Abstract
Scientific research of artificial intelligence (AI) in dermatology has increased exponentially. The objective of this study was to perform a systematic review and meta-analysis to evaluate the performance of AI algorithms for skin cancer classification in comparison to clinicians with different levels of expertise. Based on PRISMA guidelines, 3 electronic databases (PubMed, Embase, and Cochrane Library) were screened for relevant articles up to August 2022. The quality of the studies was assessed using QUADAS-2. A meta-analysis of sensitivity and specificity was performed for the accuracy of AI and clinicians. Fifty-three studies were included in the systematic review, and 19 met the inclusion criteria for the meta-analysis. Considering all studies and all subgroups of clinicians, we found a sensitivity (Sn) and specificity (Sp) of 87.0% and 77.1% for AI algorithms, respectively, and a Sn of 79.78% and Sp of 73.6% for all clinicians (overall); differences were statistically significant for both Sn and Sp. The difference between AI performance (Sn 92.5%, Sp 66.5%) vs. generalists (Sn 64.6%, Sp 72.8%), was greater, when compared with expert clinicians. Performance between AI algorithms (Sn 86.3%, Sp 78.4%) vs expert dermatologists (Sn 84.2%, Sp 74.4%) was clinically comparable. Limitations of AI algorithms in clinical practice should be considered, and future studies should focus on real-world settings, and towards AI-assistance.
Similar content being viewed by others
Introduction
Skin cancer is the most common neoplasm worldwide. Early detection and diagnosis are critical for the survival of affected patients. For skin cancer detection in early stages, a complete physical examination is of paramount importance; however, visual inspection is often not sufficient, and less than one quarter of U.S. patients will have a dermatologic examination in their lifetime1. Dermoscopy is a diagnostic tool, which allows for improved recognition of numerous skin lesions when compared to naked eye examination alone; however, this improvement depends on the level of training and experience of clinicians2. In recent years, advances have been made in noninvasive tools to improve skin cancer diagnostic performance, including the use of artificial intelligence (AI) for clinical and/or dermoscopic image diagnosis in dermatology.
Convolutional neural networks (CNN) is a type of machine learning (ML) that simulates the processing of biological neurons and is the state-of-the-art network for pattern recognition in medical image analysis1,2. As diagnosis in dermatology relies heavily on both clinical and dermoscopic image recognition, the use of CNN has the potential to collaborate or improve diagnostic performance. Studies have been published demonstrating that CNN-based AI algorithms can perform similarly or even outperform specialists for skin cancer diagnosis3. This has created an ‘AI revolution’ in the field of skin cancer diagnosis. Recently, a few dermatology AI systems have been CE (Conformité Européenne) approved by the European Union and are use in practice making of paramount importance to understand the data behind these algorithms4.
While there have been relevant systematic reviews performed in the past few years, the importance of this work which combines a high-quality systematic review with a meta-analysis is that it quantitatively asks the question of where we are with AI for skin cancer detection. The main objective of this study was to perform a systematic review and meta-analysis to critically evaluate the evidence published to date on the performance of AI algorithms in skin cancer classification in comparison with clinicians.
Methods
Guidelines followed
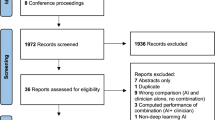
This systematic review was based on the PRISMA guidelines. A flow chart diagram is presented in Fig. 1. The present study has also been registered in the Prospective Register of Systematic Reviews (PROSPERO) System (PROSPERO ID: CRD42022368285).
Search strategy
Three electronic databases, PubMed, Embase, and Cochrane library were searched by a librarian (J.M.). Studies published up to August 2022 were included. We uploaded all the titles and abstracts retrieved by electronic searching into Rayyan and removed any duplicate. Then we collected all the full texts of the studies that met the inclusion criteria based on the title or abstract for detailed inspection. Two reviewers (M.P.S. and J.S.) independently assessed the eligibility of the retrieved papers and resolved any discrepancies through discussion.
Study population—selection
The following PICO (Population, Intervention or exposure, Comparison, Outcome) elements were applied as inclusion criteria for the systematic review: (i) Population: Images of patients with skin lesions, (ii) Intervention: Artificial intelligence diagnosis/classification, (iii) Comparator: Diagnosis/ classification by clinicians, (iv) Outcome: Diagnosis of skin lesions. Only primary studies comparing the performance of artificial intelligence versus dermatologists or clinicians were included.
Studies about diagnosis of inflammatory dermatoses, without extractable data, non-English publications, or animal studies, were excluded.
Data extraction
For studies fulfilling the inclusion criteria, two independent reviewers extracted data in a standardized and predefined form. The following data were extracted and recorded: (i) Database (ii) Title, (iii) Year of publication, (iv) Author, (v) Journal, (vi) Prospective vs retrospective study, (vii) Image database used for training and internal vs external dataset for testing (viii) Type of images included: clinical and/or dermoscopy, (ix) Histopathology confirmation of diagnosis, (x) Inclusion of clinical information, (xi) Number and expertise of participants (experts dermatologists, non-expert dermatologists, and generalists), (xii) Name and type of AI algorithm, (xiii) Included diagnosis, (xiv) Statistics on diagnostic performance (sensitivity [Sn], specificity [Sp], receiver operating characteristic [ROC] curve, area under the curve [AUC]). The main comparisons conducted were diagnostic performance of the AI algorithm compared with clinician diagnostic performance. When available, the change in diagnostic performance of dermatologists with the support of the AI algorithm was included, as well as the change in diagnostic performance after including clinical data (data in supplementary material).
Risk of bias assessment
Two review authors independently assessed the quality of the studies included and the risk of bias using QUADAS-25. Based on the questions, we classified each QUADAS-2 domain as low (0), high (1) or unknown (2) risk of bias.
Meta-analysis
Nineteen out of 53 studies were included in the meta-analysis. The studies met the following criteria: dermoscopic images only, diagnosis of skin cancer, dichotomous classification (benign/malignant, melanoma/nevus), extractable data from the original article (to calculate true positives [TP], false positives [FP], true negatives [TN], and false negatives [FN]), distinction in level of expertise of clinicians (experts dermatologists vs non-expert dermatologists vs generalists). For study purposes and to obtain a global estimate, we grouped all levels of clinical expertise as ‘overall clinicians’. During data processing, two extra analysis that were not pre-specified in the PROSPERO protocol were performed: clinician vs AI algorithms in prospective vs retrospective studies and internal vs external test (validation) sets, respectively. Internal vs external test sets were defined according to Cabitza6 and Shung et al.7. ‘Internal test set’ was defined as a non-overlapping, ‘held out’ subset of the original patient group data that was not used for AI algorithm development and training, used to test the AI model. ‘External test set’ was defined as a set of new data originating from different cohorts, facilities, or repositories other than the data used for model development and training (e.g., dataset originated in different country or institution). Two investigators classified included studies into internal vs external test sets. If both internal and external test sets were used, we classified them as external for study purposes. We decided to perform these non-pre-specified analysis given the relevance of the results for understanding of the data8.
We extracted binary diagnostic accuracy data and constructed contingency tables to calculate Sn and Sp. We conducted a meta-analysis of studies providing 2 ×2 tables to estimate the accuracy of AI and clinicians (confirmatory approach). If an included study provided various 2 ×2 tables, we assumed these data to be independent from each other. We performed a hierarchical summary receiver operating characteristic (HSROC) as well as a bivariate model of the accuracy of AI and clinicians. ROC curves were constructed to simplify the plotting of graphical summaries of fitted models. A likelihood ratio test was used to compare models. A p-value less than 0.05 was considered statistically significant. Analyses were performed using Stata 17.0 statistics software package (codes in supplementary material).
Results
A total of 53 comparative studies (since Piccolo et al. in 20029) fulfilled the inclusion criteria (Fig. 1). Most of the studies focused on dermoscopic images (n = 31), followed by clinical images (n = 14), or both (n = 8). Detailed extracted data is shown in Table 1 for dermoscopic imaging studies, Table 2 for clinical imaging studies, and Table 3 for clinical and dermoscopic imaging studies.
Regarding the risk of bias, most of the studies had an uncertain risk (58%), and 14 (26%) had a low risk of bias. Detail of QUADAS-2 score for each study included in the systematic review is in Fig. 2.
QUADAS-2 tool was used to assess the risk of bias in the included studies in terms of 4 domains (participants, index test, reference standard, and analysis). Low risk (cyan) refers to the number of studies that have a low risk of bias in the respective domain. Unclear (gray) refers to the number of studies that have an unclear risk of bias in the respective domain due to lack of information reported by the study. High risk (purple) refers to the number of studies that have a high risk of bias in the respective domain. a. Risk of Bias Assessment b. Applicability Concerns.
Databases used
Only institutional or private databases were used in 20 articles (37.7%). In all, 16 articles (30.2%) used exclusively open-source data; the most commonly used databases were ‘ISIC’ and ‘HAM10000’10,11. Eighteen studies (33.9%) used a combination of institutional and public dataset. Twenty-two studies (41.5%) used only images of lesions confirmed with histopathology, while 27 (50.9%) included images diagnosed by expert consensus as the gold standard. Four studies (7.5%) did not specify a method of diagnosis confirmation. Fourteen studies (26.4%) used an external database for testing the algorithm, 39 studies (73.6%) tested with an internal dataset (Tables 1–3).
Study type and participants included
A total of 50 studies (94.3%) were retrospective and 3 (5.7%) were prospective. Twenty-seven studies (50.9%) included only specialists, in some cases detailing the level of expertise (expert dermatologists vs non-expert dermatologists). Twenty-three studies (43.3%) included dermatologists and other non-specialist clinicians (dermatology residents and/or generalists), and 3 studies (5.6%) included only generalists.
Diagnosis included and metadata
Forty-three studies (81.1%) considered differential diagnosis between skin tumors only, while 10 (18.8%) also included inflammatory diagnosis or other pathologies (multiclass algorithms). Eighteen articles (33.9%) included clinical information on the patients (metadata), mainly age, sex, and lesion location.
Artificial intelligence assistance
Of the total number of articles included in the review, 11 (20.7%) evaluated potential changes in diagnostic performance or therapeutic decisions of clinicians with AI assistance. Nine of 11 studies showed an improvement in global diagnostic performance when using AI collaboration, 6 of which showed a higher percentage of improvement in the generalists group.
Diagnostic performance of artificial intelligence algorithm versus clinicians, from dermoscopic images of skin lesions
Thirty-one studies evaluated diagnostic performance with dermoscopic images (Table 1). In general, 61.2% (n = 19) of the studies showed a better performance of AI when compared to clinicians. A total of 29.0% (n = 9) resulted in a comparable performance, and in 9.7% (n = 3) specialists outperformed AI.
Dichotomous classification (‘benign’ vs ‘malignant’)
Eighteen studies used AI with dichotomous classification (58.0%) as ‘benign’ vs ‘malignant’. In 61.1% AI outperformed clinicians (n = 11)12,13,14,15,16,17,18,19,20,21,22,23, being statistically significant in 54.5% of them12,15,16,18,20,21. A total of 27.7% showed comparable performance between AI and clinicians (n = 5)9,24,25,26,27. In all, 11.1% resulted in a better performance for clinicians in comparison to AI (n = 2)28,29, 1 of them showing statistical significance29. Five studies16,17,18,19,28 evaluated the collaboration between AI and clinicians (‘augmented intelligence’). All of them showed improved diagnostic accuracy when evaluating clinicians with the support of AI algorithms, being more relevant for less experienced clinicians. Statistical significance was demonstrated in two16,17.
Multiclass and combined classification
Eight of the 31 studies used multiclass classification; in 4 of them, AI had a better performance30,31,32,33; in 3 studies the diagnostic accuracy was comparable34,35,36; and in 1 clinicians outperformed AI37. Two out of 8 studies evaluated AI-assistance, all of them showing improvement in diagnostic accuracy for human raters, with least experienced clinicians benefiting the most32,35. Five of the 31 dermoscopy studies developed both dichotomous and multiclass algorithms, 4 of them resulting in a better performance of AI over humans38,39,40,41.
Diagnostic performance of artificial intelligence algorithms versus clinicians, using clinical images
A total of 14 AI articles evaluating CNN-based classification approaches that used clinical images only were included (Table 2). Of these, 42.8% (n = 6) showed a better performance of AI algorithms, 28.6% (n = 4) obtained comparable results, and in 28.6% (n = 4) clinicians outperformed AI.
Dichotomous classification (‘benign’ vs ‘malignant’)
Six studies42,43,44,45,46 developed an AI algorithm with dichotomous outcomes, obtaining a performance comparable or superior to clinicians in 5 of them42,43,44,45. One study showed a better performance for clinicians46.
Multiclass and combined classification
Five studies47,48,49,50,51 incorporated AI algorithms with multiclass classification. Zhao et al.48 and Pangti et al.49 obtained superior performance of AI algorithms, while Chang et al.47, showed comparable performance between AI and specialists. In one study, clinicians outperformed AI algorithm50.
Three studies52,53,54 with clinical images used both dichotomous and multiclass algorithms. Han et al.53 observed an improvement in diagnostic Sn and Sp with the assistance of the AI algorithm for both classifications, being statistically significant for less experienced clinicians.
Diagnostic performance of artificial intelligence algorithms versus clinicians, from both clinical and dermoscopic images
Eight studies included clinical and dermoscopic images as part of their analysis21,55,56,57,58,59,60,61. Overall, 75% (n = 6) resulted in comparable performance, and 25% (n = 2) showed better performance for AI algorithms in comparison to clinicians. Only 1 study obtained statistical significance57.
Dichotomous classification
Six studies applied dichotomous classification; Haenssle et al.57 being the only study obtaining a better performance for the AI algorithm over clinicians despite the incorporation of metadata. Five remaining studies showed a comparable performance between AI and clinicians.
Multiclass and combined classification
Huang et al.61 classified into 6 categories, with AI being superior to specialists in average accuracy. Finally, Esteva et al.55 used two multiclass classifications, showing comparable performance between AI and clinicians in both.
Meta-analysis
A total of 19 studies were included in the meta-analysis. Table 4 shows the summary estimates calculated to compare performance between AI and clinicians with different levels of experience.
Only 1 prospective study met the inclusion criteria and was included in the meta-analysis.
AI vs overall clinicians’ meta-analysis
When analyzing the whole group of clinicians, not accounting for expertise level, AI obtained a Sn 87.0% (95% CI 81.7–90.9%) and Sp 77.1% (95% CI 69.8–83.0%), and overall clinicians obtained a Sn 79.8% (95% CI 73.2–85.1%) and Sp 73.6% (95% CI 66.5–79.6%), with a statistically significant difference for both Sn and Sp, according to the likelihood ratio test (p < 0.001 for both Sn and Sp). The Forest plot is available in Fig. 3a, b. The ROC curve shapes confirmed the prior differences (Fig. 4). Supplementary Fig. 1a, b shows the sub analysis adjusted for retrospective vs prospective design.
AI vs generalists clinicians’ meta-analysis
When analyzing the AI performance vs generalists, AI obtained a Sn 92.5% (95% CI 88.9–94.9%) and Sp 66.5% (95% CI 56.7–75.0%), and generalists a Sn 64.6% (95% CI 47.1–78.9%) and Sp 72.8% (95% CI 56.7–84.5%), the difference being statistically significant for both Sn and Sp, according to the likelihood ratio tests (p < 0.001 for both). The ROC curve shapes confirmed the prior differences, with higher heterogeneity and wider confidence interval for generalists (Fig. 5). Subgroup analysis comparing internal vs external test set was not possible given all included studies were performed using internal test set in this subgroup (Fig. 6a, b).
AI vs non-expert dermatologists’ meta-analysis
AI obtained a Sn 85.4% (95% CI 78.9–90.2%) and Sp 78.5% (95% CI 70.6–84.8%), while non-expert dermatologists obtained Sn 76.4% (95% CI 71.1–80.9%) and Sp 67.1% (95% CI 57.2–75.6%), with a statistically significant difference, both in Sn and Sp (p < 0.001 for both). The ROC curve shapes confirmed these results (Fig. 7). The Forest plot is available in Fig. 8a, b. In the internal vs external test set subgroup analysis (Fig. 8a, b), AI achieved better Sn in the external test set, while greater Sp with an internal test set. For non-expert dermatologists, no changes in Sn were observed; however, they achieved better Sp in the external test set. In the prospective vs. retrospective subgroup analysis (Supplementary Fig. 2), only 1 prospective study met the inclusion criteria and was included in the meta-analysis. A trend towards better Sn in retrospective versus prospective studies was observed.
AI vs expert dermatologists’ meta-analysis
AI obtained a Sn 86.3% (95% CI 80.4–90.7%) and Sp 78.4% (95% CI 71.1–84.3%), and expert dermatologists a Sn 84.2% (95% CI 76.2–89.8%) and Sp 74.4% (95% CI 65.3–81.8%), this difference was statistically significant for both Sn and Sp, according to the likelihood ratio test (p < 0.001 for both). The ROC curve shapes were comparable for both AI and expert dermatologists, with narrow confidence intervals (Fig. 9). The subgroup analysis by internal vs external test set showed that AI had better Sn in external test set while Sp was better for internal test set. For expert dermatologists there was no difference in Sn; Sp was better in external test set (Fig. 10a, b). The subgroup analysis regarding study design, retrospective vs. prospective (Supplementary Fig. 3), found only one study.
Discussion
In the present study, we found an overall Sn and Sp of 87% and 77% for AI algorithms and an overall Sn of 79% and Sp of 73% for all clinicians (‘overall clinicians’) when performing a meta-analysis of the included studies. Differences between AI and all clinicians were statistically significant. Performance between AI algorithms vs specialists was comparable between both groups. The difference between AI performance (Sn 92%, Sp 66%) and the generalists subgroup (Sn 64%, Sp 72%) was more marked when compared to the difference between AI and expert dermatologists. In studies that evaluated AI-assistance (‘augmented intelligence’), overall diagnostic performance of clinicians was found to improve significantly when using AI algorithms62,63,64. This improvement was more important for those clinicians with less experience. This is in line with this meta-analysis’ results where the difference was greater for generalist than for expert dermatologists and opens an opportunity for AI assistance in the group of less-experienced clinicians. To the best of our knowledge, this is the first systematic review and meta-analysis on the diagnostic accuracy of health-care professionals versus AI algorithms using dermoscopic or clinical images of cutaneous neoplasms. The inclusion of a meta-analysis is key to better understanding, quantitatively, the current state-of-the-art of AI algorithms for the automated diagnosis of skin cancer.
In general, the included studies presented diverse methodologies and significant heterogeneity regarding the type of images included, the different classifications, the characteristics of the participants, and the methodology for presenting the results. This is important to consider when analyzing and attempting to generalize and meta-analyze the obtained findings and should be taken into consideration when interpreting this study results. Research in AI and its potential applications in clinical practice have increased exponentially during the last few years in different areas of medicine, not only in dermatology65. Other systematic reviews have also reported that, in experimental settings, most algorithms are able to achieve at least comparable results when compared with clinicians; however, they also describe similar limitations as those described here66,67,68,69. Only a few studies have evaluated the role of AI algorithms in real clinical scenarios in dermatology. Our study confirms that only 5.7% of studies were prospective and only one of the prospective studies was suitable for meta-analysis62,63. This contrasts with recent data in other medical areas showing an increase in the clinical use of AI70 and highlights the relevance of understanding the role of AI in skin cancer and dermatology. However, prospective studies pose a real challenge for AI algorithms to become part of daily clinical practice as they face specific tests such as ‘out-of-distribution’ images or cases.
Based on this systematic review and meta-analysis results, several challenges have been evidenced when applying AI in clinical practice. First, databases are essential when training an AI algorithm. Small databases, inclusion of only specific populations, or limited variation in skin phototypes, limits the extrapolation of results71,72,73. The lack and underrepresentation of certain ethnic groups and skin types in current datasets has been mentioned as a potential source of perpetuation healthcare disparity73. Based on the results of our systematic review, we can confirm that most algorithms have been trained using the same datasets over and over in at least half of the studies. This translates into lack of representation of specific groups. The diversity of techniques and camera types (e.g. professional vs smartphones) used to capture images and their quality, possible artifacts such as pencil marks, rulers or other objects, are variables that must also be considered when evaluating the performance of AI algorithms71,72,74. A second limitation is the lack of inclusion of metadata in the AI algorithms. In the real world, we manage additional layers of information from patients, including demographic data, personal and family history, habits, evolution of the disease, and a complete physical examination, including palpation, side illumination, and not only 2-D visual examination. These elements are important to render a correct differential diagnosis and to guide clinical decision-making, and so far, very few AI models incorporate them. Therefore, real-world diagnosis is different from static 2-D image evaluations. Regarding the design of human evaluation in experimental and retrospective studies, in most cases it aims to determine whether a lesion is benign or malignant, or to provide a specific diagnosis. This differs from clinical practice in a real-life setting, in which decisions are generally behavioral, whether following up, taking a biopsy or removing a lesion, beyond exclusively providing a specific diagnosis based on the clinical evaluation. The scarce available prospective studies that account for this real-world clinical evaluation makes generalization of these positive results of AI mainly based on retrospective studies restricted. In addition, the management of patient information and privacy, and legal aspects of regulation regarding the application of AI-based software in clinical practice, also represents an emerging challenge75.
The current evidence gathered from this article supports collaboration between AI and clinicians (‘augmented intelligence’), especially for non-expert physicians. In the future, AI algorithms are likely to become a relevant tool to improve the evaluation of skin lesions by generalists in primary care centers, or clinicians with less access to specialists63. AI algorithms could also allow for prioritization of referral or triage, improving early diagnosis. Currently, there are undergoing studies evaluating the application of AI algorithms in real clinical settings, which will demonstrate the applicability of these results in clinical practice. The first prospective randomized controlled trial by Han et al.62, showed that when a group of clinicians used AI assistance, the diagnosis accuracy improved. This improvement was better for generalists. The results of this recent randomized clinical trial partially confirm the potentially positive role of AI in dermatology. These results also confirm that the benefit is more pronounced for generalists, aligning with the findings of the present meta-analysis.
With the aim of reducing the current barriers, we propose to generate and apply guidelines with standardization of the methodology for AI studies. One proposal is the Checklist for Evaluation of Image-Based Artificial Intelligence Reports in Dermatology, published by Daneshjou et al.76. These guidelines should include the complete workflow and start from the moment images are captured to protocols on databases, experience of participants, statistical data, definition on how to measure accuracy, among many others. This will allow us to compare different studies and generate better quality evidence. For example, Esteva et al.52. defined ‘overall accuracy’ as the average of individual inference class accuracies, which might differ from others. In addition, it is mandatory to collaborate with international collaborative databases (e.g. ISIC, available at www.isic-archive.com) to provide accessible public benchmarks and ensure repeatability and the inclusion of a diverse group of skin types and ethnicities to avoid for underrepresentation of certain groups. These strategies would make current datasets more diverse and generalizable.
The main strengths of the present study were the extensive and systematic search in 3 databases, encompassing studies from early AI days up to the most recently published studies, the strict criteria applied for the evaluation of studies and extraction of data, following the available guidelines for systematic reviews, and the performance of a meta-analysis, that allows for quantitatively assess the current AI data.
Limitations include the possibility of not having incorporated articles available in databases other than the ones included, or in other languages, thus constituting selection bias. Also, AI is a rapidly evolving field, and new relevant articles might have emerged while analyzing the data. To the best of our knowledge, no landmark studies were published in the meantime. Publication bias cannot be ruled out, since it is more likely that those articles with statistically significant results were to be published. Also, as shown in our results, more than half of the studies (64.1%) utilized the same public databases (e.g. ISIC and HAM10000), generating a possible overlap of the images in the training and testing group. Furthermore, most studies used the same dataset for training and testing the algorithm (73.6% used an internal test set) which might further bias the results. As observed in the subgroup analysis of the present study, there were differences in estimated Sn and Sp for both AI and clinicians depending on whether an internal vs. external test set was used. However, these were post-hoc analysis and should be interpreted with caution. External test set is key for proper evaluation of AI algorithms6 to ‘validate’ that the algorithm will retain its performance when presented with data from other datasets. Limited details regarding humans’ assessment by readers were available and could also affect the results. We also grouped all skin cancers as one group for analysis, variations in accuracy exists for different skin cancers (e.g. melanoma vs basal cell carcinoma vs squamous cell carcinoma) for humans and for AI algorithms. The application of QUADAS-2 shows a potential information bias, as it is an operator-dependent tool which generates subjectivity and qualitative results. Regarding the meta-analysis, we faced two main limitations. Firstly, the heterogeneity between studies makes it difficult to interpret or generalize the results obtained. Secondly, due to the lack of necessary data, the number of studies included in the meta-analysis was reduced when compared to the studies included in the systematic review. Finally, there was a minimal number of prospective studies included in the systematic review and only one was subjected to the meta-analysis and therefore, those results must be interpreted with caution. Nevertheless, in this post-hoc analysis prospective studies showed worse performance of AI algorithms compared to clinicians confirming the relevance of the complete physical examination and other clinical variables such as history, palpation, etc. This also shows a lack of real-world data published given most studies were retrospective reader studies.
Conclusion
This systematic review and meta-analysis demonstrated that the diagnostic performance of AI algorithms was better than generalists, non-expert dermatologists, and despite being statistically significant, AI algorithms were comparable to expert dermatologists in the clinical practice as the differences were minimal. As most studies were performed in experimental settings, future studies should focus on prospective, real-world settings, and towards AI-assistance. Our study suggests that it is time to move forward to real-world studies and randomized clinical trials to accelerate progress for the benefit of our patients. The only randomized study available has shown a better diagnosis accuracy when using AI algorithms as ‘augmented intelligence’62. We envision a fruitful collaboration between AI and humans leveraging the strengths of both to enhance diagnostic capabilities and patient care.
Data availability
All metadata are available as supplementary material.
Code availability
Codes are available as supplementary material.
Change history
24 May 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41746-024-01138-0
References
Lakhani, N. A. et al. Total body skin examination for skin cancer screening among U.S. adults from 2000 to 2010. Prev. Med. 61, 75–80 (2014).
Wu, Y. et al. Skin cancer classification with deep learning: A systematic review. Front Oncol. 12, 893972 (2022).
Jones, O. T. et al. Artificial intelligence and machine learning algorithms for early detection of skin cancer in community and primary care settings: a systematic review. Lancet Digit Health 4, e466–e476 (2022).
Sangers, T. E. et al. Position statement of the EADV Artificial Intelligence (AI) Task Force on AI-assisted smartphone apps and web-based services for skin disease. J. Eur. Acad. Dermatol Venereol https://doi.org/10.1111/jdv.19521 (2023).
Whiting, P. F. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern Med. 155, 529 (2011).
Cabitza, F. et al. The importance of being external. methodological insights for the external validation of machine learning models in medicine. Comput. Methods Prog. Biomed. 208, 106288 (2021).
Shung, D., Simonov, M., Gentry, M., Au, B. & Laine, L. Machine learning to predict outcomes in patients with acute gastrointestinal bleeding: A systematic review. Dig. Dis. Sci. 64, 2078–2087 (2019).
Steyerberg, E. W. & Harrell, F. E. Prediction models need appropriate internal, internal–external, and external validation. J. Clin. Epidemiol. 69, 245–247 (2016).
Piccolo, D. et al. Dermoscopic diagnosis by a trained clinician vs. a clinician with minimal dermoscopy training vs. computer-aided diagnosis of 341 pigmented skin lesions: a comparative study. Br. J. Dermatol 147, 481–486 (2002).
The International Skin Imaging Collaboration. https://www.isic-archive.com/
Tschandl, P. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 14, 180161 (2018).
Friedman, R. J. et al. The diagnostic performance of expert dermoscopists vs a computer-vision system on small-diameter melanomas. Arch. Dermatol 144, 476–482 (2008).
Marchetti, M. A. et al. Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J. Am. Acad. Dermatol 78, 270 (2018).
Brinker, T. J. et al. Comparing artificial intelligence algorithms to 157 German dermatologists: the melanoma classification benchmark. Eur. J. Cancer 111, 30–37 (2019).
Brinker, T. J. et al. Deep neural networks are superior to dermatologists in melanoma image classification. Eur. J. Cancer 119, 11–17 (2019).
Maron, R. C. et al. Artificial intelligence and its effect on dermatologists’ accuracy in dermoscopic melanoma image classification: Web-based survey study. J. Med. Internet Res 22, e18091 (2020).
Lee, S. et al. Augmented decision-making for acral lentiginous melanoma detection using deep convolutional neural networks. J. Eur. Acad. Dermatol Venereol. 34, 1842–1850 (2020).
Marchetti, M. A. et al. Computer algorithms show potential for improving dermatologists’ accuracy to diagnose cutaneous melanoma: Results of the International Skin Imaging Collaboration 2017. J. Am. Acad. Dermatol 82, 622–627 (2020).
Fink, C. et al. Diagnostic performance of a deep learning convolutional neural network in the differentiation of combined naevi and melanomas. J. Eur. Acad. Dermatol Venereol. 34, 1355–1361 (2020).
Tognetti, L. et al. A new deep learning approach integrated with clinical data for the dermoscopic differentiation of early melanomas from atypical nevi. J. Dermatol Sci. 101, 115–122 (2021).
Haenssle, H. A. et al. Man against machine reloaded: Performance of a market-approved convolutional neural network in classifying a broad spectrum of skin lesions in comparison with 96 dermatologists working under less artificial conditions. Ann. Oncol. 31, 137–143 (2020).
Pham, T. C., Luong, C. M., Hoang, V. D. & Doucet, A. AI outperformed every dermatologist in dermoscopic melanoma diagnosis, using an optimized deep-CNN architecture with custom mini-batch logic and loss function. Sci. Rep. 11, 17485 (2021).
Yu, Z. et al. Early melanoma diagnosis with sequential dermoscopic images. IEEE Trans. Med. Imaging 41, 633–646 (2022).
Ferris, L. K. et al. Computer-aided classification of melanocytic lesions using dermoscopic images. J. Am. Acad. Dermatol 73, 769–776 (2015).
Tschandl, P., Kittler, H. & Argenziano, G. A pretrained neural network shows similar diagnostic accuracy to medical students in categorizing dermatoscopic images after comparable training conditions. Br. J. Dermatol. 177, 867–869 (2017).
Yu, C. et al. Acral melanoma detection using a convolutional neural network for dermoscopy images. PLoS One 13, e0193321 (2018).
Phillips, M. et al. Assessment of accuracy of an artificial intelligence algorithm to detect melanoma in images of skin lesions. JAMA Netw. Open 2, e1913436 (2019).
Dreiseitl, S., Binder, M., Hable, K. & Kittler, H. Computer versus human diagnosis of melanoma: evaluation of the feasibility of an automated diagnostic system in a prospective clinical trial. Melanoma Res. 19, 180–184 (2009).
Winkler, J. K. et al. Monitoring patients at risk for melanoma: May convolutional neural networks replace the strategy of sequential digital dermoscopy? Eur. J. Cancer 160, 180–188 (2022).
Tschandl, P. et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: an open, web-based, international, diagnostic study. Lancet Oncol. 20, 938–947 (2019).
Minagawa, A. et al. Dermoscopic diagnostic performance of Japanese dermatologists for skin tumors differs by patient origin: A deep learning convolutional neural network closes the gap. J. Dermatol 48, 232–236 (2021).
Tschandl, P. et al. Human-computer collaboration for skin cancer recognition. Nat. Med. 26, 1229–1234 (2020).
Combalia, M. et al. Validation of artificial intelligence prediction models for skin cancer diagnosis using dermoscopy images: the 2019 International Skin Imaging Collaboration Grand Challenge. Lancet Digit Health 4, e330–e339 (2022).
Wang, S. Q. et al. Deep learning-based, computer-aided classifier developed with dermoscopic images shows comparable performance to 164 dermatologists in cutaneous disease diagnosis in the Chinese population. Chin. Med. J. (Engl.) 133, 2027–2036 (2020).
Lucius, M. et al. Deep neural frameworks improve the accuracy of general practitioners in the classification of pigmented skin lesions. Diagnostics (Basel) 10, 969 (2020).
Zhu, C. Y. et al. A deep learning based framework for diagnosing multiple skin diseases in a clinical environment. Front Med. (Lausanne) 8, 626369 (2021).
Van Molle, P. et al. The value of measuring uncertainty in neural networks in dermoscopy. J. Am. Acad. Dermatol 87, 1191–1193 (2022).
Hekler, A. et al. Superior skin cancer classification by the combination of human and artificial intelligence. Eur. J. Cancer 120, 114–121 (2019).
Maron, R. C. et al. Systematic outperformance of 112 dermatologists in multiclass skin cancer image classification by convolutional neural networks. Eur. J. Cancer 119, 57–65 (2019).
Winkler, J. K. et al. Collective human intelligence outperforms artificial intelligence in a skin lesion classification task. J. Dtsch Dermatol Ges. 19, 1178–1184 (2021).
Ba, W. et al. Convolutional neural network assistance significantly improves dermatologists’ diagnosis of cutaneous tumours using clinical images. Eur. J. Cancer 169, 156–165 (2022).
Han, S. S. et al. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J. Investig. Dermatol. 138, 1529–1538 (2018).
Fujisawa, Y. et al. Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumour diagnosis. Br. J. Dermatol. 180, 373–381 (2019).
Han, S. S. et al. Keratinocytic skin cancer detection on the face using region-based convolutional neural network. JAMA Dermatol 156, 29–37 (2020).
Huang, K. et al. Assistant diagnosis of basal cell carcinoma and seborrheic keratosis in Chinese population using convolutional neural network. J. Health. Eng. 2020, 1713904 (2020).
Polesie, S. et al. Discrimination between invasive and in situ melanomas using clinical close-up images and a de novo convolutional neural network. Front Med (Lausanne) 8, 723914 (2021).
Chang, W. Y. et al. Computer-aided diagnosis of skin lesions using conventional digital photography: A reliability and feasibility study. PLoS One 8, e76212 (2013).
Zhao, X. Y. et al. The application of deep learning in the risk grading of skin tumors for patients using clinical images. J. Med. Syst. 43, 283 (2019).
Pangti, R. et al. Performance of a deep learning-based application for the diagnosis of basal cell carcinoma in Indian patients as compared to dermatologists and nondermatologists. Int J. Dermatol 60, e51–e52 (2021).
Agarwala, S., Mata, D. A. & Hafeez, F. Accuracy of a convolutional neural network for dermatological diagnosis of tumours and skin lesions in a clinical setting. Clin. Exp. Dermatol 46, 1310–1311 (2021).
Kim, Y. J. et al. Augmenting the accuracy of trainee doctors in diagnosing skin lesions suspected of skin neoplasms in a real-world setting: A prospective controlled before-and-after study. PLoS One 17, e0260895 (2022).
Han, S. S. et al. Assessment of deep neural networks for the diagnosis of benign and malignant skin neoplasms in comparison with dermatologists: A retrospective validation study. PLoS Med. 17, e1003381 (2020).
Han, S. S. et al. Augmented intelligence dermatology: Deep neural networks empower medical professionals in diagnosing skin cancer and predicting treatment options for 134 skin disorders. J. Investig. Dermatol. 140, 1753–1761 (2020).
Jinnai, S. et al. The development of a skin cancer classification system for pigmented skin lesions using deep learning. Biomolecules 10, 1–13 (2020).
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017).
Tschandl, P. et al. Expert-level diagnosis of nonpigmented skin cancer by combined convolutional neural networks. JAMA Dermatol 155, 58–65 (2019).
Haenssle, H. A. et al. Man against Machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 29, 1836–1842 (2018).
Brinker, T. J. et al. A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task. Eur. J. Cancer 111, 148–154 (2019).
Li, C. X. et al. Diagnostic capacity of skin tumor artificial intelligence-assisted decision-making software in real-world clinical settings. Chin. Med. J. (Engl.) 133, 2020–2026 (2020).
Willingham, M. L. et al. The potential of using artificial intelligence to improve skin cancer diagnoses in Hawai’i’s multiethnic population. Melanoma Res. 31, 504–514 (2021).
Huang, K. et al. The classification of six common skin diseases based on xiangya-derm: Development of a chinese database for artificial intelligence. J. Med. Internet Res 23, e26025 (2021).
Han, S. S. et al. Evaluation of artificial intelligence-assisted diagnosis of skin neoplasms: a single-center, paralleled, unmasked, randomized controlled trial. J. Invest Dermatol 142, 2353–2362.e2 (2022).
Muñoz‐López, C. et al. Performance of a deep neural network in teledermatology: A single‐centre prospective diagnostic study. J. Eur. Acad. Dermatol. Venereol. 35, 546–553 (2021).
Han, S. S. et al. The degradation of performance of a state-of-the-art skin image classifier when applied to patient-driven internet search. Sci. Rep. 12, 16260 (2022).
Liu, X. et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health 1, e271–e297 (2019).
Haggenmuller, S. et al. Skin cancer classification via convolutional neural networks: Systematic review of studies involving human experts. Eur. J. Cancer 156, 202–216 (2021).
Takiddin, A., Schneider, J., Yang, Y., Abd-Alrazaq, A. & Househ, M. Artificial intelligence for skin cancer detection: Scoping review. J. Med. Internet Res. 23, e22934 (2021).
Brinker, T. J. et al. Skin cancer classification using convolutional neural networks: Systematic review. J. Med. Internet Res. 20, e11936 (2018).
Marka, A., Carter, J. B., Toto, E. & Hassanpour, S. Automated detection of nonmelanoma skin cancer using digital images: A systematic review. BMC Med. Imaging 19, 21 (2019).
Wu, K. et al. Characterizing the clinical adoption of medical AI devices through U.S. insurance claims. NEJM AI https://doi.org/10.1056/AIoa2300030 (2023).
Navarrete-Dechent, C. et al. Automated dermatological diagnosis: Hype or reality? J. Investig. Dermatol. 138, 2277–2279 (2018).
Navarrete-Dechent, C., Liopyris, K. & Marchetti, M. A. Multiclass artificial intelligence in dermatology: Progress but still room for improvement. J. Investig. Dermatol. 141, 1325–1328 (2021).
Adamson, A. S. & Smith, A. Machine learning and health care disparities in dermatology. JAMA Dermatol 154, 1247–1248 (2018).
Daneshjou, R., Smith, M. P., Sun, M. D., Rotemberg, V. & Zou, J. Lack of transparency and potential bias in artificial intelligence data sets and algorithms: A scoping review. JAMA Dermatol 157, 1362–1369 (2021).
Jobson, D., Mar, V. & Freckelton, I. Legal and ethical considerations of artificial intelligence in skin cancer diagnosis. Australas. J. Dermatol 63, e1–e5 (2022).
Daneshjou, R. et al. Checklist for evaluation of image-based artificial intelligence reports in dermatology: CLEAR derm consensus guidelines from the international skin imaging collaboration artificial intelligence working group. JAMA Dermatol 158, 90–96 (2022).
Tenenhaus, A. et al. Detection of melanoma from dermoscopic images of naevi acquired under uncontrolled conditions. Skin Res. Technol. 16, 85–97 (2010).
Brinker, T. et al. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur. J. Cancer 113, 47–54 (2019).
Acknowledgements
Partial funding was obtained from La Fondation La Roche Possay Research Awards. ANID - Millennium Science Initiative Program ICN2021_004.
Author information
Authors and Affiliations
Contributions
M.P.S, J.S., L.H, V.R., J.B., D.M., and C.N-D. designed and conceived the study; M.P.S, J.S., D.P., M.M., V.R., J.B., D.M., and C.N-D. acquired, analyzed and interpreted the data; M.P.S, J.S., L.H, D.P., M.M., P.U., V.R., J.B., D.M., and C.N-D. drafted and revised the manuscript; M.P.S, J.S., L.H, D.P., M.M., P.U., V.R., J.B., D.M., and C.N-D.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Salinas, M.P., Sepúlveda, J., Hidalgo, L. et al. A systematic review and meta-analysis of artificial intelligence versus clinicians for skin cancer diagnosis. npj Digit. Med. 7, 125 (2024). https://doi.org/10.1038/s41746-024-01103-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41746-024-01103-x