Abstract
Genetic alternation of REarranged during Transfection (RET) that leads to constitutive RET activation is a crucial etiological factor for thyroid cancer. RET is known to regulate mitochondrial processes, although the underlying molecular mechanisms remain unclear. We previously showed that the multi-kinase inhibitors vandetanib and cabozantinib increase the mitochondrial membrane potential (Δψm) in RET-mutated thyroid tumor cells and that this effect can be exploited to increase mitochondrial enrichment of Δψm-sensitive agents in the tumor cells. In this study, we hypothesized that the RET-selective inhibitor, selpercatinib, can increase Δψm and, subsequently, tumor cell uptake of the mitochondria-targeted ubiquinone (MitoQ) to the level to break the mitochondrial homeostasis and induce lethal responses in RET-mutated thyroid tumor cells. We show that selpercatinib significantly increased Δψm, and its combination with MitoQ synergistically suppressed RET-mutated human thyroid tumor cells, which we validated using RET-targeted genetic approaches. Selpercatinib and MitoQ, in combination, also suppressed CCDC6-RET fusion cell line xenografts in mice and prolonged animal survival more effectively than single treatments of each agent. Moreover, we treated two patients with CCDC6-RET or RETM918T thyroid cancer, who could not take selpercatinib at regular doses due to adverse effects, with a dose-reduced selpercatinib and MitoQ combination. In response to this combination therapy, both patients showed tumor reduction. The quality of life of one patient significantly improved over a year until the tumor relapsed. This combination of selpercatinib with MitoQ may have therapeutic potential for patients with RET-mutated tumors and intolerant to regular selpercatinib doses.
Similar content being viewed by others
Introduction
RET (REarranged during Transfection) is a proto-oncogene that encodes a cell surface receptor tyrosine kinase (RTK) and is subject to various genetic alternations1. For example, RET fusions, which result in ligand-independent dimerization and constitutive activation of RET2,3,4, have been identified in 10–20% of papillary thyroid cancer (PTC)5,6,7, 1–2% of non-small cell lung cancer8,9,10, and 0.2–1.6% of colorectal cancer11,12,13. Somatic and germline RET mutations leading to constitutive RET activation are also found in cancers, including multiple endocrine neoplasia type 2 A and 2B (MEN2A and MEN2B) syndromes and familial medullary thyroid carcinoma (MTC)14,15. Upon activation, RET promotes various cellular processes, including cell growth, proliferation, and survival, mainly through the mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK), the phosphatidylinositol-3 kinase/protein kinase B (AKT)16, and the Janus kinase/signal transducer and activator of transcription pathways2.
RET mutation is an actionable therapeutic target. While vandetanib and cabozantinib have been approved by the Food and Drug Administration (FDA) for RET-mutant MTC and RET fusion-positive differentiated thyroid cancer17,18,19, these inhibitors are not RET-selective and ineffective against RETV804 gatekeeper mutations. Selpercatinib (RETEVMO™) and pralsetinib (GAVRETO™) are highly selective RET inhibitors designed to overcome the gatekeeper mutations20,21 and approved by the FDA in 2020 for RET-mutant thyroid and lung cancers22,23,24. Moreover, in September 2022, the FDA granted accelerated approval to selpercatinib for tumor-agnostic treatment of adult patients with locally advanced or metastatic solid RET tumors25. While generally very well tolerated, these inhibitors cause adverse effects in patients, including hypertension and hepatotoxicity as the most common adverse effects23,24. These adverse effects cause permanent discontinuation of selpercatinib in about 5%, dosage interruptions in about 42%, and dose reductions in about 31% of the patients23. Therefore, it is important to design a strategy to use the drug effectively while reducing its adverse effects.
We previously showed that low-dose vandetanib and cabozantinib can synergize with mitochondria-targeted agents to suppress MEN2A and MEN2B MTC cells26. Mechanistically, these inhibitors increased mitochondrial membrane potential (Δψm) in the RET tumor cells, subsequently facilitating tumor cell uptake and retention of Δψm-sensitive triphenyl phosphonium (TPP)-conjugated carboxy-proxyl (MitoCP) and ubiquinone (MitoQ). Consequent enrichment of MitoCP and MitoQ beyond tumor cell tolerance disrupted mitochondrial homeostasis, triggering tumor cell death26,27. The TPP moiety has been used as a vehicle for mitochondria targeting of different functional moieties28,29. Of note, while MitoQ is currently used as a dietary supplement due to its beneficial effects on mitochondrial bioenergetics30,31 and vascular function32,33,34, it also suppresses tumor cells derived from different tumors, including MTC26, melanoma35, breast cancer36,37,38, and pancreatic cancer39. This tumor suppressive effect is mainly attributed to higher MitoQ accumulation in tumor cells due to higher basal Δψm in tumor cells. This information suggests that MitoQ could be a safe agent for tumor suppression if its tumor cell enrichment is selectively facilitated.
In this study, we reasoned that Δψm-inducing effects of vandetanib and cabozantinib are due to RET inhibition and hypothesized that a low dose of selpercatinib will also increase Δψm in RET-mutated tumor cells, priming the tumor cells to MitoQ sensitivity. We demonstrate that a combination of selpercatinib and MitoQ can effectively and synergistically suppress RET-mutated thyroid cancers.
Results
Selpercatinib increases Δψm in CCDC6-RET fusion mutated PTC cells
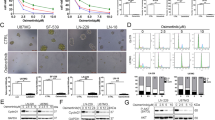
We determined whether selpercatinib can increase Δψm in TPC1 cells, the only human PTC cell line that carries a coiled-coil domain containing 6 (CCDC6)-RET fusion mutation, which is also known as RET-PTC140. As determined by a dose-dependent analysis, we found that 72-hour treatment of selpercatinib between 1 nM and 1 µM dose ranges significantly decreased TPC1 cell viability with IC50 of 15 nM (Supplementary Fig. 1a). Western blot analysis of these cells revealed that selpercatinib substantially decreased RET phosphorylation at Tyr905, an activating autophosphorylation16,41,42, and phosphorylation of ERK1/2 and AKT, the key downstream effectors of RET16, while increasing expression of the cyclin-dependent kinase inhibitor p27KIP1 but decreasing p21CIP1 expression (Supplementary Fig. 1b, c). These effects are consistent with those of other RET inhibitors in this cell line43,44. Under these conditions validating selpercatinib potency, TPC1 cells stained with tetramethyl-rhodamine methyl ester (TMRM), a Δψm-sensitive fluorescent dye, significantly increased in a selpercatinib dose-dependent manner (Fig. 1a, b). We confirmed that these selpercatinib effects are specific to the depletion of RET activity using RNA interference. Similar as selpercatinib, two short hairpin RNA (shRNA) constructs targeting different regions of RET mRNA consistently increased Δψm in TPC1 cells (Fig. 1c, d), while inducing growth inhibitory effects (Supplementary Fig. 1d–f). To further determine RET-specificity of this effect, we also used the immortalized normal rat thyrocytes PCCL3 expressing a doxycycline-inducible human nuclear receptor coactivator 4 (NCOA4)-RET45, which is the second most common RET fusion in PTC46. Indeed, selpercatinib increased Δψm in this cell line model only upon doxycycline treatment (Fig. 1e, f). These data suggest that depletion of RET activity increases Δψm in RET-mutated PTC cells.
a TPC1 cells, treated with selpercatinib (selp) in a 3.125 to 50 nM dose range for 2 days, were stained with TMRM. Cellular TMRM retention was analyzed by flow cytometry measuring yellow fluorescence. b Mean fluorescence intensities (MFI) of TMRM-stained cells in a quantified by FCS Express software. Data are mean ± SEM (N = 4). ***P < 0.001 (compared with no treatment), one-way ANOVA with Bonferroni post-tests. c TPC1 cells, infected with lentiviral pLKO.1-shRET#1 and shRET#2 for 2 days, were stained with TMRM. Cellular TMRM retention was analyzed by flow cytometry measuring yellow fluorescence. d MFI of TMRM-stained cells in c quantified by FCS Express software. Data are mean ± SEM (N = 3). *P < 0.05 (compared with pLKO.1), one-way ANOVA with Bonferroni post-tests. e PCCL3 cells stably expressing a doxycycline (Dox)-inducible NCOA4-RET were treated with different doses of selpercatinib for 48 hours in the presence or absence of 0.5 µg/mL doxycycline. MFI of TMRM-stained cells was quantified by FCS Express software. Data are mean ± SEM (N = 4). **P < 0.005, ***P < 0.001 (compared with no doxycycline), two-way ANOVA with Bonferroni post-tests. f Western blot analysis of total lysates of PCCL3 cells with or without 48-hour doxycycline treatment. g TPC1 cells pretreated with selpercatinib for 2 days were treated with 2 μM MitoCP for 1 hour. Mitochondrial lysates of these cells were analyzed by western blotting to detect the formation of TPP-adducts using an antibody specific to the TPP moiety of MitoCP. Total proteins were visualized in the stain-free gel as the control for equal protein loading. h Densitometry of the Western blot signals collected from three independent experiments performed as described in g. Signals were normalized for total protein intensity. Data are mean ± SEM (N = 3). **P < 0.005 (compared with no treatment), one-way ANOVA with Bonferroni post-tests. i Western blotting to detect TPP-adducts in the mitochondrial lysates of TPC1 cells, infected with pLKO.1-shRET#1 and shRET#2 for 2 days, and then treated with 2 μM MitoCP for 1 hour. Total proteins visualized in the stain-free gel are the control for equal protein loading. j Densitometry of the Western blot signals collected from three independent experiments performed as described in i. Data are mean ± SEM (N = 3). *P < 0.05 (compared with pLKO.1), one-way ANOVA with Bonferroni post-tests.
Next, we determined whether selpercatinib can sufficiently increase mitochondrial enrichment of a Δψm-sensitive agent, using MitoCP as a tool compound. MitoCP contains TPP and the CP moiety, a 5-membered nitroxide free radical that can form a covalent conjugate with a thiol protein, which can be detected by a TPP-specific antibody26,47. For this, TPC1 cells pretreated with selpercatinib, were treated with MitoCP for 1 hour. When mitochondria fractions extracted from selpercatinib-pretreated cells were compared with the fractions extracted from unpretreated cells, much higher levels of TPP-protein adducts were detected by Western blotting in the cells pretreated with selpercatinib (Fig. 1g, h). Consistently, RET knockdown also increased TPP-protein adducts in TPC1 cells (Fig. 1i, j). These data suggest that selpercatinib can increase mitochondrial enrichment of a Δψm-sensitive agent in RET-mutated tumor cells, similar as vandetanib and cabozantinib26.
Selpercatinib and MitoQ can synergistically suppress the viability of RET-mutant PTC and MTC cells in culture
As the first step to evaluate the efficacy of MitoQ and its combination with selpercatinib in RET-mutant tumor cells, we validated that mitochondrial enrichment of its functional moiety, ubiquinone, is critical for its growth inhibitory effects in TPC1 cells. As determined by the crystal violet staining, MitoQ treatment for 48 hours significantly decreased TPC1 cell viability, exhibiting the IC50 value at 250 nM (Fig. 2a). In contrast, the functional moiety, CoQ10, did not significantly affect the cell viability while the vehicle, TPP, decreased cell viability only mildly at higher doses when these compounds were compared with MitoQ in identical dose ranges (Fig. 2a).
a TPC1 cells in 12 well plates were treated with increasing doses of MitoQ, CoQ10, and TPP for 48 hours prior to crystal violet viability assay. Data are expressed as the percentage of untreated controls. Data are mean ± SEM (N = 3). b TPC1 cells, pretreated with different concentrations of selpercatinib, were treated with different doses of MitoQ for 48 hours prior to crystal violet viability assay. Data are mean ± SEM (N = 4). c SynergyFinder-generated plot of the viability data in b. The ZIP (zero interaction potency) score is indicated for the most synergistic area. d TPC1 cells, infected with lentiviral pLKO.1-shRET#1 for 24 hours, were treated with MitoQ for 48 hours prior to crystal violet viability assay. Data are expressed as the percentage of percentage of untreated controls. Data are mean ± SEM (N = 3). e Chou-Talalay plot of the data in d. CI was determined as a function of effect level (Fa). f Western blotting analysis of TPC1 cells cultured under hypoxia. β-actin is the control for equal protein loading. g TPC1 cells pretreated with selpercatinib were treated with MitoQ for 48 hours under the hypoxic condition prior to crystal violet viability assay. Data are mean ± SEM (N = 3). h SynergyFinder plot of the viability data in g. The ZIP score is indicated for the most synergistic area. i TT and MZ-CRC-1 cells pretreated with selpercatinib were treated with MitoQ for 48 hours prior to crystal violet viability assay. Data are mean ± SEM (N = 4). j SynergyFinder plot of the viability data in i. The ZIP score is indicated for the most synergistic area. All data are mean ± SEM (N ≥ 3). *P < 0.05, **P < 0.005, ***P < 0.001, Two-Way ANOVA with Bonferroni post-tests.
Having established the necessity of mitochondrial delivery of CoQ10 for suppressing TPC1 cell viability, we determined whether selpercatinib can synergize with MitoQ to suppress TPC1 cell viability in vitro. As determined by crystal violet staining, the combination of 24-hour selpercatinib pretreatment with a subsequent MitoQ treatment induced a robust viability loss in TPC1 cells (Fig. 2b). Of note, our analysis using SynergyFinder 2.048 suggested that this viability loss is attributed to a synergy between these two agents (Fig. 2c and Supplementary Fig. 2). Consistent with this effect, a combination of RET knockdown and MitoQ treatment also effectively suppressed TPC1 cell viability (Fig. 2d), with a combination index (CI) suggesting synergy (Fig. 2e), as determined by Compusyn49. Importantly, tumors in clinical contexts are generally under hypoxic conditions50. We therefore determined whether the selpercatinib-MitoQ combination produces similar synergistic effects in TPC1 cells in Human Plasma-Like Medium under a hypoxic culture condition, which was validated by increased HIF1α expression (Fig. 2f). Indeed, selpercatinib and MitoQ synergistically suppressed the viability of TPC1 cells under this hypoxic condition (Fig. 2g, h), which is consistent with their effects under the normoxic culture condition.
These combinatory effects were not limited to TPC1 cells. The combination of 24-hour selpercatinib pretreatment with the subsequent MitoQ treatment also synergistically suppressed the viability of the human MTC cell lines, TT (RETC634W) and MZ-CRC-1 (RETM918T), in cultures, as determined by crystal violet staining (Fig. 2i, j; Supplementary Fig. 2). Consistently, RET knockdown also increased Δψm in these cells (Supplementary Fig. 3a, b). Moreover, overexpression of wild-type RET, RETE632D/L633V/C634R, and RETM918T rendered HEK293 cells to increase Δψm upon selpercatinib treatment (Supplementary Fig. 3c, d), demonstrating that the selpercatinib on Δψm effect can be reconstituted in normal cells by increasing RET activity. These data strongly suggest that depletion of RET activity can synergize with MitoQ to suppress RET-mutant tumor cells.
A combination of selpercatinib and MitoQ effectively suppresses the growth of TPC1 xenografts in mice
We determined the preclinical efficacy of the selpercatinib and MitoQ combination using immune-compromised mice bearing TPC1 xenografts. These animals were subjected to 4 cycles of treatments wherein selpercatinib and MitoQ were orally administered singly or in combination. For the drug combination, each cycle consisted of 2-day selpercatinib treatment followed by 2-day MitoQ treatment and 1 drug holiday (Fig. 3a), which is similar to the schedule that we previously used for preclinical evaluation of the vandetanib and MitoCP combination [23]. For the comparison between mono- and combination therapies, we used selpercatinib at 0.5 mg/kg/dose, a much lower dose than the highly potent doses (16-30 mg/kg twice a day) in different preclinical models51,52, because its standard doses were too potent to determine the effect of its combination with other drugs (Supplementary Fig. 4). Likewise, we used MitoQ at 20 mg/kg/dose, which did not significantly reduce tumor volume in a preclinical breast cancer model albeit decreasing tumor metastasis37.
a Treatment schedule for a cycle in total four cycles. Detailed information is in Materials and Method. b Changes in tumor sizes at the indicated time points (N = 5). *P < 0.05, **P < 0.005, ***P < 0.001 (relative to the vehicle); #P < 0.05 (combination therapy versus selpercatinib monotherapy). Two-way ANOVA with Bonferroni post-tests. c Survival probability plot. Tumor size over 1200 mm3 was the endpoint. *P < 0.05, **P < 0.005, Log-rank test. d Body weights measured during the treatment. e Western blot analysis of tumor homogenates. β-actin is the control for equal protein loading.
Consistent with the in vitro data above, the sequential administration of selpercatinib and MitoQ most effectively suppressed the growth of TPC1 xenografts when compared with the monotherapy groups (Fig. 3b), and significantly prolonged mouse survival (Fig. 3c) without affecting body weight (Fig. 3d). In contrast, the selpercatinib monotherapy group exhibited partial tumor suppression while the MitoQ monotherapy group did not show any difference from the control group. Although we also analyzed tumors by Western blotting, we could not detect any significant changes in the cleavage of poly (ADP-ribose) polymerase (PARP) and lamin A, expression of p27KIP1, p21CIP1, and cytochrome c oxidase subunit 4 (COX IV), and the activation phosphorylation of RET, ERK1/2, and AKT (Fig. 3e), which are the surrogate markers for cell death, cell cycle arrest, mitochondrial integrity, and RET signaling activity, respectively. These data suggest that the combination effect may be independent of these proteins, although the different timing of tumor harvest complicates the interpretation of these data.
Patients with RET-mutant thyroid cancer tolerate and respond to combination therapy using selpercatinib and MitoQ
We treated two patients with RET-mutant thyroid cancer with a selpercatinib and MitoQ combination. First patient was a 55-year-old man with poorly differentiated metastatic PTC with a history of poorly controlled hypertension. His tumor progressed after surgical removal of tumor mass and 153.8 mCi of I-131 for 4 weeks (Fig. 4a, full case report in the supplemental document). While awaiting approval for next-generation sequencing (NGS), the patient was treated with lenvatinib but this therapy was held after 5 weeks due to exacerbation of pre-existing refractory hypertension, grade 3, and symptoms of right heart failure (Fig. 4b). While on lenvatinib and during the hold, his cancer continued to progress and his heart dysfunction worsened (Fig. 4b, c). Because of the deteriorating medical status of this patient and the presence of CCDC6-RET fusion mutation in his tumor, we started a weekly cycled schedule for reduced selpercatinib dose with the addition of MitoQ: selpercatinib 160 mg twice a day (BID) on days 1, 2 (off days 3, 4, 5, 6, 7) followed by MitoQ 20 mg daily (QD) on days 3, 4, 5, 6, 7 (off days 1, 2) (Fig. 4a). The patient responded well to this treatment for 22 months, with a sustained radiographic partial response evaluated by response evaluation criteria in solid tumors (RECIST) following the first four months of treatment (Fig. 4c). A chest wall skin metastasis completely resolved during treatment (Fig. 4d). However, at 23 months of treatment, neck and chest CT scan revealed a new site of oligometastatic disease in the manubrium, which progressed over the next month.
a Treatment timeline. b Blood pressure changes during the treatment. Dotted line indicates normal range of blood pressure. c CT scans of lung metastasis (orange arrows) prior to lenvatinib treatment (left), after stopping lenvatinib (middle), and after receiving 7 weeks of the selpercatinib and MitoQ combination therapy (right). Colors in b indicate the same treatment schedule shown in a. d Images of chest wall skin metastasis prior to lenvatinib treatment (left), after stopping lenvatinib (middle), and after receiving 3 weeks of the combination therapy (right).
The second patient was a 34-year-old female with RETM918T locally advanced, unresectable MTC, exhibiting elevated carcinoembryonic antigen and calcitonin levels. This patient was initially started on selpercatinib 160 mg BID for 7 weeks (Fig. 5a, full case report in the supplemental document) but this treatment was on hold due to hepatotoxicity with elevated asymptomatic alanine transferase (ALT) and aspartate transferase (AST) (Fig. 5b). Selpercatinib was resumed in accordance with the FDA-approved package insert to dose level -2, 80 mg BID after liver function tests normalized (Fig. 5a). However, she briskly developed recurrent grade 2 transaminase elevation, nearly grade 3, soon after resumption of treatment (Fig. 5b). Therapy would normally be permanently discontinued after a second occurrence of grade 3/4 hepatotoxicity. However, due to her critical situation, selpercatinib was resumed in a reduced dose schedule with the addition of MitoQ using the following weekly cycled schedule: selpercatinib 80 mg BID on days 1, 2 (off days 3–7) followed by MitoQ 10 mg QD on days 3–7 (off days 1 and 2) (Fig. 5a). Calcitonin levels decreased after full dosage of selpercatinib, elevated again at reduced dose of selpercatinib, and decreased and stabilized during the combination therapy (Fig. 5c). Follow-up imaging after 7 weeks on this regimen indicated further reduction in tumor size with achievement of partial response by RECIST criteria (Fig. 5d). The treatment continued with the plateau of response on imaging at 24 weeks of treatment. Her treatment response and reduction in tumor size allowed her to undergo curative intent surgical resection.
a Treatment timeline. b Changes in ALT and AST. c Changes in serum calcitonin levels. Dotted lines in b, c indicate normal range of tests; colors indicate the same treatment schedule shown in a. d CT scans of right 7 cm MTC with mass effects on the trachea and esophagus (orange circle) prior to initiating selpercatinib treatment (left), prior to resuming reduced dosage of selpercatinib (middle), and after 7 weeks of the selpercatinib and MitoQ combination therapy (right).
Discussion
Previous studies demonstrated that many tumor cells, mainly derived from the cancers of thyroid, skin, and breast, are more sensitive to MitoQ than normal cells35,36,38. MitoQ sensitivity of these tumor cells is mainly attributed to the elevated steady-state Δψm of tumor cells, which is required to enhance their capacity to adapt to hypoxia, escape anoikis, and increase invasiveness53,54. Because the cationic TPP moiety of MitoQ is sensitive to Δψm, TPP-conjugated agents would accumulate higher in the mitochondria of tumor cells than those of most normal cell types34, and the overenrichment of MitoQ would increase the probability of disrupting mitochondrial homeostasis. We thus reasoned that the property of RET inhibition to increase Δψm could be exploited to drive mitochondrial enrichment of MitoQ more effectively in tumor cells beyond the threshold to disrupt a bioenergetics balance (Fig. 6). Our data presented in this report strongly support this notion.
An increase in Δψm facilitates the mitochondrial accumulation of TPP cations. Because RET inhibition increases Δψm, a RET inhibitor can enhance mitochondrial enrichment of a TPP-conjugated agent beyond a level to disrupt mitochondrial homeostasis and induce cell death. The image was generated with BioRender.com.
Selpercatinib and RET-specific RNA interference consistently increased Δψm in CCDC6-RET PTC cells, which supports RET-specificity of the selpercatinib effect. Together with our previous demonstration that vandetanib and cabozantinib can increase Δψm in MEN2A and MEN2B MTC cells26, these data suggest that RET is implicated in regulating mitochondrial activity in thyroid tumor cells. In support, Ret overexpression rescued muscle degeneration of the Pink1 deficient Drosophila model of Parkinson’s disease (PD) by restoring the mitochondrial respiratory complex I activity partly through upregulating mRNA expression of its subunit, NDUFV255. Of note, complex I inhibition induced mitochondrial hyperpolarization in HEK293 cells56, while a complex I defective mutation caused increased Δψm in neurons57. Similarly, knockout of NDUFS4, another complex I subunit, also increased Δψm in mouse embryonic fibroblasts58, although NDUFS4 knockout caused mitochondrial depolarization in mouse skeletal muscle cells59. These contrasting effects suggest that complex I activity can conditionally affect Δψm, probably in a cell context-dependent manner as well as in a temporal- and magnitude-dependent manner. Indeed, short-term inhibition of the complex I by rotenone increased Δψm whereas its prolonged inhibition by the drug decreased Δψm in 143B cells60. Given this information, it remains to be answered whether different selpercatinib doses can induce differential effects on complex I activity.
By what mechanism does mitochondrial overenrichment of MitoQ induce tumor cell death? MitoQ is expected to function primarily as an antioxidant in cells. Tumor cells depend on reactive oxygen species (ROS) more than normal cells to facilitate tumor cell survival, angiogenesis, metastasis, and chemoresistance61,62. In this context, MitoQ would limit tumor cell survival by depleting ROS. Indeed, an oxidized form of ubiquinone (BPM31510) induces anti-cancer effects and is currently tested for solid tumors with high ROS burden in clinical trials63. An alternative scenario is also available. Although initially designed as an antioxidant, MitoQ can elevate ROS production in cells. Studies using mitochondria isolated from bovine aortic endothelial cells showed that MitoQ can increase ROS generation by mitochondrial complex I while mildly decreasing ROS generation by mitochondrial complex II without any effect on complex III64,65. In contrast, its functional moiety CoQ10 had no similar effects64,65. Mechanistically, MitoQ is poorly reduced by complex I because of the bulkiness of TPP moiety that sterically hinders the access of the ubiquinone group to the complex I66,67. However, TPP does not affect the access of ubiquinone moiety to the complex II, which makes MitoQ a good substrate for the complex II67. Meanwhile, MitoQ is not oxidized by Complex III66 because the positive charge of the TPP moiety limits the access of MitoQ to the active site(s) of complex III67. Of note, MitoQ treatment of endothelial cells increased superoxide production from malate- and glutamate-fueled complex I, leading to the onset of apoptosis64. Because many tumor types maintain a higher level of malate and glutamate level to fuel complex I68, MitoQ might have pro-oxidant effects in most metabolically active tumor cells. One may question that MitoQ sensitivity of certain normal cell types would limit the use of high dose MitoQ for cancer therapy and requires a strategy to selectively increase MitoQ delivery into tumor cells for effective treatment. We posit that selpercatinib preferentially sensitize RET-mutated tumor cells to MitoQ, proposing that our strategy to combine selpercatinib and MitoQ might confer an advantage in therapeutically using the agent.
We also appreciate the possibility that selpercatinib and MitoQ synergize through additional mechanisms. In a tubular injury mouse model and human kidney 2 (HK-2) cells, MitoQ partially attenuated mitophagic cell death caused by PINK/Parkin-deficiency by increasing Nrf2 level69. Similarly, MitoQ enhanced PINK/Parkin-mediated mitophagy to inhibit hepatic stellate cell activation and liver fibrosis70, and to ameliorate myocardial ischemia-reperfusion injury in type 2 diabetic rats71. Meanwhile, Ret overexpression rescued muscle degeneration of the Pink1 deficient Drosophila PD model55, and functional redundancy between RET and Parkin in maintaining mitochondrial function and morphology has been demonstrated in a mouse PD model72. Given this information, MitoQ and RET might have effects that overlap in association with PINK. Therefore, it is also possible that RET inhibition and MitoQ may synergistically target a PINK/Parkin-mediated mechanism. Lastly, MitoQ may support the general health status of patients undergoing chemotherapy, as previously suggested73,74. Indeed, MitoQ improved muscle atrophy and weakness of the mice bearing colon cancer cell line xenografts74.
Clinically, the recommended dosing of selpercatinib for a body weight of ≥50 kg is 160 mg BID, while 120 mg BID for patients weighing <50 kg23. The adverse effects of selpercatinib result in permanent discontinuation or dose reduction in about 5% and 31% of patient, respectively23. The two patients in this report did not tolerate standard doses of selpercatinib due to adverse effects. However, these patients did tolerate reduced doses of selpercatinib when combined with MitoQ. Whether MitoQ significantly enhanced the effects of selpercatinib cannot be determined in this brief analysis of only two patients. Nevertheless, these observations suggest a strategy to effectively use selpercatinib in the patients with low tolerance to this drug. MitoQ has been confirmed for its safety in humans, and its clinical benefits are currently being evaluated in Phase II trials32,33,34, although its benefit for cancer patients has not yet been tested in the setting of a clinical trial. We believe our preclinical data and patient cases are pertinent to the design of a future clinical trial to determine whether selpercatinib and MitoQ combination can provide a clinical benefit to patients with RET-mutated cancers.
Methods
Cell culture and reagents
The human thyroid cancer cell line TPC1 (ATCC, Manassas, VA) was maintained in RPMI 1640 (Invitrogen, Thermo Fisher Scientific, Waltham, MA, #11875) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco, Thermo Fisher Scientific, #16000-044) and 100 U of Penicillin-Streptomycin (Gibco, #15140) per ml. TT was maintained in DMEM supplemented with 16% heat-inactivated FBS and 100 U of Penicillin-Streptomycin per ml. MZ-CRC-1 was maintained in DMEM (Gibco, #11965) supplemented with 10% heat-inactivated FBS and 100 U of Penicillin-Streptomycin per ml. As indicated in the text, cells were also maintained in a Human Plasma-Like Medium (HPLM, Gibco, #A4899101) supplemented with heat-inactivated 10% FBS, and 100 U Penicillin-Streptomycin per ml. HEK293T was maintained in DMEM supplemented with 10% BGS (Hyclone, Cytiva, Muskegon, MI, #SH30541.03). PCCL3 was maintained in Ham F12 media (MilliporeSigma, N6658), supplemented with 5% FBS, 1 mIU/mL bovine thyroid stimulating hormone (SCRIPPS, CA, T0117), 10 μg/mL insulin (Gibco, # 41400045), 5 μg/mL apo-transferrin (MilliporeSigma, T2252), 10 nM of hydrocortisone (MilliporeSigma, #H0888) and 100 U of Penicillin-Streptomycin. Hypoxic cell culture was carried out in a humidified incubator with 1% O2, 5% CO2, and 94% N2. All experiments were performed using cells within ten passages from the acquisition point. Selpercatinib was purchased from Chemietek (Indianapolis, IN, #CT-LX292). MitoCP was obtained from Dr. Balaraman Kalyanaraman (Biophysics, Medical College of Wisconsin). MitoQ ([10-(4,5-dimethoxy-2-methyl-3,6-dioxo-1,4-cyclohexadien-1-yl) decyl] triphenyl phosphonium) used for in vitro and preclinical studies was obtained from MitoQ (Auckland, New Zealand); MitoQ for patient treatments was purchased from Amazon.com. CoQ10 (#C9538) and methyl-TPP (#130079) were purchased from MilliporeSigma (St. Louis, MO).
Cell viability and cell cycle analyses
IC50 values and combination effects of drug treatments were determined by Crystal Violet (Fisher Chemical, Thermo Fisher Scientific, #C58125) cell viability staining assay. Briefly, cells in 96 well-plates were fixed in formaldehyde (Fisher Chemical, #BP531), stained with 0.05% Crystal Violet for 30 minutes, washed with water three times, air-dried, and incubated in 200 µl of methanol (VWR Chemicals BDH®, Radnor, PA #BDH1135) for 10 min at room temperature before measuring absorbance at 540 nm. Cell viability for RET knockdown was determined by flow cytometry of cells stained with the fluorescent DNA intercalator Thiazole Red (TO-PRO®3, Invitrogen, # T3605) using Guava EasyCyte flow cytometry system (MilliporeSigma, #05005007). Data were analyzed by FCS EXPRESS software (De Novo Software, Los Angeles, California). For the cell cycle, flow cytometry of cells stained with propidium iodide (PI, Invitrogen, #P1304MP) was performed using the Guava EasyCyte flow cytometry system, as previously described75.
Detection of Δψm using TMRM
Cells were incubated with culture medium containing 2 nM TMRM (Invitrogen, #T668) in 12-well plates for 30 min at 37 °C in dark, collected by trypsinization, resuspended in phosphate-buffered saline containing 0.5% bovine serum albumin, and analyzed using Guava EasyCyte flow cytometry system Data were analyzed by FCS EXPRESS software flow, as described previously27.
Immunoblotting
Mitochondrial fractions were extracted using the Mitochondria Isolation Kit for Cultured Cells (Thermo Fisher Scientific, #89874). For total cell lysate, cells were harvested in a lysis buffer containing 62.5 mM Tris-HCl (pH 6.8), 2% sodium dodecyl sulfate (SDS), and the protease and phosphatase inhibitor cocktails 2 and 3 (MilliporeSigma, #P8340, P5726, and P0044, respectively). Protein concentration was determined using the Pierce™ BCA Protein Assay Kit (Pierce, Thermo Fisher Scientific, #23227). Protein samples were resolved on the SDS-polyacrylamide gel electrophoresis and transferred to the polyvinylidene difluoride membrane filter (Bio-Rad, Hercules, CA, #1620177). After transfer, membranes were blocked in a buffer containing 0.1 M Tris (pH 7.4), 0.9% NaCl, 0.1% Tween 20, and 5% nonfat dry milk for 1 h at 25 °C. Membranes were then incubated with an appropriate antibody overnight at 4 °C with agitation at the following dilutions: RET (Cell Signaling, Danvers, MA, #3223) 1:2000; phospho-RET Try905 (Cell Signaling, #3221) 1:1000; ERK (Cell Signaling, #9102) 1:2000; phospho-ERK Thr202/Tyr204 (Santa Cruz Biotechnology, Santa Cruz, CA, #sc-16982) 1:1000; AKT (Cell Signaling, #9272) 1:2000; phospho-AKT Ser473 (Cell Signaling, #9271); PARP (Cell Signaling, #9542) 1:2000; cleaved lamin A (Cell Signaling, #2035) 1:1000; cytochrome c oxidase subunit IV (COX IV, Cell Signaling, #4850) 1:2000; p21CIP1 (Santa Cruz Biotechnology, #sc-56335) 1:1000, p27KIP1 (Santa Cruz Biotechnology, #sc-1641) 1:1000; HIF1α (Cell Signaling, #14179) 1:1000 and β-actin (MilliporeSigma, #A2228) 1:5000. Chemiluminescence signals of immunoblots were visualized by the chemiluminescence kits SuperSignal West Pico (#34079, Pierce) and Femto (#23227, Pierce), captured by ChemiDoc XRS+ (Bio-Rad, Hercules, CA), and analyzed by Image Lab software (Bio-Rad) for densitometry. All blots were derived from the same experiment and were processed in parallel. Their original uncropped images are provided in Supplementary Fig. 5.
RNA interference and recombinant lentiviral constructs
RET was depleted using two independent lentiviral shRNA expression systems, pLKO-shRET#1 (MilliporeSigma, #NM_000323) and pLKO-shRET#2 (#NM_020629), which target CCGCTGGTGGACTGTAATAAT and CCTCATCTCATTTGCCTGGCA in human RET, respectively. Lentiviral pHAGE expressing human wild-type RET, RETE632D/L633V/C634R, and RETM918T were generated by ligating each RET encoding gene into the NheI/SphI site of the pHAGE-GFP vector76. Lentiviruses were generated from HEK293T cells and used, as we previously described76.
Tumor xenografts
A total of 5 × 106 TPC1 cells suspended in 100 µL of Hank’s balanced salt solution (Gibco, #14025) mixed with Extracellular Matrix Gel (MilliporeSigma, #E6909) at 1:1 ratio was inoculated subcutaneously into the rear flanks of 6-week-old female athymic nude (nu/nu) mice (The Jackson Laboratory, Bar Harbour, ME, #007850). Once palpable, tumors were measured using Vernier calipers twice a week. Tumor volumes were calculated using the formula: length × width × height × 0.5236. When tumor volumes reached 50 mm3, mice were randomized and sorted into 4 groups of 5 animals to achieve equal tumor size distribution in all treatment groups. Mice were treated with vehicle, MitoQ, selpercatinib, and the combination of two compounds, respectively. Drugs dissolved in 100 μl vehicle (1:12 mixture of DMSO/15% β-cyclodextrin) were orally administered by gavage daily for 4 days, followed by 1-day break. Four cycles of this treatment were conducted. The control group received only the vehicle, the MitoQ group received 20 mg drug/kg body weight/dose, the selpercatinib group received 0.5 mg drug/kg body weight/dose, and the combination group received two doses of selpercatinib followed by two doses of MitoQ in a cycle. Tumor sizes were measured, and survival probability was also plotted. Ethical endpoints were when tumor size reached 1200 mm3. At the end of the experiments, animals were euthanized by CO2 asphyxiation, and tumor tissues were harvested. All animal studies were performed according to protocols approved by the Institutional Animal Care and Use Committee at the Medical College of Wisconsin (AUA00001327).
Patients
The patients were treated at the Froedtert Hospital and Medical College of Wisconsin. Analysis and reporting of patients’ treatment and outcome were performed in accordance with the Declaration of Helsinki under an Institutional Review Board-approved study (IRB number PRO00002787) that waives the requirement for informed consent because of the use of de-identified data. Responses were assessed by RECIST 1.177. Adverse events were graded using the Common Terminology Criteria for Adverse Events version 4.0378. NGS revealed CCDC6-RET fusion; CDKN2B loss; CDKN2A loss; TERT promoter-124C>T; TP53R248Q in the tumor of the first patient, which was performed by Linda T. and John A. Mellowes Center for Genomic Sciences and Precision Medicine (Medical College of Wisconsin, Wisconsin, WI) and FoundationOne® CDx (Foundation Medicine, Cambridge, MA); and RETM918T in the tumor of the second patient, which was performed by TEMPUS (Chicago, IL).
Quantification and statistical analyses
All graphs represent the mean ± the standard deviation of biological triplicates’ mean. Measurements were taken from distinct samples. Statistical significance was determined by one-way or two-way ANOVA with Bonferroni post-tests and two-tailed unpaired Student’s t test using PRISM (Graph-Pad Software, La Jolla, CA). IC50 was determined by PRISM. P values of the Kaplan–Meier curves were determined by the Log-rank test by PRISM. P values of <0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and the Supplementary Materials. Any additional information required to reanalyze the data reported in this paper is available from the corresponding authors upon request. Reagents and materials produced in this study are available pending a completed Materials Transfer Agreement. The raw NGS data for the genomic analysis of patient tumors are not publicly available to protect patient privacy, but qualified researchers may apply for access to the datasets via a collaboration or data usage agreement.
Code availability
This paper does not report any original code.
References
Santoro, M. & Carlomagno, F. Central role of RET in thyroid cancer. Cold Spring Harb. Perspect. Biol. 5, a009233 (2013).
Qian, Y. et al. KIF5B-RET fusion kinase promotes cell growth by multilevel activation of STAT3 in lung cancer. Mol. Cancer 13, 176 (2014).
Bongarzone, I. et al. Molecular characterization of a thyroid tumor-specific transforming sequence formed by the fusion of ret tyrosine kinase and the regulatory subunit RI alpha of cyclic AMP-dependent protein kinase A. Mol. Cell Biol. 13, 358–366 (1993).
Tong, Q., Xing, S. & Jhiang, S. M. Leucine zipper-mediated dimerization is essential for the PTC1 oncogenic activity. J. Biol. Chem. 272, 9043–9047 (1997).
Grieco, M. et al. PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell 60, 557–563 (1990).
Eszlinger, M. et al. Systematic population-based identification of NTRK and RET fusion-positive thyroid cancers. Eur. Thyroid J. 11, e210061 (2022).
Pekova, B. et al. RET, NTRK, ALK, BRAF, and MET fusions in a large cohort of pediatric papillary thyroid carcinomas. Thyroid 30, 1771–1780 (2020).
Kohno, T. et al. KIF5B-RET fusions in lung adenocarcinoma. Nat. Med. 18, 375–377 (2012).
Takeuchi, K. et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 18, 378–381 (2012).
Lipson, D. et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat. Med. 18, 382–384 (2012).
Pietrantonio, F. et al. RET fusions in a small subset of advanced colorectal cancers at risk of being neglected. Ann. Oncol. 29, 1394–1401 (2018).
Kim, S. Y. et al. NCOA4-RET fusion in colorectal cancer: therapeutic challenge using patient-derived tumor cell lines. J. Cancer 9, 3032–3037 (2018).
Le Rolle, A. F. et al. Identification and characterization of RET fusions in advanced colorectal cancer. Oncotarget 6, 28929–28937 (2015).
Salvatore, D. et al. Increased in vivo phosphorylation of ret tyrosine 1062 is a potential pathogenetic mechanism of multiple endocrine neoplasia type 2B. Cancer Res. 61, 1426–1431 (2001).
Santoro, M. et al. Activation of RET as a dominant transforming gene by germline mutations of MEN2A and MEN2B. Science 267, 381–383 (1995).
Raman, R. et al. Inhibition of FGF receptor blocks adaptive resistance to RET inhibition in CCDC6-RET-rearranged thyroid cancer. J. Exp. Med. 219, e20210390 (2022).
Cabanillas, M. E., Brose, M. S., Holland, J., Ferguson, K. C. & Sherman, S. I. A phase I study of cabozantinib (XL184) in patients with differentiated thyroid cancer. Thyroid 24, 1508–1514 (2014).
Degrauwe, N., Sosa, J. A., Roman, S. & Deshpande, H. A. Vandetanib for the treatment of metastatic medullary thyroid cancer. Clin. Med. Insights Oncol. 6, 243–252 (2012).
Nagilla, M., Brown, R. L. & Cohen, E. E. Cabozantinib for the treatment of advanced medullary thyroid cancer. Adv. Ther. 29, 925–934 (2012).
Subbiah, V. et al. Structural basis of acquired resistance to selpercatinib and pralsetinib mediated by non-gatekeeper RET mutations. Ann. Oncol. 32, 261–268 (2021).
Wirth, L. J. et al. Emergence and targeting of acquired and hereditary resistance to multikinase RET inhibition in patients with RET-altered cancer. JCO Precis. Oncol. 3, PO.19.00189 (2019).
Markham, A. Selpercatinib: first approval. Drugs 80, 1119–1124 (2020).
Bradford, D. et al. FDA approval summary: selpercatinib for the treatment of lung and thyroid cancers with RET gene mutations or fusions. Clin Cancer Res. 27, 2130–2135 (2021).
Kim, J. et al. FDA approval summary: pralsetinib for the treatment of lung and thyroid cancers with RET gene mutations or fusions. Clin. Cancer Res. 27, 5452–5456 (2021).
FDA. FDA grants accelerated approval to dabrafenib in combination with trametinib for unresectable or metastatic solid tumors with BRAF V600E mutation. (2022).
Starenki, D., Hong, S. K., Wu, P. K. & Park, J. I. Vandetanib and cabozantinib potentiate mitochondria-targeted agents to suppress medullary thyroid carcinoma cells. Cancer. Biol. Ther. 18, 473–483 (2017).
Starenki, D. & Park, J. I. Mitochondria-targeted nitroxide, Mito-CP, suppresses medullary thyroid carcinoma cell survival in vitro and in vivo. J. Clin. Endocrinol. Metab. 98, 1529–1540 (2013).
Kulkarni, C. A. et al. A novel triphenylphosphonium carrier to target mitochondria without uncoupling oxidative phosphorylation. J. Med. Chem. 64, 662–676 (2021).
Zielonka, J. et al. Mitochondria-targeted triphenylphosphonium-based compounds: syntheses, mechanisms of action, and therapeutic and diagnostic applications. Chem. Rev. 117, 10043–10120 (2017).
Kelso, G. F. et al. Selective targeting of a redox-active ubiquinone to mitochondria within cells: antioxidant and antiapoptotic properties. J. Biol. Chem. 276, 4588–4596 (2001).
Murphy, M. P. & Smith, R. A. Targeting antioxidants to mitochondria by conjugation to lipophilic cations. Annu. Rev. Pharmacol. Toxicol. 47, 629–656 (2007).
Park, S. Y. et al. Acute mitochondrial antioxidant intake improves endothelial function, antioxidant enzyme activity, and exercise tolerance in patients with peripheral artery disease. Am. J. Physiol. Heart Circ. Physiol. 319, H456–H467 (2020).
Rossman, M. J. et al. Chronic supplementation with a mitochondrial antioxidant (MitoQ) improves vascular function in healthy older adults. Hypertension 71, 1056–1063 (2018).
Smith, R. A. & Murphy, M. P. Animal and human studies with the mitochondria-targeted antioxidant MitoQ. Ann. N Y Acad. Sci. 1201, 96–103 (2010).
Hong, S. K., Starenki, D., Wu, P. K. & Park, J. I. Suppression of B-Raf(V600E) melanoma cell survival by targeting mitochondria using triphenyl-phosphonium-conjugated nitroxide or ubiquinone. Cancer Biol. Ther. 18, 106–114 (2017).
Capeloa, T. et al. MitoQ inhibits human breast cancer cell migration, invasion and clonogenicity. Cancers (Basel) 14, 1516 (2022).
Capeloa, T. et al. MitoQ prevents human breast cancer recurrence and lung metastasis in mice. Cancers (Basel) 14, 1488 (2022).
Rao, V. A. et al. The antioxidant transcription factor Nrf2 negatively regulates autophagy and growth arrest induced by the anticancer redox agent mitoquinone. J. Biol. Chem. 285, 34447–34459 (2010).
Capeloa, T. et al. Inhibition of mitochondrial redox signaling with mitoq prevents metastasis of human pancreatic cancer in mice. Cancers (Basel) 14, 4918 (2022).
Ishizaka, Y. et al. Presence of aberrant transcripts of ret proto-oncogene in a human papillary thyroid carcinoma cell line. Jpn. J. Cancer Res. 80, 1149–1152 (1989).
Iwashita, T. et al. Biological and biochemical properties of Ret with kinase domain mutations identified in multiple endocrine neoplasia type 2B and familial medullary thyroid carcinoma. Oncogene 18, 3919–3922 (1999).
Hu, X., Liu, X., Khatri, U. & Wu, J. The heterogeneous transition state of resistance to RET kinase inhibitors converges on ERK1/2-driven Aurora A/B kinases. Drug Resist. Updat. 68, 100958 (2023).
Mologni, L. et al. Inhibition of RET tyrosine kinase by SU5416. J. Mol. Endocrinol. 37, 199–212 (2006).
Vitagliano, D. et al. Regulation of p27Kip1 protein levels contributes to mitogenic effects of the RET/PTC kinase in thyroid carcinoma cells. Cancer Res. 64, 3823–3829 (2004).
Knauf, J. A., Kuroda, H., Basu, S. & Fagin, J. A. RET/PTC-induced dedifferentiation of thyroid cells is mediated through Y1062 signaling through SHC-RAS-MAP kinase. Oncogene 22, 4406–4412 (2003).
Nikiforov, Y. E. RET/PTC rearrangement in thyroid tumors. Endocr. Pathol. 13, 3–16 (2002).
Lin, T. K. et al. Specific modification of mitochondrial protein thiols in response to oxidative stress: a proteomics approach. J. Biol. Chem. 277, 17048–17056 (2002).
Ianevski, A., Giri, A. K. & Aittokallio, T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res. 48, W488–W493 (2020).
Chou T, M. N. CompuSyn software. CompuSyn for drug combinations: PC software and user’s guide: a computer program for quantitation of synergism and antagonism in drug combinations, and the determination of IC50 and ED50 and LD50 values. ComboSyn Inc., Paramus, NJ. (2005).
Hompland, T., Fjeldbo, C. S. & Lyng, H. Tumor hypoxia as a barrier in cancer therapy: why levels matter. Cancers (Basel) 13, 499 (2021).
Shen, T. et al. The L730V/I RET roof mutations display different activities toward pralsetinib and selpercatinib. NPJ Precis. Oncol. 5, 48 (2021).
Subbiah, V. et al. Selective RET kinase inhibition for patients with RET-altered cancers. Ann. Oncol. 29, 1869–1876 (2018).
Mani, S., Swargiary, G. & Singh, K. K. Natural agents targeting mitochondria in cancer. Int. J. Mol. Sci. 21, 6992 (2020).
Heerdt, B. G., Houston, M. A. & Augenlicht, L. H. Growth properties of colonic tumor cells are a function of the intrinsic mitochondrial membrane potential. Cancer Res. 66, 1591–1596 (2006).
Klein, P. et al. Ret rescues mitochondrial morphology and muscle degeneration of Drosophila Pink1 mutants. EMBO J 33, 341–355 (2014).
Forkink, M. et al. Mitochondrial hyperpolarization during chronic complex I inhibition is sustained by low activity of complex II, III, IV and V. Biochim. Biophys. Acta 1837, 1247–1256 (2014).
Abramov, A. Y. et al. Mechanism of neurodegeneration of neurons with mitochondrial DNA mutations. Brain 133, 797–807 (2010).
Valsecchi, F. et al. Metabolic consequences of NDUFS4 gene deletion in immortalized mouse embryonic fibroblasts. Biochim. Biophys. Acta 1817, 1925–1936 (2012).
Alam, M. T. et al. Skeletal muscle mitochondria of NDUFS4-/- mice display normal maximal pyruvate oxidation and ATP production. Biochim. Biophys. Acta 1847, 526–533 (2015).
Barrientos, A. & Moraes, C. T. Titrating the effects of mitochondrial complex I impairment in the cell physiology. J. Biol. Chem. 274, 16188–16197 (1999).
Szatrowski, T. P. & Nathan, C. F. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 51, 794–798 (1991).
Galadari, S., Rahman, A., Pallichankandy, S. & Thayyullathil, F. Reactive oxygen species and cancer paradox: to promote or to suppress? Free Radic. Biol. Med. 104, 144–164 (2017).
Dadali, T. et al. Elevated levels of mitochondrial CoQ(10) induce ROS-mediated apoptosis in pancreatic cancer. Sci. Rep. 11, 5749 (2021).
Doughan, A. K. & Dikalov, S. I. Mitochondrial redox cycling of mitoquinone leads to superoxide production and cellular apoptosis. Antioxid. Redox Signal. 9, 1825–1836 (2007).
O’Malley, Y., Fink, B. D., Ross, N. C., Prisinzano, T. E. & Sivitz, W. I. Reactive oxygen and targeted antioxidant administration in endothelial cell mitochondria. J. Biol. Chem. 281, 39766–39775 (2006).
James, A. M., Cocheme, H. M., Smith, R. A. & Murphy, M. P. Interactions of mitochondria-targeted and untargeted ubiquinones with the mitochondrial respiratory chain and reactive oxygen species. Implications for the use of exogenous ubiquinones as therapies and experimental tools. J. Biol. Chem. 280, 21295–21312 (2005).
James, A. M. et al. Interaction of the mitochondria-targeted antioxidant MitoQ with phospholipid bilayers and ubiquinone oxidoreductases. J. Biol. Chem. 282, 14708–14718 (2007).
Zhong, X. et al. Complex metabolic interactions between ovary, plasma, urine, and hair in ovarian cancer. Front. Oncol. 12, 916375 (2022).
Xiao, L. et al. The mitochondria-targeted antioxidant MitoQ ameliorated tubular injury mediated by mitophagy in diabetic kidney disease via Nrf2/PINK1. Redox Biol. 11, 297–311 (2017).
Dou, S. D. et al. MitoQ inhibits hepatic stellate cell activation and liver fibrosis by enhancing PINK1/parkin-mediated mitophagy. Open Med. (Wars) 16, 1718–1727 (2021).
Ji, Y. et al. The mitochondria-targeted antioxidant MitoQ ameliorates myocardial ischemia-reperfusion injury by enhancing PINK1/Parkin-mediated mitophagy in type 2 diabetic rats. Cell Stress Chaperones 27, 353–367 (2022).
Meka, D. P. et al. Parkin cooperates with GDNF/RET signaling to prevent dopaminergic neuron degeneration. J. Clin. Invest. 125, 1873–1885 (2015).
Drisko, J. A., Chapman, J. & Hunter, V. J. The use of antioxidants with first-line chemotherapy in two cases of ovarian cancer. J. Am. Coll. Nutr. 22, 118–123 (2003).
Pin, F., Huot, J. R. & Bonetto, A. The mitochondria-targeting agent MitoQ improves muscle atrophy, weakness and oxidative metabolism in C26 tumor-bearing mice. Front. Cell Dev. Biol. 10, 861622 (2022).
Wu, P. K. et al. A mortalin/HSPA9-mediated switch in tumor-suppressive signaling of Raf/MEK/extracellular signal-regulated kinase. Mol. Cell Biol. 33, 4051–4067 (2013).
Hong, S. K., Yoon, S., Moelling, C., Arthan, D. & Park, J. I. Noncatalytic function of ERK1/2 can promote Raf/MEK/ERK-mediated growth arrest signaling. J. Biol. Chem. 284, 33006–33018 (2009).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009).
U.S.-Department-of-Health-and-Human-Services. Common Terminology Criteria for Adverse Events (CTCAE) Version 4.03. (2010).
Acknowledgements
We thank MitoQ (Auckland, New Zealand) for MitoQ, Dr. Barry Nelkin (Johns Hopkins Medical Institute) for TPC1 and TT, Dr. Robert Gagel (MD Anderson) for MZ-CRC-1, Dr. James A. Fagin for PCCL3 expressing dox-NCOA4-RET, Dr. Gilbert Cote (MD Anderson) for RET constructs, Dr. Rozita Yarmand (MD Anderson) for pLKO-shRET#1, Dr. Michael Murphy (Medical Research Council Dunn Human Nutrition Unit) for the TPP antibody, Drs. Balaraman Kalyanaraman and Gang Cheng (Medical College of Wisconsin) for MitoCP, and Dr. Yiliang Chen (Medical College of Wisconsin) for HIF1α antibody. This work was supported by NIH/National Cancer Institute grants (R01CA269452 and R01CA138441) to J.P.
Author information
Authors and Affiliations
Contributions
W.C. performed in vitro and preclinical studies, analyzed data, and wrote the paper; S.D. performed clinical studies, analyzed data, and wrote the paper; P.L. performed in vitro studies; P.W. developed methodology; S.W. performed and supervised clinical studies, analyzed data, and wrote the paper; J.P. conceived the project, supervised in vitro and preclinical studies, wrote the paper, and secured funding. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, W., Dream, S., Leung, PY. et al. Selpercatinib combination with the mitochondria-targeted antioxidant MitoQ effectively suppresses RET–mutant thyroid cancer. npj Precis. Onc. 8, 39 (2024). https://doi.org/10.1038/s41698-024-00536-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-024-00536-7