppNL-G-F (App knock-in mice harboring Swedish mutation) brain59. We thus speculate that this event may be cell type/tissue specific, and OB-lineage cells may be more sensitive to APPswe than neurons in its induction of ER stress.
Are senescence and SASPs induced by osteoblastic APPswe involved in the behavior changes observed in TgAPPsweOCN mice? Our results suggest that they are likely contributors to anxiety and depression, but insufficient to cause cognitive decline. In addition to the temporal association between the increased SASPs and the behavior changes, inhibition of senescence and SASPs by D + Q diminishes nearly all the behavior changes in TgAPPsweOCN mice at 6-MO (Fig. 7). Among the SASPs induced by APPswe, IL-1β is noteworthy, because IL-1β is found to mediate bi-functions in regulating spatial learning and memory71,72,73. Expressing IL-1β in the brain (in particular, the cortex) exhibits enhanced spatial learning and memory in young adult, but not aged, mice74, a similar behavioral phenotype examined in the TgAPPsweOCN mice (Figs. 2g and 4 and Supplementary Fig. 5g). This IL-1β’s function is also in agreement with numerous reports, that IL-1β is upregulated by long term potentiation (LTP) (an event critical for learning and memory)75,76,77. The overexpression of IL-1ra, an endogenous IL-1R antagonist or IL-1R KO (knock-out), blocks spatial memory78,79 as well as LTP73,80. In the light of these reports, we speculate that the osteoblastic APPswe, via increasing IL-1β, a key SASP, may improve hippocampal/cortex-dependent spatial learning and memory function in an age-dependent manner. We are also aware of controversial reports, which claim that IL-1β plays a detrimental role in regulating learning and memory81,82. While IL-1β plays a role in modulating learning and memory, its precise function appears to strongly depend on the site of IL-1β injection/increase, timing, and dosage73,79. Notice that Il1b was increased in the hippocampus but not the cortex of 12-MO TgAPPsweOCN mice (Supplementary Fig. 5g–h); and such IL-1β increase was accompanied by elevated Aβ42 and glial activation in the hippocampus, and cognitive decline behaviors (Supplementary Figs. 1b and 5 and Fig. 4g–i). We thus speculate that the hippocampal inflammation phenotype may be induced by the weak APPswe/Aβ42 expression in dDG hippocampal neurons, which may also impair cognitive function in 12-MO TgAPPsweOCN mice (Fig. 10b). It would be of interest to further test this view in future experiments.
Finally, it is highly possible that complex mechanisms underlie APPswe regulation of brain and behavior phenotypes in TgAPPsweOCN mice. In addition to IL-1β and TNFα, other SASPs and growth factors may also contribute to the brain pathology. In addition to senescence and SASPs, the weak expression of APPswe/Aβ42 in the OCN-Cre+ dDG neurons may be exacerbated by systemic inflammation and be responsible for the hippocampal pathology and cognitive decline in aged (e.g., 12-MO) TgAPPsweOCN mice. It is also noteworthy that while chronic inflammation is believed to be one of the environmental risk factors for AD development30,83, our studies suggest that the chronic systemic inflammation associated with AD patients (either EOAD or LOAD) may be induced by a combination of AD genetic risk gene(s), a primary hit, and environmental risk factors (e.g., aging, infection), a secondary hit, in line with the two-hit hypothesis84. Further investigations that address how chronic inflammation is induced, how it promotes the brain pathology and behavior changes, and what is the function/contribution of APPswe/Aβ42 in dDG neurons to the AD development may gain more insights into the two-hit hypothesis and AD pathogenesis.
Methods
Mice
The LSL-APPswe mice were generated using the pCCALL2 plasmid as described previously23. In brief, the transcription of hAPPswe in LSL-APPswe mice is controlled by the CAG promoter, but its translation is blocked by a loxP-stop-loxP sequence23. Thus, the expression of hAPPswe is controlled by both the CAG promoter and the Cre-dependent removal of LSL. The OCN–Cre mice were kindly provided by Tom Clemens (Johns Hopkins Medical School). OCN-Cre; Ai9 and TgAPPsweOCN; Ai9 mice were generated by crossing Ai9 mice (from the Jackson Laboratory, donated by Dr. Hongkui Zeng, Allen Institute for Brain Science) with OCn-Cre and TgAPPsweOCN mice, respectively. Ai9 mice have a loxP-flanked STOP cassette preventing the translation of a CAG promoter-driven red fluorescent protein variant (tdTomato). Thus, tdTomato is expressed following Cre-mediated recombination. The Tg2576 mice were purchased from Taconic, Hudson, NY, USA, which express human APP695 with Swedish double mutations at KM670/671NL (APPswe) under the control of a hamster prion promoter24. 5xFAD transgenic mice were obtained from The Jackson Laboratory (MMRRC stock #34 840-JAX)25 which express human APP and PSEN1 transgenes with five AD-linked mutations (the Swedish [K670N/M671L], Florida [I716V], and London [V717I] mutation in APP, and the M146L and L286V mutation in PSEN1) under the control of mouse Thy1 promoter. All mouse lines were backcrossed into C57BL/6 background and housed in a room with a 12 h light/dark cycle and ad libitum access to water and rodent chow diet (Harlan Tekled S-2335). Control littermates were used in parallel for each experiment. All experimental procedures were approved by the Institutional Animal Care and Use Committee at Case Western Reserve University (IACUC, 2017–0121), according to the United States National Institutes of Health guidelines.
Antibodies and chemicals
The following primary antibodies were used and purchased as indicated below: Anti-hAPP (6E10, 803001, mouse) and anti-6E10 (Alexa Fluor® 647 anti-β-Amyloid, 1-16 Antibody, cat#803021) from biolegend (San Diego, California, USA); Anti-Amyloid Fibrils OC antibody (AB2286, rabbit) from EMD Millipore (Temecula, California, USA); Anti-DCX (SC-8066, goat) from Santa Cruz Biotech (Santa Cruz, California, USA); Anti-Ctip2 (ab18465, Rat), anti-IBA1 (ab178846, rabbit and ab5076, goat), anti-P16ink4a (ab211542, rabbit), and anti-P53 (ab26, mouse) from Abcam (Cambridge, Massachusetts, USA); Anti-S100β (287004, Guinea pig) from Synaptic System (Göttingen, Germany); Anti-Olig2 (p21954, rabbit), anti-GRP78 (PA1-014A, rabbit) and anti-EEA1 (PA1-063A, rabbit) from Invitrogen (Carlsbad, California, USA); Anti-ATF6 (NBP1-40256, mouse) from Novus biologicals (Centennial, CO, USA); anti-GM130 (610822, mouse) from BD biosciences (San Jose, CA, USA) and Anti-P21 (2947S, rabbit), anti-NEUN) (12943S, rabbit), anti-GFAP (12389S, rabbit), and anti-GAPDH (97166S, mouse) from cell signaling (Danvers, Massachusetts, USA). Secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, Pennsylvania, USA). Dasatinib was from LC Laboratories (Woburn, MA, USA). Quercetin, polyethylene glycol 400, 4-PBA, DMSO, DAPI, and d 5-ethynyl-2’-deoxyuridine (EdU, a modified thymidine analogue that is incorporated into the DNA of dividing cells) were from Sigma Aldrich (St. Louis, MO, USA). All chemicals and reagents used in this study were of analytical grade.
Immunofluorescence staining and image analysis
Immunostaining was performed as described previously29. In brief, mice were anesthetized with isoflurane and were transcardially perfused with PBS (50 mL) followed by 4% (w/v) paraformaldehyde (PFA) in phosphate buffer (PBS) (pH 7.4) (50 ml) to remove intravascular plasma proteins. The dissected brains were post-fixed in 4% PFA at 4 °C overnight. Coronal sections (40 μm) were washed 3 times with PBS (10 min each) and treated with blocking reagent (10% Donkey Serum + 0.5% Triton 100×) for 1 h, then incubated overnight at 4 °C with the primary antibody. Sections were washed 3 times and incubated with corresponding conjugated secondary antibody for 1 h. DAPI was used for nucleus counter staining. Stained sections were imaged by confocal microscope at room temperature. Fluorescent quantification was performed using ZEN software according to the manufacturer’s instructions (Carl Zeiss).
Western blotting
Western blotting was performed as described previously85. Brain tissues and cultured BMSCs were homogenized in modified RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA,) containing 0.5% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF, 1 mM DTT, and protease inhibitor cocktail (Millipore, 539134). Lysates were centrifuged at 10,000 x g for 10 min at 4 °C to remove debris and to obtain homogenates. Samples were resolved by SDS-PAGE and transferred to a nitrocellulose membrane (1620112, Bio-Rad Laboratories). After incubation with 5% milk in TBST (10 mM Tris, 150 mM NaCl, and 0.5% Tween 20, pH 8.0) for 1 h, membranes were immunoblotted with indicated antibodies overnight at 4 °C. Membranes were washed with TBST three times and incubated with a 1:2000 dilution of horseradish peroxidase–conjugated anti–mouse or anti–rabbit antibodies for 1 h. Blots were washed with TBST three times and immunoreactive bands were visualized using the LI-COR Odyssey infrared imaging system. Intensity of immunoreactive bands were quantitated by using ImageJ (NIH).
EdU injection and labeling
Control (LSL-APPswe) and TgAPPsweOCN mice were given four intraperitoneal injections of EdU (50 mg/kg/time, 1 time/4 h) within 12 h. 12 hours after their last injection, mice were euthanized and transcardially perfused first with 50 ml of cold PBS and then with 50 ml of 4% PFA. The dissected brains were post-fixed in 4% PFA at 4 °C overnight. Coronal sections (40 μm) were obtained for staining. Cultured BMSCs were incubated with 10 µM EdU for 2 hours, and then cells were fixed with 4%PFA for 10 min. EdU staining was performed using a Clik-iT EdU imaging kit with Alexa-Fluor 488 (Invitrogen) following the manufacturer’s instructions.
Behavioral tests
Mice (male) at ages of 3-, 6- or 12-MO (month old) were subjected to behavioral studies. Behavioral tests were done blind to genotypes or treatments. For all behavioral experiments, mice were transferred to the testing room 4 h before any test to acclimate to the environment. All behavioral instruments were cleaned with 70% ethanol prior to each trial.
Open field test (OFT), Elevated plus maze test (EPMT), and Light/dark transition test (LDT) were performed as described previously29. In brief, for OFT, each mouse was placed in a chamber (L × W × H = 50 × 50 × 20 cm) and its movement was monitored for 10 min using an overhead camera. Light intensity was about 150 lux. The video was analyzed by a tracking software (Etho Vision, Noldus). The total distance and center (25 × 25 cm) duration time were quantified. For elevated plus maze test (EPMT), the EPM was placed 50 cm above the ground. Each mouse was initially placed in the center square facing one of the open arms (L × W = 60 × 5 cm). Light intensity was about 100 lux. Mice movement was recorded for 5 min using an overhead camera and tracking software (Etho Vision, Noldus). The time spent in the open arms and the number of open arm entries were quantified. For light/dark transition test (LDT), mouse was firstly placed in the dark compartment, overhead camera was turned on, and the door between lit and dark chambers was opened. Light intensity was about 200 lux in the lit chamber. 10 min of movement was recorded using a tracking software ((Etho Vision, Noldus). The time spent in the lit chamber and the number of transitions were quantified.
The tail suspension test (TST), forced swimming test (FST), and sucrose preference test (SPT) were performed as described previously86. For the TST and FST, the last 4-min of a 6-min test were analyzed, and the immobility time was measured directly. The sucrose preference test was carried out using a two-bottle choice procedure. Single housed mice were habituated to drink 2% (wt/vol) sucrose solution (dissolved in water) for 3 days, then mice were given access to the two pre-weighed bottles, one containing water and the other containing 2% sucrose solution. Bottle positions were changed every day and water and sucrose solution consumption was assessed daily for 4 days. The consuming ratio of sucrose over total solution consumed was used for measuring the sucrose preference.
The Morris water maze (MWM) was performed as previously described87. Specifically, a 120 cm pool and 10 cm platform were used for water maze and nontoxic bright white gel (Soft Gel Paste Food Color, AmeriColor) was added to the water to make the surface opaque and to hide the escape platform (1 cm below the surface). Mice were trained for 5 days, four trials per day with 20 min interval between trials and 60 s per trial to locate the hidden platform. Eight spatial cues were placed on the pool wall, visible for mice to find the hidden platform. On the 6th day, the platform was removed, and mice were placed into the pool at a new starting position. The time spent in each platform quadrant and the number of platform-crossing within 60 s were analyzed. The swim speed and the amount of time spent in each quadrant were quantified using the video tracking system (Noldus). The investigators were blind to genotype during data acquisition and analysis.
The Novel Object Recognition Task (NOR) was based on a previous published procedure88. It consists of a habituation phase followed by a testing phase. During the habituation phase, each mouse was allowed to freely explore the empty arena over two days. On the third day, the testing phase begun. Habituation consisted of one ten-minute session administered one per day. The testing phase consisted of a (1) familiarization trial followed by a (2) test trial. During the familiarization trial, a single mouse was placed in the arena containing two identical objects and released against the center of the opposite wall with its back to the objects. This was done to prevent coercion to explore the objects. Object interaction is defined as entrance into the object-containing zone resulting in direct or nearly direct object contact with the nose or whiskers. The test trials were administered after delays of 1-hour post-familiarization. The test trial was administered in the aforementioned way except that one sample object from the familiarization trial and one novel object were presented. During the test trials, time spent with novel object per total time with both objects as the novel object preference was quantified.
AAV virus injection
AAV9-CamkII-GFP (105541-AAV9) and AAV9-CamkII-Cre (105551-AAV9) were purchased from Addgene. Virus injection was performed as described previously29. In brief, male LSL-APPswe mice (4-month-old) were anesthetized with Ketamine/Xylazine (HENRY SCHEIN #056344), and the head was fixed in a stereotaxic device (David Kopf Instruments). After the antiseptic treatment, the skull was exposed and cleaned using 1% H2O2. Holes were drilled into the skull and viruses were bilaterally injected into DG at the coordinates relative to bregma: caudal: −2.06 mm; lateral:±1.3 mm; ventral: −1.75 mm. After injection, the needle was left in place for 5 min to allow for diffusion of injected viruses before being slowly withdrawn. For the following 5 days, mice were daily injected with Meloxicam to reduce pain. Injection locations were validated in each mouse after experiments.
In vitro primary OB-progenitor (BMSCs) cultures
OB-progenitor (BMSCs) culture was carried out following a standard protocol as described previously28,85. In brief, the whole bone marrow cells flushed out from long bones of mice with DMEM were filtered through a 70-mm filter mesh, washed, re-suspended, and then plated in 100-mm dishes with growth medium (DMEM plus 10% FBS), which were incubated at 37°C with 5% CO2. 3 days later, the non-adherent cells were removed. The attached bone marrow cells were cultured with the growth medium for 7 days. These cells were passaged and cultured for another 3–6 days with the same growth medium. These cells, so called BMSCs, were used for Western blot, RT-PCR, and SA-β-gal staining.
Flow cytometry analysis
Flow cytometry analysis was done as previously described28. BMSCs were flushed from femurs and tibias of 6-MO OCN-Cre; Ai9 and TgAPPsweOCN; Ai9 mice, the attached bone marrow cells were cultured with the growth medium for 7 days. These cells were passaged and cultured for another 3 days with the same growth medium. Then cell media were removed from culture dishes and cells were rinsed with PBS. Trypsin solution was added to incubate at 37 °C for 2 min. The detached adherent cells were centrifuged, and the pellet cells were washed with 1 ml cold PBS, and finally resuspended in 0.5 ml PBS with 1% FBS for flow cytometry analysis. Flow cytometric analysis was performed by use of a flow cytometer in CWRU core facility. Acquisition and analysis were performed by using FACSDiva 8.0.1 software (BD).
Generation of plasmids of APPWT-YFP, APPswe-YFP, and APPlon-YFP
YFP-APPswe mutation (K670N/M671L, AAG ATG - AAC TTG) and YFP-APP-London point mutation (V717I, GTC - ATC) from the YFP-APPWT construct by using the Q5 Site-Directed Mutagenesis Kit (E0554S, New England Biolabs, Inc). The primers ‘CTGAAGTGAACTTGGATGCAGAATTCCGACATG’ and ‘AGATCTCCTCCGTCTTGATATTTG’ were used to generate the K670N/M671L mutation, and the primers ‘CATCACCTTGGTGATGCTGAAG’ and ‘ATGATCACTGTCGCTATGACAAC’ were used to generate the V717I point mutation.
MC3T3 cell culture and transfections of YFP, APPWT-YFP, APPswe-YFP, and APPlon-YFP plasmids
MC3T3-E1 cells were grown in DMEM containing 10% (vol/vol) FBS, and 50 units/ml penicillin and streptomycin. Cells plated at 1 × 104/well onto 12-wells coverslips the day before transfection. Cells were transfected with control-YFP Vector, APPWT-YFP, APPswe-YFP, and APPlon-YFP by Lipofectamine 3000 (Invitrogen). Forty-eight hours after transfection, cells were subjected to SA-β-gal staining and immunofluorescence staining.
SA-β -gal staining
Cultured OB progenitors and MC3T3 cells SA-β-gal staining was performed as previously reported85. SA-β-gal staining was performed using a SA-β-gal staining kit (Cell Signaling, #9860) according to the manufacturer’s instructions.
Elisa assay for IL1β, IL6, human Aβ40, and human Aβ42
Blood samples were collected, allowed to clot for 30 min, and centrifuged for 10 min at 3000 rpm. Serum was frozen and aliquot at −80 °C until use. Serum IL1β was measured with Mouse IL-1 beta ELISA Kit (KE10003, Proteintech), following the manufacturers’ instruction. Serum IL-6 was measured with mouse IL-6 ELISA kit (550950, BD Biosciences), following the manufacturers’ instruction. Serum, Brain and BMSCs homogenization was obtained for human Aβ40/42 Elisa assay. Brain tissues were homogenized as previously described87. Human Aβ40 and Aβ42 level in serum, brain (300 µg in total protein) and BMSCs (50 µg in total protein) homogenates were measured by the Aβ40 human ELISA kit (Invitrogen, catalog #KHB3481) and the Aβ42 human ELISA kit (Millipore, catalog #EZHS42), respectively. Their concentrations were determined by comparing readings against their standard curves.
L-Series label-multiplex antibody arrays
Mice blood samples were collected and allowed to clot for 30 min at room temperature and centrifuged for 15 min at 3000 rpm. Serum was frozen and aliquot at −80 °C until use. The antibody arrays were performed using an L-Series Glass Slide antibody arrays kit (AAM-SERV-LG, Raybiotech, USA) according to the manufacturer’s instructions. In brief, the serum was dialyzed before the biotin-labeling step. The primary amine of the proteins in the sample was biotinylated, followed by dialysis to remove free biotin. The newly biotinylated sample was added onto the glass slide and incubated at room temperature. After incubation with Fluorescent Dye-Strepavidin, the signals were visualized by fluorescence.
Mouse cytokine array
Serum samples were collected as described above. Cytokines were measured with Mouse Cytokine Array Panel A (ARY006, R&D Systems). In Brief, the serum was mixed with a cocktail of biotinylated detection antibodies. The sample/antibody mixture was then incubated with the Mouse Cytokine Array membrane. Any cytokine/detection antibody complex present was bound by its cognate immobilized capture antibody on the membrane. Following a wash to remove unbound material, streptavidin–horseradish peroxidase and chemiluminescent detection reagents were added sequentially. Light was produced at each spot in proportion to the amount of cytokine bound.
RNA isolation and qPCR
Total RNA was isolated from brain tissues and BMSCs by using the RNeasy Mini Kit (QIAGEN, Cat No. 74104), and purified RNA (1–5 μg) was used for cDNA synthesis with the Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific, # K1621). cDNA products were subjected for subsequent quantitative PCR (qPCR) using the QuantiFast SYBR Green PCR Kit (204057; QIAGEN) with a qPCR System (StepOne Plus). Primers used were as follows: hAPP, 5′-GCCCTTCTCGTTCCTGAC-3′ and 5′-TCGCAAACATCCATCCTC-3′; OCN-Cre, 5′-CAAATAGCCCTGGCAGATTC-3′ and 5′-TGATACAAGGGACATCTTCC-3′; mAPP, 5′-TCCGTGTGATCTACGAGCGCAT-3′ and 5′-GCCAAGACATCGTCGGAGTAGT-3′; Vps35, 5′-GACTTCGCTGATGAACAGAGCC-3′ and 5′-CAGTGTGAAGCGAATCCGCTGA-3′; Trem2, 5′-CTACCAGTGTCAGAGTCTCCGA-3′ and 5′-CCTCGAAACTCGATGACTCCTC-3′; Apoe, 5′-GAACCGCTTCTGGGATTACCTG-3′ and 5′-GCCTTTACTTCCGTCATAGTGTC-3′; Sorl1, 5′-GAACACCTGTCTCCGAAACCAG-3′ and 5′-CGGAACTGAGTGTCTGCATCAC-3′; Ptk2b, 5′-CTGGAGAGCATCAACTGTGTGC-3′ and 5′-GATGGGTAGACGTGTCACAGAG-3′; Msx2, 5′-AAGACGGAGCACCGTGGATACA-3′ and 5′-CGGTTGGTCTTGTGTTTCCTCAG-3′; Runx2, 5′- CCTGAACTCTGCACCAAGTCCT-3′ and 5′- TCATCTGGCTCAGATAGGAGGG-3′; Sp7 (Osterix), 5′- GGCTTTTCTGCGGCAAGAGGTT-3′ and 5′- CGCTGATGTTTGCTCAAGTGGTC-3′; Nfatc1, 5′- GGTGCCTTTTGCGAGCAGTATC-3′ and 5′- CGTATGGACCAGAATGTGACGG-3′; Col1a1, 5′- CCTCAGGGTATTGCTGGACAAC-3′ and 5′- CAGAAGGACCTTGTTTGCCAGG-3′; Spp1 (Osteopontin), 5′- GCTTGGCTTATGGACTGAGGTC-3′ and 5′- CCTTAGACTCACCGCTCTTCATG-3′; Bglap (Osteocalcin), 5′- GCAATAAGGTAGTGAACAGACTCC-3′ and 5′- CCATAGATGCGTTTGTAGGCGG-3′; Sparc (Osteonectin), 5′- CACCTGGACTACATCGGACCAT-3′ and 5′- CTGCTTCTCAGTGAGGAGGTTG-3′; Bmp2, 5′- TGTGAGGATTAGCAGGTCTTTGC-3′ and 5′- CTCGTTTGTGGAGCGGATGT-3′; Lrp4, 5′- GTGTGGCAGAACCTTGACAGTC-3′ and 5′- TACGGTCTGAGCCATCCATTCC-3′; Ctnnb1(beta-catenin), 5′- GTTCGCCTTCATTATGGACTGCC-3′ and 5′- ATAGCACCCTGTTCCCGCAAAG-3′; Tnfsf11 (Rankl), 5′-ATCCCATCGGGTTCCCATAA-3′ and 5′-TCCGTTGCTTAACGTCATGTTAG-3′; Tnfrsf11b (Opg), 5′-GGCCTGATGTATGCCCTCAA-3′ and 5′-GTGCAGGAACCTCATGGTCTTC-3′; Alpl, 5′- CCAGAAAGACACCTTGACTGTGG-3′ and 5′- TCTTGTCCGTGTCGCTCACCAT-3′; Il1b, 5′- TGGACCTTCCAGGATGAGGACA-3′ and 5′- GTTCATCTCGGAGCCTGTAGTG-3′; Il6, 5′- CTTGGGACTGATGCTGGTG-3′ and 5′- TTGGGAGTGGTATCCTCTGTGA-3′; Il10, 5′- CGGGAAGACAATAACTGCACCC-3′ and 5′- CGGTTAGCAGTATGTTGTCCAGC-3′; Tnfa, 5′- GGCGGTGCCTATGTCTCA-3′ and 5′- CCTCCACTTGGTGGTTTGT-3′; Cxcl1, 5′- TCCAGAGCTTGAAGGTGTTGCC-3′ and 5′- AACCAAGGGAGCTTCAGGGTCA-3′; Ccl5, 5′- ACCACTCCCTGCTGCTTT-3′ and 5′- ACACTTGGCGGTTCCTTC-3′; Tgfb1, 5′-ACCGCAACAACGCCATCT-3′ and 5′-GGGCACTGCTTCCCGAAT-3′; Csf2, 5′- AACCTCCTGGATGACATGCCTG-3′ and 5′- AAATTGCCCCGTAGACCCTGCT-3′; Mmp3, 5′- CTCTGGAACCTGAGACATCACC-3′ and 5′- AGGAGTCCTGAGAGATTTGCGC-3′; Grp78, 5’-TGTCTTCTCAGCATCAAGCAAGG-3’ and 5’-CCAACACTTCCTGGACAGGCTT-3’; Atf6, 5’-GTCCAAAGCGAAGAGCTGTCTG-3’ and 5’-AGAGATGCCTCCTCTGATTGGC-3’; Hsp90b1, 5’-GTTTCCCGTGAGACTCTTCAGC-3’ and 5’-ATTCGTGCCGAACTCCTTCCAG-3’; Eif2ak3, 5’-CCGATGTCAGTGACAACAGCTG-3’ and 5’-AAGACAACGCCAAAGCCACCAC-3’; Ern1, 5’-GGCTACCATTATCCTGAGCACC-3’ and 5’-CTCCTTCTGGAACTGTTGGTGC-3’; Hsp90aa1, 5’-GCTTTCAGAGCTGTTGCGGTAC-3’ and 5’-AAAGGCGGAGTTAGCAACCTGG-3’; Hspa2, 5’-GCACCTTCGATGTGTCCATCCT-3’ and 5’-TGGCTGACCATACGGTTGTCGA-3’; Sirt3, 5’-GCTACATGCACGGTCTGTCGAA-3’ and 5’-CAATGTCGGGTTTCACAACGCC-3’; GAPDH, 5′-AAGGTCATCCCAGAGCTGAA-3′ and 5′-CTGCTTCACCACCTTCTTGA-3′. Each sample was repeated at least 3 times, and the mRNA level was normalized to GAPDH using the 2-△△Ct method.
Bulk RNA-sequencing
Total RNAs were extracted from purified Td+ OB-progenitor cells from OCN-Cre; Ai9 and TgAPPsweOCN; Ai9 mice by flow cytometer. RNA Integrity Number (RIN) was accessed for every sample, and the samples were considered qualified with RIN > 2. These RNA samples were then subjected to RNA-seq analyses by BGI America (Cambridge, MA) using the DNBseq platform. Firstly, we removed the reads mapped to rRNA and obtained the raw data with 52.47 Mb reads. After filtering low-quality, adaptor-polluted and high content of unknown base reads in the sequencing reads, 51.9 Mb clean reads were obtained per sample on average. Then clean reads were mapped to reference genome using HISAT2. On average 92.91% reads were mapped and the uniformity of the mapping result for each sample suggests that the samples were comparable. Comparisons to RNAseq were normalized to fpkm values. DEseq2 was used and the PossionDis algorithms detected the differential expression genes (DEGs). The Benjamini and Hochberg (BH) correction was applied to adjust p-value. DEGs were determined with adj.P.value ≤ 0.05 and |Log2 fold change | ≥ 1. Normalized RNA-seq data were provided in Supplementary Data 1. Heatmap was generated by TBtools software. Gene expression profiles were Z-transferred. Secreted protein database was obtained from http://proteomics.ysu.edu/secretomes/animal/index.php. Gene Ontology (GO) enrichment analysis was performed by GO database (http://www.geneontology.org/). GO terms with p-value ≤ 0.05 were defined as significantly enriched.
Statistics and reproducibility
All data were expressed as means ± SD. For in vivo studies, three to ten male mice per genotype per assay were used. For in vitro cell biological and biochemical studies, each experiment was repeated at least three times. Statistical analyses were performed using GraphPad Prism 7.0. Mann–Whitney U test or unpaired Student’s t test was used to compare data from two groups. For multiple comparisons of three or more groups of samples, ANOVA was used. The significance level was set at P < 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Source data of figures are provided in Supplementary Data 1. Sequencing data that support the findings in this study have been assigned Gene Expression Omnibus accession number GSE186827. The uncropped Western blots are provided in Supplementary Figs. 15–22. The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Leng, F. & Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: where do we go from here? Nat. Rev. Neurol. 17, 157–172 (2020).
Wang, J., Gu, B. J., Masters, C. L. & Wang, Y. J. A systemic view of Alzheimer disease—insights from amyloid-beta metabolism beyond the brain. Nat. Rev. Neurol. 13, 612–623 (2017).
Basgoz, B., Ince, S., Safer, U., Naharci, M. I. & Tasci, I. Low bone density and osteoporosis among older adults with Alzheimer’s disease, vascular dementia, and mixed dementia: a cross-sectional study with prospective enrollment. Turkish J. Phys. Med. Rehabil. 66, 193–200 (2020).
Mjoberg, B., Hellquist, E., Mallmin, H. & Lindh, U. Aluminum, Alzheimer’s disease and bone fragility. Acta Orthopaedica Scandinavica 68, 511–514 (1997).
Frame, G., Bretland, K. A. & Dengler-Crish, C. M. Mechanistic complexities of bone loss in Alzheimer’s disease: a review. Connect. Tissue Res. 61, 4–18 (2020).
Zhou, R., Zhou, H., Rui, L. & Xu, J. Bone loss and osteoporosis are associated with conversion from mild cognitive impairment to Alzheimer’s disease. Curr. Alzheimer Res. 11, 706–713 (2014).
O’Mahony, D. et al. Bone aluminium content in Alzheimer’s disease. Dementia 6, 69–72 (1995).
Loskutova, N., Honea, R. A., Vidoni, E. D., Brooks, W. M. & Burns, J. M. Bone density and brain atrophy in early Alzheimer’s disease. J. Alzheimer’s Dis. 18, 777–785 (2009).
Zhou, R., Deng, J., Zhang, M., Zhou, H. D. & Wang, Y. J. Association between bone mineral density and the risk of Alzheimer’s disease. J. Alzheimers Dis. 24, 101–108 (2011).
Sinha, U. K., Saadat, D., Linthicum, F. H. Jr., Hollen, K. M. & Miller, C. A. Temporal bone findings in Alzheimer’s disease. Laryngoscope 106, 1–5 (1996).
Paloneva, J. et al. DAP12/TREM2 deficiency results in impaired osteoclast differentiation and osteoporotic features. J. Exp. Med. 198, 669–675 (2003).
Takahashi, K., Rochford, C. D. & Neumann, H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 201, 647–657 (2005).
Yeh, F. L., Hansen, D. V. & Sheng, M. TREM2, microglia, and neurodegenerative diseases. Trends Mol. Med. 23, 512–533 (2017).
Otero, K. et al. TREM2 and beta-catenin regulate bone homeostasis by controlling the rate of osteoclastogenesis. J. Immunol. 188, 2612–2621 (2012).
Ulland, T. K. & Colonna, M. TREM2 - a key player in microglial biology and Alzheimer disease.Nat. Rev. Neurol. 14, 667–675 (2018).
Belloy, M. E., Napolioni, V. & Greicius, M. D. A quarter century of APOE and Alzheimer’s disease: progress to date and the path forward. Neuron 101, 820–838 (2019).
Zajickova, K., Zofkova, I., Hill, M., Horinek, A. & Novakova, A. Apolipoprotein E 4 allele is associated with low bone density in postmenopausal women. J. Endocrinol. Investig. 26, 312–315 (2003).
Peter, I. et al. Associations of APOE gene polymorphisms with bone mineral density and fracture risk: a meta-analysis. Osteoporos. Int. 22, 1199–1209 (2011).
Hardy, J. A hundred years of Alzheimer’s disease research. Neuron 52, 3–13 (2006).
Hardy, J. Has the amyloid cascade hypothesis for Alzheimer’s disease been proved? Curr. Alzheimer Res. 3, 71–73 (2006).
Hardy, J. Alzheimer’s disease: the amyloid cascade hypothesis: an update and reappraisal. J. Alzheimers Dis. 9, 151–153 (2006).
Cui, S. et al. APPswe/Abeta regulation of osteoclast activation and RAGE expression in an age-dependent manner. J. Bone Min. Res. 26, 1084–1098 (2011).
Xia, W. F. et al. Swedish mutant APP suppresses osteoblast differentiation and causes osteoporotic deficit, which are ameliorated by N-acetyl-L-cysteine. J. Bone Min. Res. 28, 2122–2135 (2013).
Hsiao, K. et al. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science 274, 99–102 (1996).
Oakley, H. et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006).
Pan, J. X. et al. APP promotes osteoblast survival and bone formation by regulating mitochondrial function and preventing oxidative stress. Cell Death Dis. 9, 1077 (2018).
Zhang, J. & Link, D. C. Targeting of mesenchymal stromal cells by Cre-recombinase transgenes commonly used to target osteoblast lineage cells. J. Bone Miner. Res. 31, 2001–2007 (2016).
Pan, J. X. et al. YAP promotes osteogenesis and suppresses adipogenic differentiation by regulating beta-catenin signaling. Bone Res. 6, 18 (2018).
Sun, D. et al. Critical roles of embryonic born dorsal dentate granule neurons for activity-dependent increases in BDNF, adult hippocampal neurogenesis, and antianxiety-like behaviors. Biol. Psychiatry 89, 600–614 (2020).
Akiyama, H. et al. Inflammation and Alzheimer’s disease. Neurobiol. Aging 21, 383–421 (2000).
Fakhoury, M. Microglia and astrocytes in Alzheimer’s Disease: implications for therapy. Curr. Neuropharmacol. 16, 508–518 (2018).
Wirths, O. Altered neurogenesis in mouse models of Alzheimer disease. Neurogenesis 4, e1327002 (2017).
Miller, A. H. & Raison, C. L. The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nat. Rev. Immunol. 16, 22–34 (2016).
Singhal, G. & Baune, B. T. Microglia: an interface between the loss of neuroplasticity and depression. Front. Cell. Neurosci. 11, 270 (2017).
Hill, A. S., Sahay, A. & Hen, R. Increasing adult hippocampal neurogenesis is sufficient to reduce anxiety and depression-like behaviors. Neuropsychopharmacology 40, 2368–2378 (2015).
Revest, J. M. et al. Adult hippocampal neurogenesis is involved in anxiety-related behaviors. Mol. Psychiatry 14, 959–967 (2009).
Hanson, N. D., Owens, M. J. & Nemeroff, C. B. Depression, antidepressants, and neurogenesis: a critical reappraisal. Neuropsychopharmacology 36, 2589–2602 (2011).
Rossi, S. et al. Neuroinflammation drives anxiety and depression in relapsing-remitting multiple sclerosis. Neurology 89, 1338–1347 (2017).
Bailey, K. R. & Crawley, J. N. in Methods of Behavior Analysis in Neuroscience (eds. nd & Buccafusco, J. J.) (Boca Raton, 2009).
Belovicova, K., Bogi, E., Csatlosova, K. & Dubovicky, M. Animal tests for anxiety-like and depression-like behavior in rats. Interdiscip. Toxicol. 10, 40–43 (2017).
Nie, L. et al. Melatonin ameliorates anxiety and depression-like behaviors and modulates proteomic changes in triple transgenic mice of Alzheimer’s disease. BioFactors 43, 593–611 (2017).
Nyarko, J. N. K., Quartey, M. O., Baker, G. B. & Mousseau, D. D. Can animal models inform on the relationship between depression and Alzheimer disease? Can. J. Psychiatry Rev. Canadienne de. Psychiatr. 64, 18–29 (2019).
Espana, J. et al. Intraneuronal beta-amyloid accumulation in the amygdala enhances fear and anxiety in Alzheimer’s disease transgenic mice. Biol. Psychiatry 67, 513–521 (2010).
Cerejeira, J., Lagarto, L. & Mukaetova-Ladinska, E. B. Behavioral and psychological symptoms of dementia. Front. Neurol. 3, 73 (2012).
Starkstein, S. E., Jorge, R., Mizrahi, R. & Robinson, R. G. The construct of minor and major depression in Alzheimer’s disease. Am. J. Psychiatry 162, 2086–2093 (2005).
Ismail, Z. et al. Neuropsychiatric symptoms as early manifestations of emergent dementia: provisional diagnostic criteria for mild behavioral impairment. Alzheimer’s Dement. 12, 195–202 (2016).
Chapman, P. F. et al. Impaired synaptic plasticity and learning in aged amyloid precursor protein transgenic mice. Nat. Neurosci. 2, 271–276 (1999).
Xia, D. et al. Presenilin-1 knockin mice reveal loss-of-function mechanism for familial Alzheimer’s disease. Neuron 85, 967–981 (2015).
Vorhees, C. V. & Williams, M. T. Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat. Protoc. 1, 848–858 (2006).
Antunes, M. & Biala, G. The novel object recognition memory: neurobiology, test procedure, and its modifications. Cogn. Process. 13, 93–110 (2012).
Reger, M. L., Hovda, D. A. & Giza, C. C. Ontogeny of rat recognition memory measured by the novel object recognition task. Dev. Psychobiol. 51, 672–678 (2009).
Young, A. R. & Narita, M. SASP reflects senescence. EMBO Rep. 10, 228–230 (2009).
Martinez, D. E. et al. SASP, a Senescence-Associated Subtilisin Protease, is involved in reproductive development and determination of silique number in Arabidopsis. J. Exp. Bot. 66, 161–174 (2015).
Mirzayans, R., Andrais, B., Scott, A., Paterson, M. C. & Murray, D. Single-cell analysis of p16(INK4a) and p21(WAF1) expression suggests distinct mechanisms of senescence in normal human and Li-Fraumeni Syndrome fibroblasts. J. Cell. Physiol. 223, 57–67 (2010).
Kang, C. et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 349, aaa5612 (2015).
Kirkland, J. L. & Tchkonia, T. Senolytic drugs: from discovery to translation. J. Intern. Med. 288, 518–536 (2020).
Zhu, Y. et al. The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell 14, 644–658 (2015).
Pao, H. P. et al. Suppression of endoplasmic reticulum stress by 4-PBA protects against hyperoxia-induced acute lung injury via up-regulating claudin-4 expression. Front. Immunol. 12, 674316 (2021).
Hashimoto, S. et al. Endoplasmic reticulum stress responses in mouse models of Alzheimer’s disease: overexpression paradigm versus knockin paradigm. J. Biol. Chem. 293, 3118–3125 (2018).
Zhang, M. et al. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J. Biol. Chem. 277, 44005–44012 (2002).
McCormick, A. & Campisi, J. Cellular aging and senescence. Curr. Opin. Cell Biol. 3, 230–234 (1991).
Raskin, N. & Ehrenberg, R. Senescence, senility, and Alzheimer’s disease. Am. J. Psychiatry 113, 133–137 (1956).
Boccardi, V., Pelini, L., Ercolani, S., Ruggiero, C. & Mecocci, P. From cellular senescence to Alzheimer’s disease: the role of telomere shortening. Ageing Res Rev. 22, 1–8 (2015).
Charlesworth, B. Evolution of senescence: Alzheimer’s disease and evolution. Curr. Biol. 6, 20–22 (1996).
Han, X., Zhang, T., Liu, H., Mi, Y. & Gou, X. Astrocyte senescence and Alzheimer’s disease: a review. Front. Aging Neurosci. 12, 148 (2020).
Bryant, A. G. et al. Cerebrovascular senescence is associated with Tau pathology in Alzheimer’s disease. Front. Neurol. 11, 575953 (2020).
Farr, J. N. et al. Targeting cellular senescence prevents age-related bone loss in mice. Nat. Med. 23, 1072–1079 (2017).
Musi, N. et al. Tau protein aggregation is associated with cellular senescence in the brain. Aging Cell 17, e12840 (2018).
Zhang, P. et al. Senolytic therapy alleviates Abeta-associated oligodendrocyte progenitor cell senescence and cognitive deficits in an Alzheimer’s disease model. Nat. Neurosci. 22, 719–728 (2019).
Haass, C. et al. The Swedish mutation causes early-onset Alzheimer’s disease by beta-secretase cleavage within the secretory pathway. Nat. Med. 1, 1291–1296 (1995).
Huang, Z. B. & Sheng, G. Q. Interleukin-1beta with learning and memory. Neurosci. Bull. 26, 455–468 (2010).
Donzis, E. J. & Tronson, N. C. Modulation of learning and memory by cytokines: signaling mechanisms and long term consequences. Neurobiol. Learn. Mem. 115, 68–77 (2014).
Goshen, I. et al. A dual role for interleukin-1 in hippocampal-dependent memory processes. Psychoneuroendocrinology 32, 1106–1115 (2007).
Takemiya, T., Fumizawa, K., Yamagata, K., Iwakura, Y. & Kawakami, M. Brain Interleukin-1 facilitates learning of a water maze spatial memory task in young mice. Front. Behav. Neurosci. 11, 202 (2017).
Balschun, D. et al. Hippocampal interleukin-1 beta gene expression during long-term potentiation decays with age. Ann. N. Y. Acad. Sci. 992, 1–8 (2003).
del Rey, A., Balschun, D., Wetzel, W., Randolf, A. & Besedovsky, H. O. A cytokine network involving brain-borne IL-1beta, IL-1ra, IL-18, IL-6, and TNFalpha operates during long-term potentiation and learning. Brain, Behav., Immun. 33, 15–23 (2013).
Schneider, H. et al. A neuromodulatory role of interleukin-1beta in the hippocampus. Proc. Natl Acad. Sci. USA 95, 7778–7783 (1998).
Spulber, S. et al. Impaired long term memory consolidation in transgenic mice overexpressing the human soluble form of IL-1ra in the brain. J. Neuroimmunol. 208, 46–53 (2009).
Yirmiya, R., Winocur, G. & Goshen, I. Brain interleukin-1 is involved in spatial memory and passive avoidance conditioning. Neurobiol. Learn. Mem. 78, 379–389 (2002).
Ross, F. M., Allan, S. M., Rothwell, N. J. & Verkhratsky, A. A dual role for interleukin-1 in LTP in mouse hippocampal slices. J. Neuroimmunol. 144, 61–67 (2003).
Moore, A. H., Wu, M., Shaftel, S. S., Graham, K. A. & O’Banion, M. K. Sustained expression of interleukin-1beta in mouse hippocampus impairs spatial memory. Neuroscience 164, 1484–1495 (2009).
Loscher, C. E., Mills, K. H. & Lynch, M. A. Interleukin-1 receptor antagonist exerts agonist activity in the hippocampus independent of the interleukin-1 type I receptor. J. Neuroimmunol. 137, 117–124 (2003).
Kinney, J. W. et al. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimer’s Dement. 4, 575–590 (2018).
Zhu, X., Raina, A. K., Perry, G. & Smith, M. A. Alzheimer’s disease: the two-hit hypothesis. Lancet Neurol. 3, 219–226 (2004).
Xiong, L. et al. Linking skeletal muscle aging with osteoporosis by lamin A/C deficiency. PLoS Biol. 18, e3000731 (2020).
Sun, D. et al. Neogenin, a regulator of adult hippocampal neurogenesis, prevents depressive-like behavior. Cell Death Dis. 9, 8 (2018).
Zhang, H. et al. A role of Low-Density Lipoprotein Receptor-Related Protein 4 (LRP4) in astrocytic abeta clearance. J. Neurosci. 40, 5347–5361 (2020).
Ennaceur, A. & Delacour, J. A new one-trial test for neurobiological studies of memory in rats. 1: behavioral data. Behav. Brain Res. 31, 47–59 (1988).
Acknowledgements
We thank members in MeiXiong laboratories for helpful discussions, suggestions, and technical help. This study is supported in part by the National Institutes of Health (AG045781, AG051773, and AG051510 to W.-C.X.) and the Department of Veterans Administration (to W.-C.X). We also thank Meisel Family and InMotion at Cleveland, Ohio.
Author information
Authors and Affiliations
Contributions
J.P. and W.-C.X. designed the project and wrote the manuscript. J.P. and D.S. performed behavioral tests, virus injection, immunostaining, Western blot, and data analysis. D.L. performed MWM tests and data quantification. X.R. performed RNA-seq data analysis. L.X. generated the plasmids encoding APPlon-YFP and APPswe-YFP and did transfection of MC3T3 cells and data analysis; H.G. did L-Series label-multiplex antibody arrays. L.Y. helped with virus injection and data analysis. Y.L. performed mice genotyping and kept mouse lines. C.J. performed BMSCs culture. W.-C.X. helped data analysis, interpretation, and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Martina Rauner and Karli Montague-Cardoso. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pan, JX., Sun, D., Lee, D. et al. Osteoblastic Swedish mutant APP expedites brain deficits by inducing endoplasmic reticulum stress-driven senescence. Commun Biol 4, 1326 (2021). https://doi.org/10.1038/s42003-021-02843-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-021-02843-2
This article is cited by
-
Attenuation of Alzheimer’s brain pathology in 5XFAD mice by PTH1-34, a peptide of parathyroid hormone
Alzheimer's Research & Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.