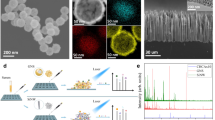
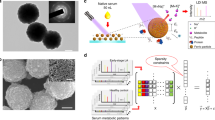
Abstract
Over a billion people across the world experience a high rate of missed disease diagnosis, an issue that highlights the need for diagnostic tools showing increased accuracy and affordability. In addition, such tools could be used in ecologically fragile and energy-limited regions, pointing to the need for developing solutions that can maximize health gains under limited resources for enhanced sustainability. Metabolic diagnosis holds promise but faces challenges due to the applicability of biospecimens and limited robustness of analytical tools. Here we present a diagnostic method coupling dried serum spots (DSS) and nanoparticle-enhanced laser desorption/ionization mass spectrometry (NPELDI MS). Our approach allows diagnosis of multiple cancers within minutes at affordable cost, environmental friendliness, serum-equivalent precision and user-friendly protocol. Our assessment shows that the implementation of this tool in less-developed regions could reduce the estimated proportion of undiagnosed cases of colorectal cancer from 84.30% to 29.20%, gastric cancer from 77.57% to 57.22% and pancreatic cancer from 34.56% to 9.30%—an overall reduction in the range of 20.35–55.10%. This work provides insights into delivering more sustainable metabolic diagnosis with maximum health gains.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The verification of the metabolites in this study was achieved by comparing the m/z features with the human metabolome database (HMDB, http://www.hmdb.ca/). The data that support the findings of this study are available within the Article and its Supplementary Information or from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
The computer codes utilized during the current study are available from the corresponding author upon reasonable request, due to competing financial interests.
References
Global Atlas of Medical Devices 2022 (WHO, 2022); https://www.who.int/publications/i/item/9789240062207
Cancer Stat Facts: Colorectal Cancer (National Cancer Institute, 2020); https://seer.cancer.gov/statfacts/html/colorect.html
Bar, N. et al. A reference map of potential determinants for the human serum metabolome. Nature 588, 135–140 (2020).
Ricotti, V. et al. Wearable full-body motion tracking of activities of daily living predicts disease trajectory in Duchenne muscular dystrophy. Nat. Med. 29, 95–103 (2023).
Chen, W. et al. Early detection of visual impairment in young children using a smartphone-based deep learning system. Nat. Med. 29, 493–503 (2023).
Carreño, J. M. et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 602, 682–688 (2022).
Adhikari, A. N. et al. The role of exome sequencing in newborn screening for inborn errors of metabolism. Nat. Med. 26, 1392–1397 (2020).
Cohen, J. D. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018).
Meng, F. et al. Ratiometric electrochemical OR gate assay for NSCLC-derived exosomes. J. Nanobiotechnol. 21, 104 (2023).
Newborn Screening Laboratory Bulletin (Centers for Disease Control and Prevention, 2014); https://archive.cdc.gov/www_cdc_gov/nbslabbulletin/bulletin_next_generation.html
Organization, W. H. Every Newborn Progress Report 2019 (WHO, 2020); https://www.who.int/publications/m/item/every-newborn-progress-report-2019
Huang, L. et al. A multifunctional platinum nanoreactor for point-of-care metabolic analysis. Matter 1, 1669–1680 (2019).
Tsesses, S. et al. Tunable photon-induced spatial modulation of free electrons. Nat. Mater. 22, 345–352 (2023).
Nachtigall, F. M., Pereira, A., Trofymchuk, O. S. & Santos, L. S. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nat. Biotechnol. 38, 1168–1173 (2020).
Yin, Q. et al. A TLR7-nanoparticle adjuvant promotes a broad immune response against heterologous strains of influenza and SARS-CoV-2. Nat. Mater. 22, 380–390 (2023).
Chen, H., Huang, C., Wu, Y., Sun, N. & Deng, C. Exosome metabolic patterns on aptamer-coupled polymorphic carbon for precise detection of early gastric cancer. ACS Nano 16, 12952–12963 (2022).
Unger, M. S., Blank, M., Enzlein, T. & Hopf, C. Label-free cell assays to determine compound uptake or drug action using MALDI-TOF mass spectrometry. Nat. Protoc. 16, 5533–5558 (2021).
Montefusco, L. et al. Acute and long-term disruption of glycometabolic control after SARS-CoV-2 infection. Nat. Metab. 3, 774–785 (2021).
Chen, Y. et al. Plasma metabolic fingerprints for large-scale screening and personalized risk stratification of metabolic syndrome. Cell Rep. Med. 4, 101109 (2023).
Li, X. et al. Circulating metabolite homeostasis achieved through mass action. Nat. Metab. 4, 141–152 (2022).
Wang, J. et al. New insights into the structure–performance relationships of mesoporous materials in analytical science. Chem. Soc. Rev. 47, 8766–8803 (2018).
Xu, Z. et al. Efficient plasma metabolic fingerprinting as a novel tool for diagnosis and prognosis of gastric cancer: a large-scale, multicentre study. Gut 72, 2051–2067 (2023).
Beckonert, O. et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2, 2692–2703 (2007).
Morin, S., Bazarova, N., Jacon, P. & Vella, S. The manufacturers’ perspective on World Health Organization prequalification of in vitro diagnostics. Clin. Infect. Dis. 66, 301–305 (2018).
Meng, L. et al. Development of an automatic ultrasonic matrix sprayer for matrix-assisted laser desorption/ionization mass spectrometry imaging. Anal. Chem. 94, 6457 (2022).
Harvey, D. J. Analysis of carbohydrates and glycoconjugates by matrix‐assisted laser desorption/ionization mass spectrometry: an update for 2019–2020. Mass Spectrom. Rev. 42, 1984 (2023).
Zhang, M. et al. Ultra‐fast label‐free serum metabolic diagnosis of coronary heart disease via a deep stabilizer. Adv. Sci. 8, 2101333 (2021).
Dominique, N. L. et al. Giving gold wings: ultrabright and fragmentation free mass spectrometry reporters for barcoding, bioconjugation monitoring, and data storage. Angew. Chem. Int. Ed. Engl. 62, e202219182 (2023).
Guan, M. et al. Silver nanoparticles as matrix for MALDI FTICR MS profiling and imaging of diverse lipids in brain. Talanta 179, 624–631 (2018).
Bastús, N. G., Merkoçi, F., Piella, J. & Puntes, V. Synthesis of highly monodisperse citrate-stabilized silver nanoparticles of up to 200 nm: kinetic control and catalytic properties. Chem. Mater. 26, 2836–2846 (2014).
Yagnik, G. B. et al. Large scale nanoparticle screening for small molecule analysis in laser desorption ionization mass spectrometry. Anal. Chem. 88, 8926–8930 (2016).
Chen, W. et al. Comprehensive metabolic fingerprints characterize neuromyelitis optica spectrum disorder by nanoparticle-enhanced laser desorption/ionization mass spectrometry. ACS Nano 17, 19779–19792 (2023).
Müller, W. H., Verdin, A., De Pauw, E., Malherbe, C. & Eppe, G. Surface-assisted laser desorption/ionization mass spectrometry imaging: a review. Mass Spectrom. Rev. 41, 373–420 (2022).
Prysiazhnyi, V. et al. Fate of gold nanoparticles in laser desorption/ionization mass spectrometry: toward the imaging of individual nanoparticles. J. Am. Soc. Mass Spectrom. 34, 570–578 (2023).
Poulos, R. C. et al. Strategies to enable large-scale proteomics for reproducible research. Nat. Commun. 11, 3793 (2020).
Cajka, T. & Fiehn, O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Analyt. Chem. 61, 192–206 (2014).
Saini, R. K., Prasad, P., Shang, X. & Keum, Y.-S. Advances in lipid extraction methods—a review. Int. J. Mol. Sci. 22, 13643 (2021).
Yang, Y. et al. Solubility of benzoin in three binary solvent mixtures and investigation of intermolecular interactions by molecular dynamic simulation. J. Mol. Liq. 243, 472–483 (2017).
Adeboyejo, K. et al. Simultaneous determination of HCV genotype and NS5B resistance associated substitutions using dried serum spots from São Paulo state, Brazil. Access Microbiol. 4, 326 (2022).
Okai, C. A. et al. Profiling of intact blood proteins by matrix‐assisted laser desorption/ionization mass spectrometry without the need for freezing – dried serum spots as future clinical tools for patient screening. Rapid Commun. Mass Spectrom. 35, e9121 (2021).
Verstraete, J. & Stove, C. Patient-centric assessment of thiamine status in dried blood volumetric absorptive microsamples using LC–MS/MS analysis. Anal. Chem. 93, 2660–2668 (2021).
Platt, F. M. Emptying the stores: lysosomal diseases and therapeutic strategies. Nat. Rev. Drug Discov. 17, 133–150 (2018).
Shan, D. et al. N-protein presents early in blood, dried blood and saliva during asymptomatic and symptomatic SARS-CoV-2 infection. Nat. Commun. 12, 1931 (2021).
Wang, G. X. et al. Metabolic detection and systems analyses of pancreatic ductal adenocarcinoma through machine learning, lipidomics, and multi-omics. Sci. Adv. 7, eabh2724 (2021).
Su, H. et al. Plasmonic alloys reveal a distinct metabolic phenotype of early gastric cancer. Adv. Mater. 33, 2007978 (2021).
Pang, Z. et al. Using MetaboAnalyst 5.0 for LC HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 17, 1735–1761 (2022).
Zhang, Z. et al. DHHC9-mediated GLUT1 S-palmitoylation promotes glioblastoma glycolysis and tumorigenesis. Nat. Commun. 12, 5872 (2021).
Galstyan, A. et al. Blood–brain barrier permeable nano immunoconjugates induce local immune responses for glioma therapy. Nat. Commun. 10, 3850 (2019).
Taibl, K. R. et al. Newborn metabolomic signatures of maternal per- and polyfluoroalkyl substance exposure and reduced length of gestation. Nat. Commun. 14, 3120 (2023).
Jiang, X. et al. Development of a bile acid-based newborn screen for Niemann–Pick disease type C. Sci. Transl. Med. 8, 337ra63 (2016).
Pickhardt, P. J., Hassan, C., Halligan, S. & Marmo, R. Colorectal cancer: CT colonography and colonoscopy for detection—systematic review and meta-analysis. Radiology 259, 393–405 (2011).
Duarte, R. B. et al. Computed tomography colonography versus colonoscopy for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Ther. Clin. Risk Manage. 14, 349–360 (2018).
Chen, H. et al. Participation and yield of a population-based colorectal cancer screening programme in China. Gut 68, 1450–1457 (2019).
Zheng, R. et al. Cancer incidence and mortality in China, 2016. J. Natl Cancer Cent. 2, 1–9 (2022).
Bokhorst, L. P. et al. Compliance rates with the prostate cancer research international active surveillance (PRIAS) protocol and disease reclassification in noncompliers. Eur. Urol. 68, 814–821 (2015).
Alferink, L. J. M. et al. Microbiomics, metabolomics, predicted metagenomics, and hepatic steatosis in a population-based study of 1,355 adults. Hepatology 73, 968–982 (2021).
May, T. et al. One-carbon metabolism in children with marasmus and kwashiorkor. EBioMedicine 75, 103791 (2022).
Yin, C. et al. Ultrabroadband photodetectors up to 10.6 µm based on 2D Fe3O4 nanosheets. Adv. Mater. 32, 2002237 (2020).
Park, H. et al. Magnetite nanoparticles as efficient materials for removal of glyphosate from water. Nat. Sustain. 3, 129–135 (2019).
Huang, L. et al. Machine learning of serum metabolic patterns encodes early-stage lung adenocarcinoma. Nat. Commun. 11, 3556 (2020).
Ríos-Ocampo, J. P. et al. Thermal districts in Colombia: developing a methodology to estimate the cooling potential demand. Renew. Sustain. Energy Rev. 165, 112612 (2022).
Letang, E. et al. Minimally invasive tissue sampling: a tool to guide efforts to reduce AIDS-related mortality in resource-limited settings. Clin. Infect. Dis. 73, 343–350 (2021).
Buscail, L., Bournet, B. & Cordelier, P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 17, 153–168 (2020).
Sun, Y. et al. Noninvasive urinary protein signatures associated with colorectal cancer diagnosis and metastasis. Nat. Commun. 13, 2757 (2022).
Guo, X. et al. Circulating exosomal gastric cancer-associated long noncoding RNA1 as a biomarker for early detection and monitoring progression of gastric cancer: a multiphase study. JAMA Surg. 155, 572–579 (2020).
Verstraete, J., Boffel, L. & Stove, C. Dried blood microsample-assisted determination of vitamins: recent developments and challenges. Trends Analyt. Chem. 132, 116057 (2020).
Ng, A. H. C. et al. A digital microfluidic system for serological immunoassays in remote settings. Sci. Transl. Med. 10, eaar6076 (2018).
Kvaskoff, D. et al. Minimizing matrix effects for the accurate quantification of 25-hydroxyvitamin D metabolites in dried blood spots by LC–MS/MS. Clin. Chem. 62, 639–646 (2016).
Sammut, S.-J. et al. Multi-omic machine learning predictor of breast cancer therapy response. Nature 601, 623–629 (2022).
Myszczynska, M. A. et al. Applications of machine learning to diagnosis and treatment of neurodegenerative diseases. Nat. Rev. Neurol. 16, 440–456 (2020).
Dufresne, M., Patterson, N. H., Norris, J. L. & Caprioli, R. M. Combining salt doping and matrix sublimation for high spatial resolution MALDI imaging mass spectrometry of neutral lipids. Anal. Chem. 91, 12928–12934 (2019).
Szunerits, S. et al. The impact of chemical engineering and technological advances on managing diabetes: present and future concepts. Chem. Soc. Rev. 50, 2102–2146 (2021).
Sang, L. et al. Mitochondrial long non-coding RNA GAS5 tunes TCA metabolism in response to nutrient stress. Nat. Metab. 3, 90–106 (2021).
Li, W.-Q. et al. Beneficial effects of endoscopic screening on gastric cancer and optimal screening interval: a population-based study. Endoscopy 54, 848–858 (2022).
Cai, Q. et al. Development and validation of a prediction rule for estimating gastric cancer risk in the Chinese high-risk population: a nationwide multicentre study. Gut 68, 1576–1587 (2019).
Chen, R. et al. Effectiveness of one-time endoscopic screening programme in prevention of upper gastrointestinal cancer in China: a multicentre population-based cohort study. Gut 70, 251–260 (2021).
Zhang, T. et al. Changing trends of disease burden of gastric cancer in China from 1990 to 2019 and its predictions: findings from global burden of disease study. Chin. J. Cancer Res. 33, 11–26 (2021).
Jin, G. et al. Genetic risk, incident gastric cancer, and healthy lifestyle: a meta-analysis of genome-wide association studies and prospective cohort study. Lancet Oncol. 21, 1378–1386 (2020).
Xia, R. et al. Estimated cost-effectiveness of endoscopic screening for upper gastrointestinal tract cancer in high-risk areas in China. JAMA Netw. Open 4, e2121403 (2021).
Canto, M. I. et al. Frequent detection of pancreatic lesions in asymptomatic high-risk individuals. Gastroenterology 142, 796–804 (2012).
Pereira, S. P. et al. Early detection of pancreatic cancer. Lancet Gastroenterol. Hepatol. 5, 698–710 (2020).
US Preventive Services Task Force et al. Screening for pancreatic cancer: US preventive services task force reaffirmation recommendation statement. JAMA 322, 438–444 (2019).
Godec, P. et al. Democratized image analytics by visual programming through integration of deep models and small-scale machine learning. Nat. Commun. 10, 4551 (2019).
Acknowledgements
We acknowledge financial support from the National Key R&D Program of China (Nos. 2021YFF0703500 (K.Q.), 2022YFE0103500 (K.Q.), 2022YFC2502800 (J. Wan)), the Medical-Engineering Joint Funds of Shanghai Jiao Tong University (Nos. YG2024ZD07 and YG2023ZD08 (K.Q.)), the National Natural Science Funds (Grant Nos. 82372148 (L.H.), 81971771 (K.Q.), 22074044 (J. Wan), 22122404 (J. Wan)) and the Shanghai Institutions of Higher Learning (No. 2021-01-07-00-02-E00083 (K.Q.)). This work was also sponsored by the Innovative Research Team of High-Level Local Universities in Shanghai (SHSMU-ZDCX20210700 (K.Q.)), the Innovation Group Project of Shanghai Municipal Health Commission (2019CXJQ03 (K.Q.)), the Innovation Research Plan by the Shanghai Municipal Education Commission (ZXWF082101 (K.Q.)), the National Research Center for Translational Medicine Shanghai (Nos. TMSK-2021-124 and NRCTM(SH)-2021-06 (K.Q.)), the Science and Technology Commission of Shanghai Municipality (20DZ2220400 (K.Q.)) and Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Contributions
J. Wan, J. Wang, L.H. and K.Q. conceived the study. R.W. and S.Y. carried out the study and wrote the initial article. M.W. and S.Y. constructed the disease model and calculated the missed diagnosis rate. R.W., Y.Z., X.L., W.C. and W.L. analysed the MS data with help from Y.H., J. Wu, J.C. and L.F. All authors provided comments and contributed to the final version of the paper.
Corresponding authors
Ethics declarations
Competing interests
R.W., S.Y., L.H. and K.Q. have filed patents (CN116519778A, China, 2023) for both the technology and the use of the technology to detect bio-samples. These patents are owned and managed by Shanghai Jiao Tong University. The other authors declare no competing interests.
Peer review
Peer review information
Nature Sustainability thanks Carsten Hopf and Jian Liu for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–18 and Tables 1–10.
Supplementary Data 1
Source data for supplementary figures.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, R., Yang, S., Wang, M. et al. A sustainable approach to universal metabolic cancer diagnosis. Nat Sustain (2024). https://doi.org/10.1038/s41893-024-01323-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41893-024-01323-9