Abstract
Deep neural networks have been integrated into the whole clinical decision procedure which can improve the efficiency of diagnosis and alleviate the heavy workload of physicians. Since most neural networks are supervised, their performance heavily depends on the volume and quality of available labels. However, few such labels exist for rare diseases (e.g., new pandemics). Here we report a medical multimodal large language model (Med-MLLM) for radiograph representation learning, which can learn broad medical knowledge (e.g., image understanding, text semantics, and clinical phenotypes) from unlabelled data. As a result, when encountering a rare disease, our Med-MLLM can be rapidly deployed and easily adapted to them with limited labels. Furthermore, our model supports medical data across visual modality (e.g., chest X-ray and CT) and textual modality (e.g., medical report and free-text clinical note); therefore, it can be used for clinical tasks that involve both visual and textual data. We demonstrate the effectiveness of our Med-MLLM by showing how it would perform using the COVID-19 pandemic “in replay”. In the retrospective setting, we test the model on the early COVID-19 datasets; and in the prospective setting, we test the model on the new variant COVID-19-Omicron. The experiments are conducted on 1) three kinds of input data; 2) three kinds of downstream tasks, including disease reporting, diagnosis, and prognosis; 3) five COVID-19 datasets; and 4) three different languages, including English, Chinese, and Spanish. All experiments show that our model can make accurate and robust COVID-19 decision-support with little labelled data.
Similar content being viewed by others
Introduction
Recently, the rapid development of deep neural networks has enabled their wide applications in clinics1,2. To process clinical data of different modalities, different neural networks have been employed accordingly. For processing visual data such as dermoscopy images, Convolutional Neural Network (CNN) based frameworks3 have been applied to classify the type of skin lesion4; For textual input such as Electronic Medical Record (EMR), Transformer based frameworks5 have been be applied to estimate the mortality or re-hospitalisation probabilities6; For multi-modal data such as radiology image-report pairs, the encoder-decoder based frameworks7,8,9,10,11 have been applied to generate textual reports from medical images.
Deep neural networks can assist physicians in the diagnosis process and relieve their heavy burden. Most deep neural networks exploit supervised training, and therefore their performance heavily relies on the volume and quality of labelled data. However, the labelling process of clinical data is usually costly and time-consuming. For rare diseases, it is difficult to collect and label sufficient data in a timely manner to train a deep learning model (with some studies taking over one year to collect sufficient data12,13), thus delaying the rapid deployment of deep learning models needed for combating rare diseases promptly.
Take the recent pandemic SARS-CoV-2/COVID-19 for example, which not only leads to multi-organ failures and death but also threatens to affect global health for the foreseeable future14. Although early COVID-19 incurred a high mortality rate, its most recent variants are not life-threatening for the young healthy population. It is still uncertain whether a new variant in the future would pose a life-threatening risk again. Considering the large volume of the vulnerable population for COVID-19, three common types of AI-based decision-support tools can be developed to support accurate diagnosis and prognosis:
-
COVID-19 radiology reporting: Given radiology images, physicians need to write textual reports to address the clinical findings7,11,15,16,17. Given the large number of COVID-19 patients, writing medical reports is a heavy burden for physicians who could otherwise concentrate on patient care18,19. The overly-heavy workload of physicians is well-documented20,21, and using deep learning methods to automatically generate reports that can be modified and approved by physicians can partly automate routine tasks1,2,22,23.
-
COVID-19 diagnosis: Currently, the Reverse Transcription Polymerase Chain Reaction (RT-PCR) is recognised as the gold standard for COVID-19 diagnosis24. Due to the high false-negative rate of RT-PCR and shortage of equipment25,26, different diagnosis models that use medical data across different modalities1,27 to generate more timely results than RT-PCR can work as an alternative in COVID-19 diagnosis.
-
COVID-19 prognosis: A prognosis model2 can support better triage on who to admit to the hospital or intensive care, who to isolate, predicting whom and when to recover, and who is at the highest risk of deterioration.
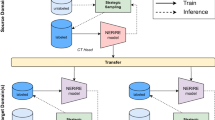
Training common neural networks for the above three tasks requires labels on visual, textual and multi-modal data. However, collecting labelled data for a rare disease is expensive and time-consuming. To this end, inspired by the great success of large-scale pre-training28,29,30,31, as shown in Fig. 1, we present the Medical Multimodal Large Language Model (Med-MLLM) framework for radiograph representation learning31,32,33,34. Our framework deals with the situation where labelled data are scarce, and shortens the time-frame of model deployment, allowing rapid response to rare diseases in the future.
As shown in Fig. 2, our framework adopts multimodal medical data across visual and textual modalities to learn the following comprehensive thorax knowledge. 1) Visual data: for medical images such as Chest X-rays (CXR) and Computed Tomography (CT), we pre-train an image encoder with two types of losses: patient-level contrastive learning loss and image-level contrastive loss. 2) Textual data: for medical texts such as medical reports and clinical notes, we pre-train a text encoder with three types of losses: masked language modelling loss, sentence reconstruction loss, and findings-impression alignment loss. 3) Multi-modal data: for unpaired radiology images and reports, we introduce a soft image-text alignment loss to further pre-train the visual encoder and text encoder. In this manner, Med-MLLM handles visual, textual and multi-modal input, and therefore can be applied to COVID-19 reporting (i.e., medical report generation), diagnosis (i.e., disease classification), and prognosis (i.e., survival prediction) tasks with limited labels for training1,2,12,13,15,27,35.
It consists of three main components: a Image-only pre-training which incorporates the patient-level contrastive learning (PCL); b Text-only pre-training which incorporates three training objectives: the masked language modelling (MLM), the sentence reconstruction (SR) loss, and the findings-impression alignment (FIA) loss; and c Image-text pre-training which incorporates a knowledge base and a pre-training objective: soft image-text alignment (SITA).
The retrospective and prospective experiments across different modalities, languages, and regions assess the effectiveness of our Med-MLLM for clinical decision-making when using limited labelled data. Besides COVID-19, the framework can be readily applied to other 14 common thorax diseases and tuberculosis as well with 1% labelled data, demonstrating the scalability of our framework in assisting physicians when encountering a rare disease.
Overall, the contributions of our work are as follows:
-
With the goal of quick deployment of tools for rapid response to rare diseases, we present the medical multimodal large language model (Med-MLLM) framework. We evaluate the effectiveness of Med-MLLM using the COVID-19 pandemic “in replay”, showing that Med-MLLM is able to accomplish accurate COVID-19 decision-support tasks with limited labelled data. In contrast, existing efforts usually require thousands, or even more, labelled data to achieve similar performance.
-
Med-MLLM is able to handle image-only, text-only, and image-text data, addressing multiple medical tasks including reporting, diagnosis, and prognosis. To demonstrate the effectiveness of Med-MLLM, we conduct both retrospective and prospective (i.e., pre-training model from the early COVID-19 and making a prediction for COVID-19-Omicron) experiments across different modalities, languages, and regions.
-
To evaluate the scalability of Med-MLLM, we investigate other 14 common thorax diseases and tuberculosis. Our results show that Med-MLLM achieves competitive performances w.r.t. previous works with 1% of the labelled training data, and comparable performance when the full training set is used.
Overall framework
As shown in Fig. 1, we develop a Medical Multimodal Large Language Model (Med-MLLM) for rare diseases to deal with the situation where the labelled data is scarce. An example is the early stages of a new pandemic, for which we will have very little data. Med-MLLM (i) adopts the unlabelled medical image data from existing public image datasets, e.g., chest radiology images36,37, COVID chest X-ray images38,39,40,41,42, and COVID CT images40,42,43,44 to perform image-only pre-training45,46 to learn visual characteristics, capturing the rich diagnostic information in medical images1,2,27; (ii) adopts the unlabelled medical text data from existing public text datasets, e.g., PubMed47, MIMIC-CXR medical reports37, and MIMIC-III clinical notes48, to perform text-only pre-training49,50,51 to learn text semantics and clinical findings in medical texts52; (iii) adopts an existing large knowledge base, i.e., Unified Medical Language System (UMLS)53, to perform image-text pre-training54 to unify the learned knowledge from unpaired images and texts, capturing accurate disease phenotypes and clinical presentations.
Figure 2 shows the detailed structure of the Med-MLLM framework. For a fair comparison, we adopt the ResNet-5055 as the image encoder and the Transformer5 as the text encoder/decoder. In detail, Med-MLLM (i) adopts contrastive learning46,56 to perform image-only pre-training, which is improved by a patient-level contrastive learning, image augmentation, and regularisation; (ii) builds a large language model (LLM)49, which adopts self-supervised learning49,50, to perform text-only pre-training. The LLM is further improved by the radiology-specific vocabulary, two pre-training objectives, and a text augmentation method; (iii) adopts contrastive learning54 to perform image-text pre-training, improved by the UMLS knowledge base53 and a pre-training objective. In this way, our framework could capture comprehensive medical knowledge to provide a solid basis for the diagnosis of rare diseases, including COVID-19 and its variant–Omicron. As a result, our framework can be taken as a “warm start" algorithm to provide an accurate and efficient diagnosis of rare diseases using limited labels. Our extensive experiments show that the framework yields encouraging performance for a wide range of downstream tasks.
Fine-tuning
Figure 3 illustrates the details of fine-tuning the Med-MLLM for downstream COVID-19 decision-support tasks. (i) We adopt the image encoder and an additional text decoder to fine-tune (cross-entropy optimisation) the pre-trained Med-MLLM on the COVID-19 reporting (medical report generation) task. (ii) For the task of COVID-19 diagnosis (disease classification), we add a classification layer on the output of image and/or text encoders, and the Med-MLLM is fine-tuned using a binary cross-entropy loss. (iii) For the task of COVID-19 prognosis (survival prediction), we adopt the same fine-tuning strategy as the COVID-19 diagnosis task above, because these two tasks differ solely in the output results. Both tasks can accept three types of input medical data: image-only, text-only, and image-text.
Results
In this section, we conduct experiments on COVID-19 reporting, diagnosis, and prognosis tasks. We first describe five COVID-19 datasets used for the experiments. Then, we present the results of our framework on COVID-19 decision-support across modalities, languages, and regions, using limited labels (e.g. 1% labelled data).
Datasets
We evaluate the performance of our framework on five COVID-19 datasets across different modalities, languages, and regions, i.e., COVIDx-CXR-2 dataset40, COVID-CXR dataset39,41, COVID-19 CT dataset44, BIMCV-COVID-19 dataset42, and COVID-HCH dataset16. The COVIDx-CXR-2 dataset includes 29,986 medical images of 16,648 patients from 51 countries; The COVID-CXR dataset contains over 900 chest X-rays of 412 patients from 26 countries, where 361 patients have survival/death labels. The COVID-19 CT dataset contains 1104 medical images associated with 368 medical reports in Chinese from 96 patients. The dataset was collected from the First Affiliated Hospital of Jinan University Guangzhou and the Fifth Affiliated Hospital of Sun Yat-sen University, Zhuhai, China. The BIMCV-COVID-19 dataset is a large dataset consisting of over 20k CXR and CT images from over 1000 COVID-19 patients along with their radiographic reports in Spanish. The COVID-HCH dataset includes 5115 COVID-19 records and 4112 non-COVID-19 records of viral and bacterial pneumonia from 91 patients, resulting in a total of 9227 records associated with radiographic reports in Chinese. Specifically, the 5115 COVID-19 records are composed of 3577 COVID-19-Delta records and 1538 COVID-19-Omicron records. Meanwhile, we invite clinical professionals to translate 100 reports into English. Each English report is associated with multiple (> 10) medical images, and different images serve as different samples. We adopt the Omicron data to perform simulated prospective studies. In detail, we pre-train the model on Delta data and fine-tune the model on Omicron data.
To pre-process the datasets, we randomly split them into training, validation and test sets with a ratio of 8:1:1, respectively. The training, validation, and test sets are used to train the model, select the optimal modules and hyper-parameters, and evaluate the performance, respectively. All protected health information (e.g., patient name and date of birth) was de-identified for all datasets used in our experiments. Several previous works57,58,59,60 construct a balanced test set to minimise the effect of dataset bias on model performance. The reason is that a balanced test set provides a genuine reflection of the models’ ability to correctly distinguish between positive and negative cases, i.e., their capability to accurately identify COVID-19 cases. Thus, the models are prevented from exploiting biases in the data distribution to achieve high overall performance. To this end, we constructed balanced validation and test sets by randomly sampling 10% of the dataset, with 5% from the positive cases and the other 5% from the negative cases (i.e. the ratio of COVID-19 records to non-COVID records is 1:1). The remaining 80% samples are used as the training set. Therefore, our models are trained on the unbalanced set, but validated and tested on the balanced set. For all experiments, we conduct multiple runs with different seeds and report the average performances for baselines and our model.
Experimental settings
In our work, we conduct both prospective and retrospective studies. In the retrospective studies, we perform the experiments by directly pre-training and evaluating the model on the COVID-19 data. For the prospective studies, we perform the experiments by pre-training the model from early COVID-19 and making predictions for COVID-19-Omicron. For example, we have observed the Delta variant but have no data for Omicron, so our prospective studies can test Med-MLLM to see how it adapts to the new variant (i.e., Omicron) from the old variant (i.e., Delta).
COVID-19 reporting
Our COVID-19 reporting task aims to automatically generate a comprehensive and coherent medical report of a given medical image. In clinical practice, writing reports for numerous images from routine imaging exams can be time-consuming and tedious for even experienced radiologists7. Given the large volume of medical images, automatically generating reports can improve current clinical practice in diagnostic radiology and assist radiologists in clinical decision-making. Therefore, automatic report generation is receiving remarkable attention in both communities of artificial intelligence and clinical medicine7,11,15,16,61,62,63,64. To measure the performance of COVID-19 reporting, we select the widely-used natural language generation metrics, including BLEU-2, -3, -465, ROUGE-L66, and CIDEr67, which are computed by a standard evaluation toolkit68 automatically. These metrics measure the match between the generated reports and reference reports annotated by professional physicians.
Retrospective studies
We further select existing methods, including R2Gen61, KGAE62, and XProNet63, for comparison. We conduct retrospective studies on the COVID-19-CT dataset in Chinese and the BIMCV-COVID-19 dataset in Spanish. We randomly select 1% labelled data for training. The results in Table 1 show that with 1% of training data, our method achieves competitive performance w.r.t. the previous models trained on the full training set across Chinese and Spanish. It shows that our approach can be efficiently trained and deployed with limited labels to combat rare diseases promptly. Using the full training set as used in previous methods, our method achieves the best results across different languages and regions. In detail, our framework outperforms previous best results by up to 4.3%/3.8% in BLEU-4, 9.1%/4.3% in ROUGE-L, and 10.9%/9.8% in CIDEr scores in Chinese/Spanish scenarios. The improvement demonstrates the effectiveness of our framework in providing a solid basis for COVID-19 reporting.
Prospective studies
We perform prospective studies on the COVID-19-Omicron data from the COVID-HCH dataset. Specifically, we adopt the Delta data for pre-training the model and adopt the Omicron data for evaluation. As shown in Table 1, our method Med-MLLM outperforms previous methods trained on full training data on most metrics. Compared with retrospective studies, our method achieves better results on COVID-19-Omicron reporting. The results of prospective studies evaluated on COVID-19-Omicron data show that our method shortens the time for data acquisition, allowing us to respond quickly in future to rare diseases across different languages and regions. We further validate it on the following COVID-19 diagnosis and prognosis tasks. It is worth noting that the performance of our method can be further improved by using more training data, achieving improved performances when it is trained with the full training set as used in previous methods.
COVID-19 diagnosis
In the retrospective setting, the COVID-19 diagnosis task (i.e., disease classification) aims to distinguish COVID-19 from non-COVID-19 cases. In the prospective setting, the aim is to identify COVID-19-Omicron. We conduct retrospective studies on the COVIDx-CXR-2 and COVID-19-Delta data and conduct prospective studies on the COVID-19-Omicron data. In our experiments, we report the widely-used AUC for assessing the diagnosis accuracy.
Retrospective studies
We utilise the COVIDx-CXR-2 dataset to perform the image-only COVID-19 diagnosis task, and adopt the COVID-19-Delta data labelled in English to perform the text-only and image-text medical diagnosis tasks. We further select self-supervised learning and contrastive learning methods for comparison, i.e., CLIP54, ConVIRT34, and BioViL30. Since previous models had not attempted to deal with image-only, text-only and image-text tasks simultaneously, we re-implement these methods for evaluation.
Table 2 shows the diagnosis accuracy of our framework and the previous methods on COVID-19 classification, where our Med-MLLM achieves superior performance on all tasks and datasets. It not only achieves competitive results compared to previous methods with 1% training data, but also outperforms them when using 100% training data. The results demonstrate the validity of our method in relaxing the dependency on the high quality of labelled data for training, while making an accurate COVID-19 diagnosis.
Prospective studies
We pre-train the model on Delta data and fine-tune the model on Omicron data. As shown in Table 2, with 1% of Omicron data, our method can outperform several previous works (e.g., CLIP). More encouragingly, with 100% training labels, Med-MLLM surpasses the previous method by up to 10.6%, 3.6%, and 5.9% in diagnosis accuracy on image-only, text-only, and image-text classification tasks, respectively. The performance of prospective studies assesses the good generalisation capability of our approach in dealing with situations where the training data are scarce. Therefore our Med-MLLM is suitable for new pandemics caused by rapidly developing pathogens, improving the practical value of AI-based decision-support tools in clinical practice.
COVID-19 prognosis
The COVID-19 prognosis task aims at predicting the survival of COVID-19 patients, i.e., predicting whether the patients will survive after treatment in the hospital. In this experiment, we evaluate the performance of prognosis on COVID-CXR and COVID-HCH datasets.
Retrospective studies
We conduct the image-only task on the COVID-CXR dataset and conduct the text-only and image-text tasks on the COVID-19-Delta data from the COVID-HCH dataset. Similar to the COVID-19 diagnosis task, we also re-implement the existing methods for COVID-19 prognosis. The results of COVID-19 prognosis are reported in Table 3, showing that our Med-MLLM is comparable to the previous approaches with 1% training data. Using the full training data, our method outperforms previous methods by up to 4.6%, 1.1%, and 1.5% in AUC on image-only, text-only, and image-text COVID-19 prognosis tasks, respectively.
Prospective studies
We adopt the Omircon data to report the results of prospective studies. In implementations, we pre-train the model on Delta and predict for Omicron. The results illustrated in Table 3 indicate that when it comes to COVID-19 Omircon prognosis, with 1% of data for fine-tuning, our Med-MLLM surpasses existing methods by substantial margins demonstrating the effectiveness of our method in making an accurate and fast COVID-19 diagnosis with limited labelled data. With 100% training data, our method surpasses existing self-supervised learning and contrastive learning methods, which is in accordance with the results of COVID-19 reporting and diagnosis.
Discussion
In addition to COVID-19, our Med-MLLM can be readily applied to other chest/respiratory diseases. Table 4 shows the performances of Med-MLLM on the CheXpert36, NIH ChestX-ray69, RSNA Pneumonia70, SIIM-ACR Pneumothorax71, and Shenzhen Tuberculosis72 benchmark datasets for common disease classification tasks. We follow previous works30,31,32,34,73 to pre-process the datasets and perform the evaluation. As we can see from Table 4, with limited labels (i.e., 1% of CheXpert, NIH ChestX-ray, RSNA Pneumonia, SIIM-ACR Pneumothorax datasets, and 10% of Shenzhen Tuberculosis), our method can achieve competitive results with previous fully-supervised methods trained on full labels. In particular, our Med-MLLM with 1% training data outperforms previous methods trained with 100% data on the CheXpert and RSNA datasets. Then, in Table 5, we further evaluate the performance of our method on 14 common thorax diseases. The t-tests between the results from Med-MLLM and the best-performing baseline REFERS indicate that the improvement is significant with p < 0.01. As we can see, our approach Med-MLLM (1%) achieves up to 0.4%, 0.5%, 0.1%, and 0.2% absolute improvements upon the current best results trained with full data for diseases–consolidation, effusion, infiltration, and pneumonia, respectively. More encouragingly, with all training labels as in previous works, our Med-MLLM (100%) can outperform these methods across all datasets and diseases. The promising results assess the generalisation capabilities of our approach.
To further evaluate the effectiveness of our framework for rare diseases, we assess the diagnosis performances of existing LLMs, i.e., GPT-2, GPT-3, ChatGPT (GPT-3.5 version), and GPT-428,74 that are released by OpenAI. Since LLMs only accept the text as input, we perform the text-only COVID-19 diagnosis task, which aims to distinguish COVID-19 from non-COVID-19 cases. To obtain the diagnosis accuracy (i.e., disease classification performance) from the LLMs, we take the following text as input: ‘Original Clinical Text’ + ‘Is this a COVID-19 case?’. Then, we sample the probabilities of ‘Yes’ (Pyes) and ‘No’ (Pno) from the next predicted token by GPT. Finally, if Pyes > Pno, we take the ‘Yes’ as the output of LLMs; if Pyes < Pno, we take the ‘No’ as the output of LLMs. In this way, we can obtain the COVID-19 diagnosis accuracy of LLMs. For the ChatGPT and GPT-4, we follow previous works75,76 to incorporate the few-shot prompting28 and chain-of-thought prompting77 strategies. It means that we incorporate five examples, which cover both COVID-19 and non-COVID-19 cases, and instructions as input to request them to generate the response. Therefore, the full input is:
This is just a text classification test. Analyze the report first, then provide the final answer here based on the following examples, which must be either “Yes" or “No".
Report: ‘Original Clinical Text’;
Question: Is this a COVID-19 case?
Answer: Provide the final answer here, which must be either “Yes" or “No".
At last, due to the potential variation in output from ChatGPT, we conduct five runs for each enquiry and select the answer that appears most frequently as the final answer. In addition, it can also be considered as an ensemble approach to achieve better results. Table 6 reports the performances of our method and existing strong LLMs. As we can observe, our approach performs better than several strong LLMs, i.e., GPT-2, GPT-3, and ChatGPT, and achieves a competitive result w.r.t. GPT-4. It is worth noting that although these LLMs have shown great success in natural text understanding, we cannot directly adopt the results provided by ChatGPT in the medical domain78,79.
We perform a robustness analysis to examine whether our method can aid in the COVID-19 diagnosis of new regions by predicting the COVID-19 cases in new regions. To this end, we conduct a cross-region prediction by training the methods on patient data from one region and evaluating the methods on patient data from other regions. In implementations, the BIMCV-COVID-19 dataset collected in Spain, the COVID-HCH dataset collected in China, and the COVID-CXR dataset collected in over 20 countries (excluding Spain and China) are used for the validation. The image-only COVID-19 diagnosis accuracy of our method and previous methods are summarised in Table 7. It shows that our approach consistently outperforms previous methods and achieves solid performances in COVID-19 diagnosis in new regions. In particular, when transferring our approach trained on patient data from Spain to China, we observe an encouraging performance, i.e., 90.1% AUC, which is competitive with the region-specific results of previous works CLIP (80.7% AUC) and BioViL (90.4% AUC), which were obtained by training and testing on the data collected from the same region. Similarly, the cross-region performance of the Spain region (84.8% AUC) of our method, which is trained on China, is competitive with the region-specific result of CLIP (85.4% AUC). These results highlight the transferability and robustness of our approach, leading to a higher-quality diagnosis of rare diseases in new regions than the current methods.
To further assess the effectiveness of our approach in diagnosis, we present to use more labels to conduct continuous learning to train the model continuously. It can evaluate whether the model can continue to be improved when more labelled data are collected as the disease evolves. It is particularly useful in real-world settings. To this end, in Fig. 4, we evaluate the performance of Med-MLLM with respect to the increasing quantity of training labels. Specifically, we evaluate the results on the BIMCV-COVID-19 and COVID-Omicron data for COVID-19 reporting, diagnosis, and prognosis tasks across modalities, languages, and regions. For comparison, we also re-implement the state-of-the-art (SOTA) models (i.e., XProNet63 for reporting and ConVIRT34 for diagnosis and prognosis) using the same training labels to better understand the strengths of our method. We conduct multiple runs with different seeds and report the average performance. As we can see in Fig. 4, for different COVID-19 decision-support tasks, our method Med-MLLM consistently outperforms SOTA with the different numbers of training labels. With more training labels, our method can be continuously improved. It is worth noting that, under the low label setting, e.g., 1% of training labels, our approach surpasses the SOTA by large margins, up to 21.8%, 6.7%, and 6.6% absolute improvements on COVID-19 reporting, diagnosis, and prognosis tasks, respectively. More importantly, with 10% labelled data for training, our method can outperform previous SOTA methods trained with 100% training data. It demonstrates the effectiveness of our approach in relaxing the reliance on the annotations to provide a solid basis for COVID-19 decision-support, which is particularly useful for rare diseases, where the labels are scarce at the early stage.
We provide two intuitive examples to illustrate our approach. Figure 5 shows that our method Med-MLLM can simultaneously generate useful and informative reports across different languages. More importantly, Med-MLLM is able to accurately report important abnormalities, e.g., ‘multiple patchy-like ground glass density shadow’ in the first example, and ‘a lamellar ground glass shadow is seen in the lower lobe of the left lung’ in the second example. It is encouraging that our approach can accurately report abnormalities. Overall, with limited labels, our Med-MLLM can generate informative and “believable” reports for different languages, demonstrating its capability for combating rare diseases.
We further detect the hallucinations and missing facts in the generated reports. To successfully assist physicians and reduce their workloads of writing medical reports, it is important to generate accurate reports (faithfulness or precision), such that the model does not generate hallucinations that “do not exist”. It is also necessary to provide comprehensive facts (comprehensiveness or recall), i.e., the model does not leave out the true findings. To this end, we first employ a medical natural language processing (NLP) tool from the work of CheXpert36, to label the ground truth reports, e.g., [Abnormality_A, Abnormality_B]. Then, we again employ the NLP tool to label the generated reports, e.g., [Abnormality_B, Abnormality_C]. We can find that the model generates a hallucination, i.e., [Abnormality_C], and misses a fact, i.e., [Abnormality_A]. Therefore, we can use this method to calculate the ‘Precision’ and ‘Recall’ scores to preliminary detect the hallucinations and missing facts, respectively. At last, we further calculate the F1 score to obtain the overall performance. Since the NLP tool can extract abnormalities from the English text, we conduct the evaluation on English report generation. For comparison, we also calculate the Precision, Recall, and F1 scores of previous methods, i.e., R2Gen61, KGAE62, and XProNet63. For a fair comparison, both previous methods and our method are trained on 100% of training data. The results are reported in Table 8, showing that our Med-MLLM method surpasses previous methods on all metrics by 5.5%, 3.6%, and 4.6% in terms of Precision, Recall, and F1 scores, respectively. It shows that our approach can generate more faithful reports (i.e., fewer hallucinations) and more comprehensive reports (i.e., fewer missing facts) than previous methods, demonstrating that our method can better assist physicians in reducing their workload.
To better understand the effectiveness of each introduced component, we provide a thorough ablation study of our Med-MLLM in Table 9. It shows that all of our introduced components can bring improvements to downstream tasks. In detail, as image-only pre-training can enable the model to learn broad thorax knowledge, e.g., the diagnostic information, from visual images, removing it would impair the performances (i.e., 73.4% → 57.8% in CIDEr on reporting, 78.2% → 69.4% in AUC on diagnosis, 92.0% → 82.3% AUC on prognosis). The impaired performances assess the effectiveness of learning the important visual characteristics from medical images to support accurate diagnosis and prognosis. Besides, we find that removing the patient-level contrastive learning (PCL) impairs performance across all tasks. By comparing settings (c-e), we notice that, among the introduced three modules in text-only pre-training, the sentence reconstruction module (SR), which can help the model efficiently learn to generate reports, brings the most improvements on reporting. In contrast, the other two modules, MLM and FIA, result in more improvements on diagnosis and prognosis. The image-text pre-training aims to unify the learned medical knowledge from medical images and text. The performance across all tasks decreases when it is removed, showing that unifying visual and textual information can boost the representation of medical data. Overall, the ablation study demonstrates the effectiveness of the Med-MLLM, where all the components can contribute to performance.
At last, to explore the effect of scaling up the number of model parameters, we introduce a larger version of the language model (i.e., Med-MLLM-Large) with 8.9 billion parameters initialized with GatorTron80, where the number of layers is 56, the number of attention heads is 56, and the dimensionality is 3584. For comparison, we perform the evaluation on the text-only COVID-19 diagnosis and prognosis tasks to evaluate the performance of different language models. The results in Table 10 show that the Med-MLLM-Large has better performance than the Med-MLLM-Base by 1.7 ~ 3.6 in AUC values. It not only shows that more model parameters can lead to further improvements, but also demonstrates the potential of LLM that can be further improved in the future by directly scaling up the models.
Methods
In this section, we describe in detail the three main components of our deep learning model.
Image-only pre-training
We first introduce Patient-level Contrastive Learning (Fig. 2a) and then present the image augmentation and regularisation.
Patient-level contrastive learning
We conduct image-only pre-training to learn medical knowledge from the large-scale unlabelled image-only data. Several existing works based on self-supervised learning or contrastive learning81,82,83 have shown the effectiveness of training models on large-scale image-only medical data. In this work, inspired by the success of contrastive learning in natural images45,46,56, we introduce Image-level Contrastive Learning (ICL) and Patient-level Contrastive Learning (PCL) for medical image understanding.
In implementations, for a fair comparison, we choose ResNet-5055 as our basic model to perform the image-only training, while several works84 are based on more powerful models, i.e., Vision Transformer (ViT)85. During training, we first sample a mini-batch of N medical images. Then, for each input medical image, we randomly select the image augmentation functions, e.g., affine transformations (shearing and rotation), colour jittering (contrast and brightness), and random Gaussian blurring30,34,45,46,56, to transform the current medical image into two correlated views of the same image, encoded by ResNet-50 as Vi and Vj, which we consider as a positive pair. As a result, we can obtain a training batch with 2N images. We treat the other 2(N − 1) augmented images as negative examples to Vi. The image-level contrastive learning aims to minimise the distance between positive examples, e.g., Vi and Vj, while maximising the distance between negative examples, e.g., Vi and Vk (k ≠ i, j). To this end, we adopt the ICL loss to train our approach, defined as follows:
where the 〈 ⋅ , ⋅ 〉 denotes the cosine similarity and τ is a temperature hyperparameter46.
While conventional (image-level) contrastive learning can enable the model to understand the input medical images by training the model to distinguish whether the inputted medical images are from the same image or not, it is plausible that this could result in a model that is primarily learning to distinguish images based on the appearance of images, instead of the pathology. To incorporate the characteristics of medical images, we further introduce patient-level contrastive learning (PCL)86,87. In detail, PCL takes two images with completely different views, i.e., Anteroposterior (AP) and Posteroanterior (PA), as input. This approach prevents the model from distinguishing the input images by learning to capture the appearance. Instead, it forces the model to pay more attention to capturing the pathologies in medical images.
In implementations, PCL considers two medical images, e.g., AP and PA views, which are encoded by ResNet-50 as \({V}_{i}^{{\prime} }\) and \({V}_{j}^{{\prime} }\) from the same patient as a positive pair, and the remaining images from other patients in the mini-batch as negative examples. The PCL is defined as follows:
The full training objective of image-only pre-training is defined as: ℓImage = ℓPCL + ℓICL. As we can see, the training of our method does not rely on labelled data, thus, the image-only pre-training could be unsupervised. During training, we exploit the image-only data from several public datasets, including CheXpert36, MIMIC-CXR37, COVID-CXR39,41, COVID-19-CT-CXR43, COVIDx-CXR-240, BIMCV-COVID-1942, RSNA Pneumonia70, and COVID-19 CT44, to conduct the image-only pre-training. As a result, we can learn comprehensive thorax knowledge from image-only data. In particular, when we evaluate the Med-MLLM on a dataset, we will exclude it from the pre-training set.
Image augmentation and regularisation
Since the size of medical image datasets is usually smaller than the size of natural image datasets, such as ImageNet88, we adopt image augmentation strategies to further improve the performance and robustness of our framework. In implementations, we apply random cropping, rotation (−10 to 10 degrees), brightness and contrast adjustment with ratios randomly sampled from [0.8, 1.2], horizontal flipping with 50% probability, and Gaussian blurring with σ ∈ [0.1, 3.0], as used in previous works30,32,34,89. Besides augmentation, we introduce several regularisation methods into our framework. First, we re-write the full training loss of the image-only pre-training as follows:
where λ ∈ [0, 1] is the hyperparameter that controls the regularisation. We set λ = 0.2 according to the performance on the validation set. Meanwhile, the global batch normalisation46, layer normalisation90 and dropout91 are used in regularisation. The experiments show that all the introduced modules contribute to improved performances.
Text-only pre-training
As shown in Fig. 1, we adopt a specialised medical large language model (LLM) with a radiology-specific vocabulary. Meanwhile, as shown in Fig. 2b, we present two training objectives and a text augmentation method to enhance the performance of our LLM.
Large language model (LLM)
In recent years, several efforts75,76,80,92 have been invested to build medical large language models, which have shown great success in processing medical text, such as BioBERT93, ClinicalBERT94, BlueBERT95, and PubMedBERT92. In detail, BioBERT is pre-trained on PubMed47, ClinicalBERT is pre-trained on MIMIC-III48, while BlueBERT combines both corpora for pre-training. All these methods use a vocabulary defined on open-domain text (i.e., Wiki + Books) as in original BERT49. For comparison, PubMedBERT is pre-trained on PubMed47 with a medical vocabulary designed on medical text from PubMed.
As we can see, among the above models, only PubMedBERT designed a domain-specific vocabulary for training; none of the existing LLMs designed a radiology-specific vocabulary. For example, the radiology-specific term ‘cardiomegaly’ will be broken into multiple sub-words (word pieces), i.e., ‘card-io-me-gal-y’ and ‘cardio-me-gal-y’ in the ClinicalBERT and PubMedBERT, respectively. Since most sub-words have no medical relevance, it hinders the LLMs from accurately understanding the radiology-specific medical terms30.
To resolve this, we introduce a radiology-specific vocabulary30 based on the medical texts from PubMed47, MIMIC-III clinical notes48, and MIMIC-CXR medical reports37. Based on the designed radiology-specific vocabulary that includes the whole-word radiology-specific terms (e.g., ‘cardiomegaly’), we perform pre-training of our model on the text-only data from PubMed + MIMIC-III + MIMIC-CXR corpora. In the following, we will introduce the training objectives of our framework in detail.
Training objectives
This section introduces the training objectives used in our method. In implementations, we adopt three training objectives, i.e., Masked Language Modelling (MLM), Sentence Reconstruction (SR), and medical-report-specific Findings-Impression Alignment (FIA).
Masked Language Modelling (MLM). Given a mini-batch of N medical text sequences, following conventional BERT49,50, for each medical text sequence, we randomly mask out the input words with 15% probability, resulting in N sequences of masked words and unmasked words (wm, w\m). The training objective of MLM is to predict the randomly masked words wm based on the remaining unmasked words w\m. Therefore, the MLM loss is defined as:
where p denotes the predicted probability. The masked tokens are predicted as a classification problem by selecting one token from the vocabulary.
Sentence Reconstruction (SR). We further introduce a training objective, sentence reconstruction, to boost the understanding and generation of medical text. As shown in Fig. 2b, we introduce an additional text decoder to reconstruct the input medical text in the auto-encoding pipeline. It means that the decoder takes the input medical text as the ground truth, i.e., W = {w1, w2, …, wM}, for sentence reconstruction. Therefore, the sentence reconstruction loss is defined as:
The training objective is to reconstruct the same input sentence, and it is straightforward for our model to be trained51,62,96 to learn the necessary domain knowledge from the unlabelled medical texts.
Findings-Impression Alignment (FIA). We observe that a medical report contains rich structural information. Typically, it contains a section for “findings” and another section for “impression”, where the former is a paragraph of multiple sentences describing both the normal and abnormal findings in detail, and the latter summarizes a diagnostic conclusion from the findings section. We therefore introduce the training objective FIA30 to exploit the structural information of medical reports.
In implementations, we adopt self-supervised learning and contrastive loss46. We first sample a batch of N medical reports, including N pairs of “Findings” and “Impression” sections. Then, we denote the encoded “Findings” and “Impression” sections of the ith input medical report as (\({T}_{i}^{F},{T}_{i}^{I}\)), which we consider as a positive pair. “Findings” and “Impression” from different medical reports are used as negative pairs. The training loss of FIA is defined as follows:
where the 〈 ⋅ , ⋅ 〉 denotes the cosine similarity and τ is a temperature hyperparameter46. We note that the numerators in both two losses are equal, representing the similarity between \({T}_{i}^{F}\) and \({T}_{i}^{I}\) for the ith positive pair of “Findings” and “Impression”. However, their denominators differ. For the first loss \({\ell }_{i}^{(F\to I)}\), the denominator measures the similarity between the ith “Findings” \(\left({T}_{i}^{F}\right)\) and all other “Impressions”. For \({\ell }_{i}^{(I\to F)}\), the denominator measures the similarity between the ith “Impression” \(\left({T}_{i}^{I}\right)\) and all other “Findings”. Therefore, the two Equations are distinct and respectively reflect the similarity of “Findings” relative to “Impression” (F → I) and “Impression” relative to “Findings” (I → F).
Finally, we obtain the full training objective of FIA by combining the \({\ell }_{i}^{(F\to I)}\) and \({\ell }_{i}^{(I\to F)}\), as follows:
Through the above operation, our method exploits the structural information to improve the understanding of medical texts, and thus boost the performance.
Text augmentation and regularisation
To further improve the performance of our method, we present a text augmentation method and several regularisation methods.
For the text augmentation, we observe that each medical text is composed of multiple sentences, which are usually permutation-invariant97. Therefore, we can randomly shuffle the sentences to augment the medical texts to boost performance.
Meanwhile, we introduce α and β for better regularisation. The full training objective of text-only pre-training ℓText is defined as follows:
In implementations, the α and β are set to 0.5 and 0.1, respectively, according to the performances on the validation set. In detail, our framework is first trained using MLM (ℓMLM), then is trained using the combination of MLM and FIA, and finally is trained on the full training objective ℓText.
Image-text pre-training
Most recently, several image-text pre-training methods30,32,34,44 have been proposed to demonstrate the importance of unifying the images and texts to improve the understanding of medical data. However, all existing methods mainly adopt supervised training and heavily rely on large-scale coupled image-report pairs for training, while collecting labelled and paired medical data across different modalities is typically very costly and time-consuming. To this end, we introduce the image-text pre-training to relax the reliance on the labelled image-text pairs89.
Soft image-text alignment (SITA)
As shown in Fig. 2 (c), we incorporate a knowledge base and a pre-training objective, i.e., Soft Image-Text Alignment (SITA)89,98. In particular, given a mini-batch of N randomly sampled pairs of images and texts, we adopt MetaMap99 to extract entities defined in the Unified Medical Language System (UMLS)53 from the ith medical text. Following previous works36,37,62,89,100,101, we focus on the 14 common radiographic entities (Atelectasis, Cardiomegaly, Consolidation, Edema, Enlarged Cardiomediastinum, Fracture, Lung Lesion, Lung Opacity, No Finding, Pleural Effusion, Pleural Other, Pneumonia, Pneumothorax, Support Devices). As a result, given the medical text, e.g., "A right pleural effusion. Heart size is enlarged. No evidence of pneumothorax”, we can extract two entities, pleural effusion and cardiomegaly. Then, we construct a multi-hot vector \({H}_{i}^{T}\) of dimension 14 from the extracted entities, where 1/0 denotes the presence/absence of the radiographic entity. Similarly, for the jth medical image with diagnosis labels, we again adopt MetaMap99 to extract radiographic entities by mapping the raw diagnosis labels of medical images to UMLS concepts, e.g., “Normal” will be mapped to “No Findings”. As a result, the images and the texts can share the same radiographic entities. Then, we can construct a multi-hot vector \({H}_{j}^{V}\) of dimension 14 for the image. At last, we calculate the cosine similarity of \({H}_{i}^{T}\) and \({H}_{j}^{V}\) to measures the similarity of the ith text and the jth image. In this way, we measure the similarity between any text and image. The target similarity score \({s}_{ij}^{(T\to V)}\) between the ith text and the jth image is calculated as:
where 〈 ⋅ , ⋅ 〉 denotes the cosine similarity and τ is a temperature parameter. Similarly, we can obtain the target similarity score \({s}_{ji}^{(V\to T)}\) between the jth image and the ith text:
\({s}_{ij}^{(T\to V)}\) and \({s}_{ji}^{(V\to T)}\) are used as the soft target labels of image-text alignment in the image-text pre-training, which will be introduced as follows.
To perform the image-text pre-training, we first use the BERT49 and ResNet-5055 to encode the ith text and jth image, resulting in Ti and Vj, respectively. Therefore, the predicted similarity score \({{s}_{ij}^{{\prime} }}^{(T\to V)}\) between the ith text and the jth image and the predicted similarity score \({{s}_{ji}^{{\prime} }}^{(V\to T)}\) between the jth image and the ith text are calculated by:
At last, the soft image-text alignment (SITA) loss is implemented by the cross entropy loss:
Through the SITA, our method performs image-text pre-training to exploit unpaired medical images and texts to efficiently and accurately align medical data across modalities89.
Data augmentation and regularisation
Similarly, we introduce image augmentation in image-only pre-training and text augmentation in text-only pre-training to further boost the robustness and thus improve the performance of our method.
More importantly, during the regularisation, we incorporate the MLM loss for joint training, resulting in the full training objective of image-text pre-training as follows:
In implementations, γ controls the regularisation and is set to 2, according to the performances on the validation set. Our preliminnarly experiments show the effectiveness of performing continuous MLM optimisation.
Experiment settings
For a fair comparison, we adopt the ResNet-5055 as the image encoder and the BERT5,102 as the text encoder. The number of encoder layers is set to 6 and the dimension of the latent states is 768 unless otherwise stated. Meanwhile, we also explored a larger version of the language model49,80 with 8.9 billion parameters, where the number of layers is 56, the number of attention heads is 56, and the dimensionality of the latent states is 3584. We adopt the AdamW optimiser103 for training. We train our model in the order of image-only, text-only, and image-text pre-training. During image-only/text-only/image-text pre-training: the hyper-parameter τ is set to 0.5/0.5/0.1 according to the average performances on the validation sets; we use a learning rate of 10−3/2 × 10−5/5 × 10−5 and a batch size of 256/256/100. During fine-tuning, we use a batch size of 32/64/16 and a learning rate of 10−4 for parameter optimisation on the COVID-19 reporting/diagnosis/prognosis task. Our code is implemented in PyTorch104. During testing, we add a text decoder, i.e., Transformer5, to perform the reporting task, and add a fully connected layer to perform the diagnosis and prognosis tasks.
Ethical considerations
Our study was conducted on thirteen datasets, in which all Protected Health Information (PHI), e.g., patient name, sex, gender, and date of birth, is officially de-identified for all datasets used in our experiments. It means that the deletion of PHI from structured data sources (e.g., database fields that provide age, genotypic information, past and current diagnosis and treatment categories) is performed in compliance with the Health Insurance Portability and Accountability Act (HIPAA) standards in order to facilitate public access to the datasets.
Recruitment statement
We do not recruit any new human research participants for this study. For the public data, all necessary patient/participant consent has been obtained and the appropriate institutional forms have been officially archived.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data used in our work may be available for research purposes from the corresponding authors upon reasonable request.
1) CheXpert is available at https://stanfordmlgroup.github.io/competitions/chexpert/.
2) COVIDx-CXR-2 is available at https://alexswong.github.io/COVID-Net/.
3) MIMIC-CXR is available at https://physionet.org/content/mimic-cxr/2.0.0/.
4) COVID-19-CT-CXR is available at https://github.com/ncbi-nlp/COVID-19-CT-CXR.
5) COVID-19 CT is available at https://covid19ct.github.io/.
6) COVID-CXR is available at https://github.com/ieee8023/covid-chestxray-dataset.
7) BIMCV-COVID-19 is available at https://bimcv.cipf.es/bimcv-projects/bimcv-covid19/.
8) PubMed is available at https://pubmed.ncbi.nlm.nih.gov/download/.
9) MIMIC-III is available at https://physionet.org/content/mimiciii/1.4/.
10) SIIM-ACR is available at https://www.kaggle.com/c/siim-acr-pneumothorax-segmentation.
11) RSNA is available at https://www.kaggle.com/c/rsna-pneumonia-detection-challenge.
12) NIH ChestX-ray is available at https://nihcc.app.box.com/v/ChestXray-NIHCC.
13) Shenzhen Tuberculosis is available at: https://www.kaggle.com/raddar/tuberculosis-chest-xrays-shenzhen.
Code availability
The code that supports the findings of this study is available from the corresponding authors upon reasonable request.
References
Roberts, M. et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for covid-19 using chest radiographs and ct scans. Nat. Mach. Intell. 3, 199–217 (2021).
Driggs, D. et al. Machine learning for covid-19 diagnosis and prognostication: lessons for amplifying the signal while reducing the noise. Radiol. Artif. Intell. 3, e210011 (2021).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems (Curran Associates, Inc., 2012).
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017).
Vaswani, A. et al. Attention is all you need. In Advances in Neural Information Processing Systems (Curran Associates, Inc., 2017).
Lyu, W. et al. A multimodal transformer: Fusing clinical notes with structured EHR data for interpretable in-hospital mortality prediction. In American Medical Informatics Association Annual Symposium (AMIA, 2022).
Jing, B., Xie, P. & Xing, E. P. On the automatic generation of medical imaging reports. In Annual Meeting of the Association for Computational Linguistics (Association for Computational Linguistics, 2018).
Liu, G. et al. Clinically accurate chest x-ray report generation. In Machine Learning for Healthcare Conference (PMLR, 2019).
Li, Y., Liang, X., Hu, Z. & Xing, E. P. Hybrid retrieval-generation reinforced agent for medical image report generation. In Advances in Neural Information Processing Systems (Curran Associates, Inc., 2018).
Liu, F., Ge, S. & Wu, X. Competence-based multimodal curriculum learning for medical report generation. In Annual Meeting of the Association for Computational Linguistics (Association for Computational Linguistics, 2021).
Liu, F., Wu, X., Ge, S., Fan, W. & Zou, Y. Exploring and distilling posterior and prior knowledge for radiology report generation. In IEEE Conference on Computer Vision and Pattern Recognition (IEEE, 2021).
Bhattacharya, S. et al. Deep learning and medical image processing for coronavirus (covid-19) pandemic: a survey. Sustain. Cities Soc. 65, 102589 (2021).
Soomro, T. A. et al. Artificial intelligence (ai) for medical imaging to combat coronavirus disease (covid-19): a detailed review with direction for future research. Artif. Intell. Rev. 55, 1409–1439 (2022).
El-Sadr, W. M., Vasan, A. & El-Mohandes, A. Facing the new covid-19 reality. N. Engl. J. Med. 388, 385–387 (2023).
Carlile, M. et al. Deployment of artificial intelligence for radiographic diagnosis of covid-19 pneumonia in the emergency department. J. Am. Coll. Emerg. Phys. Open 1, 1459–1464 (2020).
Wu, X. et al. Deltanet: Conditional medical report generation for COVID-19 diagnosis. In International Conference on Computational Linguistics (International Committee on Computational Linguistics, 2022).
You, D. et al. Aligntransformer: Hierarchical alignment of visual regions and disease tags for medical report generation. In Medical Image Computing and Computer Assisted Intervention, 72–82 (Springer, 2021).
Sinsky, C. et al. Allocation of physician time in ambulatory practice: a time and motion study in 4 specialties. Ann. Internal Med. 165, 753–760 (2016).
Weiner, M. & Biondich, P. The influence of information technology on patient-physician relationships. J. General Internal Med. 21, 35–39 (2006).
Tawfik, D. S. et al. Physician burnout, well-being, and work unit safety grades in relationship to reported medical errors. In Mayo Clinic Proceedings, 1571–1580 (Elsevier, 2018).
West, C. P., Dyrbye, L. N. & Shanafelt, T. D. Physician burnout: contributors, consequences and solutions. J. Internal Med. 283, 516–529 (2018).
Zhou, S. K. et al. A review of deep learning in medical imaging: imaging traits, technology trends, case studies with progress highlights, and future promises. Proc. IEEE 109, 820–838 (2021).
Liu, F. et al. Retrieve, reason, and refine: Generating accurate and faithful patient instructions. In Advances in Neural Information Processing Systems (Curran Associates, Inc., 2022).
Zu, Z. Y. et al. Coronavirus disease 2019 (covid-19): a perspective from china. Radiology 296, E15–E25 (2020).
Fang, Y. et al. Sensitivity of chest ct for covid-19: comparison to rt-pcr. Radiology 296, E115–E117(2020).
Ng, M.-Y. et al. Imaging profile of the covid-19 infection: radiologic findings and literature review. Radiology: Cardiothoracic Imaging 2, e200034 (2020).
Rubin, G. D. et al. The role of chest imaging in patient management during the covid-19 pandemic: a multinational consensus statement from the fleischner society. Radiology 296, 172–180 (2020).
Brown, T. et al. Language models are few-shot learners. In Advances in Neural Information Processing Systems (Curran Associates, Inc., 2020).
OpenAI. Gpt-4 technical report. Preprint at https://arxiv.org/abs/2303.08774 (2023).
Boecking, B. et al. Making the most of text semantics to improve biomedical vision–language processing. In European Conference on Computer Vision, 1–21 (Springer, 2022).
Zhou, H.-Y., Lian, C., Wang, L. & Yu, Y. Advancing radiograph representation learning with masked record modeling. In The Eleventh International Conference on Learning Representations (OpenReview.net, 2023).
Zhou, H. et al. Generalized radiograph representation learning via cross-supervision between images and free-text radiology reports. Nat. Mach. Intell. 4, 32–40 (2022).
Tiu, E. et al. Expert-level detection of pathologies from unannotated chest x-ray images via self-supervised learning. Nat. Biomed. Eng. 6, 1399–1406 (2022).
Zhang, Y., Jiang, H., Miura, Y., Manning, C. D. & Langlotz, C. P. Contrastive learning of medical visual representations from paired images and text. In Machine Learning for Healthcare Conference, 2–25 (2022).
Desai, S. B., Pareek, A. & Lungren, M. P. Deep learning and its role in covid-19 medical imaging. Intell. Based Med. 3, 100013 (2020).
Irvin, J. et al. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In Association for the Advancement of Artificial Intelligence (AAAI Press, 2019).
Johnson, A. E. et al. Mimic-cxr, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data 6, 317 (2019).
Rahman, T. et al. Exploring the effect of image enhancement techniques on COVID-19 detection using chest x-ray images. Comput. Biol. Med. 132, 104319 (2021).
Cohen, J. P. et al. Covid-19 image data collection: prospective predictions are the future. Mach. Learn. Biomed. Imaging 1, 1–10 (2020).
Pavlova, M. et al. Covid-net cxr-2: an enhanced deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Front. Med. 9, 861680 (2022).
Cohen, J. P., Morrison, P. & Dao, L. COVID-19 image data collection. Preprint at https://arxiv.org/abs/2003.11597 (2020).
Vayá, M. D. L. I. et al. Bimcv covid-19+: a large annotated dataset of rx and ct images from covid-19 patients. Preprint at https://arxiv.org/abs/2006.01174 (2020).
Peng, Y. et al. COVID-19-CT-CXR: A freely accessible and weakly labeled chest x-ray and CT image collection on COVID-19 from biomedical literature. IEEE Trans. Big Data 7, 3–12 (2021).
Liu, G. et al. Medical-vlbert: Medical visual language BERT for COVID-19 CT report generation with alternate learning. IEEE Trans. Neural Networks Learn. Syst. 32, 3786–3797 (2021).
He, K., Fan, H., Wu, Y., Xie, S. & Girshick, R. B. Momentum contrast for unsupervised visual representation learning. In IEEE Conference on Computer Vision and Pattern Recognition (IEEE, 2020).
Chen, T., Kornblith, S., Norouzi, M. & Hinton, G. E. A simple framework for contrastive learning of visual representations. In International Conference on Machine Learning (PMLR, 2020).
National Institutes of Health. PubMed Corpora, https://pubmed.ncbi.nlm.nih.gov/download/ (2022).
Johnson, A. E. W. et al. MIMIC-III, a freely accessible critical care database. Sci. Data 3 160035 (2016).
Devlin, J., Chang, M., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (Association for Computational Linguistics, 2019).
Liu, Y. et al. Roberta: A robustly optimized BERT pretraining approach. Preprint at https://arxiv.org/abs/1907.11692 (2019).
Tschannen, M., Bachem, O. & Lucic, M. Recent advances in autoencoder-based representation learning. Preprint at https://arxiv.org/abs/1812.05069 (2018).
Casey, A. et al. A systematic review of natural language processing applied to radiology reports. BMC Med. Inf. Decis. Mak. 21, 179 (2021).
Bodenreider, O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res. 32, D267–D270 (2004).
Radford, A. et al. Learning transferable visual models from natural language supervision. In International conference on machine learning, 8748–8763 (PMLR, 2021).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In IEEE Conference on Computer Vision and Pattern Recognition (IEEE, 2016).
Chen, X., Fan, H., Girshick, R. B. & He, K. Improved baselines with momentum contrastive learning. Preprint at https://arxiv.org/abs/2003.04297 (2020).
Narin, A., Kaya, C. & Pamuk, Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal. Appl. 24, 1207–1220 (2021).
Ozturk, T. et al. Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 121, 103792 (2020).
Irmak, E. A novel deep convolutional neural network model for covid-19 disease detection. In Medical Technologies Congress, 1–4 (IEEE, 2020).
Hall, L. O., Paul, R., Goldgof, D. B. & Goldgof, G. M. Finding covid-19 from chest x-rays using deep learning on a small dataset. Preprint at https://arxiv.org/abs/2004.02060 (2020).
Chen, Z., Song, Y., Chang, T. & Wan, X. Generating radiology reports via memory-driven transformer. In Conference on Empirical Methods in Natural Language Processing (Association for Computational Linguistics, 2020).
Liu, F. et al. Auto-encoding knowledge graph for unsupervised medical report generation. In Advances in Neural Information Processing Systems (Curran Associates, Inc., 2021).
Wang, J., Bhalerao, A. & He, Y. Cross-modal prototype driven network for radiology report generation. In European Conference on Computer Vision (Springer, 2022).
Liu, F. et al. Contrastive attention for automatic chest x-ray report generation. In Findings of the Association for Computational Linguistics: ACL-IJCNLP 2021, 269-280 (Association for Computational Linguistics, 2021).
Papineni, K., Roukos, S., Ward, T. & Zhu, W. BLEU: a method for automatic evaluation of machine translation. In Annual Meeting of the Association for Computational Linguistics (Association for Computational Linguistics, 2002).
Lin, C.-Y. ROUGE: A package for automatic evaluation of summaries. In Annual Meeting of the Association for Computational Linguistics (Association for Computational Linguistics, 2004).
Vedantam, R., Zitnick, C. L. & Parikh, D. Cider: Consensus-based image description evaluation. In IEEE Conference on Computer Vision and Pattern Recognition (IEEE, 2015).
Chen, X. et al. Microsoft COCO captions: data collection and evaluation server. Preprint at https://arxiv.org/abs/1504.00325 (2015).
Wang, X. et al. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In IEEE Conference on Computer Vision and Pattern Recognition (IEEE, 2017).
Shih, G. et al. Augmenting the national institutes of health chest radiograph dataset with expert annotations of possible pneumonia. Radiol. Artif. Intell. 1 e180041 (2019).
Society for Imaging Informatics in Medicine (SIIM). Siim-acr pneumothorax segmentation. In Kaggle, https://www.kaggle.com/c/siim-acr-pneumothorax-segmentation (2019).
Jaeger, S. et al. Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Medm Surg. 4, 475 (2014).
Huang, S., Shen, L., Lungren, M. P. & Yeung, S. Gloria: A multimodal global-local representation learning framework for label-efficient medical image recognition. In International Conference on Computer Vision, 3922–3931 (IEEE, 2021).
Radford, A. et al. Language models are unsupervised multitask learners. OpenAI Blog 1, 9 (2019).
Singhal, K. et al. Large language models encode clinical knowledge. Nature 620, 172–180 (2023).
Singhal, K. et al. Towards expert-level medical question answering with large language models. Preprint at https://arxiv.org/abs/2305.09617 (2023).
Wei, J. et al. Chain-of-thought prompting elicits reasoning in large language models. Adv. Neural Inf. Proces. Syst. 35, 24824–24837 (2022).
Shen, Y. et al. Chatgpt and other large language models are double-edged swords. Radiology 307, e230163 (2023).
Kitamura, F. C. Chatgpt is shaping the future of medical writing but still requires human judgment. Radiology 307, 230171 (2023).
Yang, X. et al. A large language model for electronic health records. NPJ Digital Med. 5, 194 (2022).
Haghighi, F., Taher, M. R. H., Zhou, Z., Gotway, M. B. & Liang, J. Transferable visual words: Exploiting the semantics of anatomical patterns for self-supervised learning. IEEE Trans. Med. Imaging 40, 2857–2868 (2021).
Zhou, Z., Sodha, V., Pang, J., Gotway, M. B. & Liang, J. Models genesis. Med. Image Anal. 67, 101840 (2021).
Zhou, H. et al. Comparing to learn: Surpassing imagenet pretraining on radiographs by comparing image representations. In International Conference on Medical Image Computing and Computer Assisted Intervention, 398–407 (Springer, 2020).
Shamshad, F. et al. Transformers in medical imaging: a survey. Medical Image Anal. 88, 102802 (2023).
Dosovitskiy, A. et al. An image is worth 16x16 words: transformers for image recognition at scale. In International Conference on Learning Representations (OpenReview.net, 2021).
Vu, Y. N. T. et al. Medaug: Contrastive learning leveraging patient metadata improves representations for chest x-ray interpretation. In Machine Learning for Healthcare Conference, 755–769 (PMLR, 2021).
Kiyasseh, D., Zhu, T. & Clifton, D. A. Clocs: Contrastive learning of cardiac signals across space, time, and patients. In International Conference on Machine Learning, 5606–5615 (PMLR, 2021).
Deng, J. et al. Imagenet: a large-scale hierarchical image database. In IEEE Conference on Computer Vision and Pattern Recognition (IEEE, 2009).
Wang, Z., Wu, Z., Agarwal, D. & Sun, J. Medclip: contrastive learning from unpaired medical images and text. In Conference on Empirical Methods in Natural Language Processing, 3876–3887 (Association for Computational Linguistics, 2022).
Ba, L. J., Kiros, R. & Hinton, G. E. Layer normalization. Preprint at https://arxiv.org/abs/1607.06450 (2016).
Srivastava, N., Hinton, G. E., Krizhevsky, A., Sutskever, I. & Salakhutdinov, R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014).
Gu, Y. et al. Domain-specific language model pretraining for biomedical natural language processing. ACM Trans. Comput. Heal. 3, 2:1–2:23 (2022).
Lee, J. et al. Biobert: a pre-trained biomedical language representation model for biomedical text mining. Bioinform. 36, 1234–1240 (2020).
Alsentzer, E. et al. Publicly available clinical BERT embeddings. In Proceedings of the 2nd Clinical Natural Language Processing Workshop (Association for Computational Linguistics, 2019).
Peng, Y., Yan, S. & Lu, Z. Transfer learning in biomedical natural language processing: an evaluation of BERT and elmo on ten benchmarking datasets. In BioNLP@Annual Meeting of the Association for Computational Linguistics, 58–65 (Association for Computational Linguistics, 2019).
Wang, Y., Yao, H. & Zhao, S. Auto-encoder based dimensionality reduction. Neurocomputing 184, 232–242 (2016).
Preechakul, K. et al. Set prediction in the latent space. In Advances in Neural Information Processing Systems, 25516–25527 (Curran Associates, Inc., 2021).
Liu, F., Liu, Y., Ren, X., He, X. & Sun, X. Aligning visual regions and textual concepts for semantic-grounded image representations. In Adv. Neural Inf. Proces. Syst. (Curran Associates, Inc., 2019).
Aronson, A. R. & Lang, F.-M. An overview of metamap: historical perspective and recent advances. J. Am. Med. Inf. Assoc. 17, 229–236 (2010).
Peng, Y. et al. Negbio: a high-performance tool for negation and uncertainty detection in radiology reports. AMIA Summits Transl. Sci. Proc. 2018, 188 (2018).
Wang, X., Peng, Y., Lu, L., Lu, Z. & Summers, R. M. Tienet: Text-image embedding network for common thorax disease classification and reporting in chest x-rays. In IEEE Conference on Computer Vision and Pattern Recognition (IEEE, 2018).
Wolf, T. et al. Huggingface’s transformers: State-of-the-art natural language processing. Preprint at https://arxiv.org/abs/1910.03771 (2019).
Loshchilov, I. & Hutter, F. Decoupled weight decay regularization. In International Conference on Learning Representations (OpenReview.net, 2019).
Paszke, A. et al. Pytorch: An imperative style, high-performance deep learning library. In Advances in Neural Information Processing Systems (Curran Associates, Inc., 2019).
Chen, L. et al. Self-supervised learning for medical image analysis using image context restoration. Med. Image Anal. 58, 101539 (2019).
Acknowledgements
DAC was supported by an NIHR Research Professorship and a Royal Academy of Engineering Research Chair; the NIHR Oxford Biomedical Research Centre; the Hong Kong Centre for Cerebro-cardiovascular Engineering (COCHE); and the Pandemic Sciences Institute, University of Oxford, Oxford, UK. F.L. gratefully acknowledges funding from the Clarendon Fund and the Magdalen Graduate Scholarship. T.Z. was supported by the Royal Academy of Engineering under the Research Fellowship scheme. C.W. gratefully acknowledges funding from the Clarendon Fund and the Sloane Robinson Scholarship. Y.Y. was supported by the Science and Technology Committee of Shanghai Municipality (23ZR1436400). We sincerely thank all the anonymous reviewers and editors for their constructive comments and suggestions that substantially improved this paper. D.A.C. and F.L. are the corresponding authors of this paper.
Author information
Authors and Affiliations
Contributions
D.A.C. conceived the project. F.L. conceived and designed the study, performed the data analysis, and prepared the manuscript. T.Z., X.W. and B.Y. contributed to the implementations, experiments, and results interpretation. All authors contributed to the final manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, F., Zhu, T., Wu, X. et al. A medical multimodal large language model for future pandemics. npj Digit. Med. 6, 226 (2023). https://doi.org/10.1038/s41746-023-00952-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41746-023-00952-2