Abstract
Pancreatic acinar cell carcinoma (PACC) is a rare form of pancreatic cancer that commonly harbors targetable alterations, including activating fusions in the MAPK pathway and loss-of-function (LOF) alterations in DNA damage response/homologous recombination DNA repair-related genes. Here, we describe a patient with PACC harboring both somatic biallelic LOF of NBN and an activating NTRK1 fusion. Upon disease progression following 13 months of treatment with folinic acid, fluorouracil, irinotecan, and oxaliplatin (FOLFIRINOX), genomic analysis of a metastatic liver biopsy revealed the emergence of a novel reversion mutation restoring the reading frame of NBN. To our knowledge, genomic reversion of NBN has not been previously reported as a resistance mechanism in any tumor type. The patient was treated with, but did not respond to, targeted treatment with a selective NTRK inhibitor. This case highlights the complex but highly actionable genomic landscape of PACC and underlines the value of genomic profiling of rare tumor types such as PACC.
Similar content being viewed by others
Introduction
Pancreatic acinar cell carcinomas (PACCs) are rare forms of pancreatic cancer, accounting for approximately 1–2% of pancreatic tumors in adults and 15% of those in children1. PACCs are more prevalent in men, with a male to female ratio of 2:1. Histologically, these neoplasms are characterized by tumor cells displaying features of acinar cell differentiation, with moderate amounts of eosinophilic cytoplasm containing periodic acid-Schiff (PAS)-positive diastase-resistant zymogen granules and uniform nuclei with distinct nucleoli, arranged in acinar, glandular, solid, or trabecular patterns1. Genomically, PACCs have a repertoire of alterations distinct from that of pancreatic ductal carcinomas, rarely displaying KRAS, TP53, CDKN2A, and SMAD4 somatic alterations, and more frequently harboring recurrent activating fusions affecting BRAF, RET, RAF1, and NTRK1/2/3, which are present in up to 30% of patients2,3,4,5,6,7. In addition, a subset of patients with PACCs harbor germline pathogenic alterations affecting DNA damage response (DDR)/homologous recombination (HR) DNA repair-related genes, including ATM, BRCA1, BRCA2, and PALB22,3,8.
The mainstay of treatment for patients with PACCs is surgical resection with negative margins followed by chemotherapy. Currently, no targeted therapies are approved by the US Food and Drug Administration specifically for patients with PACC. Patients whose tumors carry oncogenic fusions may be treated using therapies with tumor-agnostic labels, such as entrectinib or larotrectinib for NTRK fusions or selpercatinib for RET fusions9,10,11. Inhibitors of poly (ADP-ribose) polymerase (PARP) have been approved for patients with pancreatic cancer (including PACC) harboring germline BRCA1 or BRCA2 pathogenic mutations, and further studies are investigating the benefit of PARP inhibitors in pancreatic cancers with HR deficiency (HRD) due to alterations affecting other canonical HR-related genes (NCT04858334), and in unselected PACC (NCT05286827).
The nibrin gene (NBN, also known as NBS1 [Nijmegen breakage syndrome 1]) is an integral component of the DDR/HR DNA repair machinery12,13. Germline NBN loss-of-function (LOF) alterations have not only been causally implicated in the Nijmegen breakage syndrome, but have also been shown to result in an increased risk of multiple cancer types13. The impact of somatic pathogenic NBN alterations on the development and progression of pancreatic cancer development is uncertain.
In this report, we describe a patient with PACC whose tumor harbored both an activating SEL1L::NTRK1 fusion and somatic biallelic inactivation of NBN. Upon disease progression on treatment with folinic acid, fluorouracil, irinotecan, and oxaliplatin (FOLFIRINOX), a novel reversion mutation restoring the NBN reading frame was detected, suggesting a potential link between NBN LOF, PACC progression, and the evolution of therapeutic resistance to mainstay treatment. This novel reversion mutation affecting the NBN gene adds to the growing list of resistance mechanisms to DNA damaging- and DDR-targeted therapies.
Results
Clinical case
A 65-year-old White male patient presented with abdominal pain. Computed tomography showed a mild distension of the common bile duct, and further evaluation with an endoscopic ultrasound revealed a mass in the pancreatic head. Fine needle aspiration of this mass was performed, and cytology was consistent with adenocarcinoma (Fig. 1a). Staging evaluation indicated pT1c, pTN2, pM0 (AJCC 8th edition14), and the patient underwent pancreaticoduodenectomy. Pathologic examination revealed an acinar cell carcinoma measuring 2 cm with varied architecture, including solid trabecular and acinar formations that invaded the duodenum and intrahepatic common bile duct, with 4 of 16 regional lymph nodes involved. At the cellular level, cytoplasms were granular with monomorphic nuclei containing prominent nucleolei. Consistent with the diagnosis, immunohistochemical analysis were negative for synaptophysin, chromogranin, and MOC-31/EpCAM, while periodic acid-Schiff staining with diastase digestion (PAS-D) were positive for cytoplasmic granules. Given the rarity of the tumor type, the diagnosis was confirmed and agreed upon by multiple pathologists. Furthermore, the screening tissue used to determine molecular eligibility was retrospectively found to be BCL-10 positive consistent with the diagnosis1.
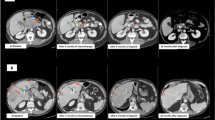
a Therapy history and CA19-9 kinetics. Representative micrographs of the primary resection and metastatic liver biopsy specimens revealed the presence of acinar structures comprising neoplastic cells with eosinophilic and granular cytoplasm, and atypical nuclei, consistent with a diagnosis of pure acinar cell carcinoma of the pancreas (scale bar: 20 µm). Representative CT scans pre- and on-treatment with FOLFIRINOX. b NBN reversion mutation detected in the metastatic liver biopsy. The reference (top), primary mutation (middle), and secondary mutation (bottom) cDNA and amino acid sequences are shown. The combination of the primary and secondary mutations created a 3-amino acid deletion and 2-amino acid insertion resulting in a novel in-frame indel (c.1146_1155delinsGAAATCA; p.383_385delinsKS). c B allele frequency/log R ratio plots derived from WGS analysis of the metastatic liver biopsy. d Genomic and transcriptomic structure of the SEL1L::NTRK1 fusion. Skipping of NTRK1 exon 7 was observed in the targeted RNA sequencing data, reflective of the coding potential of different splice configurations. e Chromosome-based circos plot derived from WGS of the metastatic liver biopsy. f Distribution of mutational signatures as determined by WGS of the metastatic liver biopsy. CA cancer antigen, cDNA complementary DNA, CT computed tomography, FOLFIRINOX folinic acid, fluorouracil, irinotecan, and oxaliplatin, indel insertion and deletion, Mb megabase, WGS whole genome sequencing.
Subsequently, the patient received adjuvant treatment with gemcitabine and capecitabine for 6 months, followed by radiation concurrent with capecitabine. After completion of this treatment, the patient was on surveillance for 16 months, at which time he was diagnosed with metastatic disease to the liver, histopathologically confirmed as metastatic acinar cell carcinoma (Fig. 1a).
At that time, a tissue specimen from the primary pancreaticoduodenectomy (herein referred to as the primary resection) was sent for clinical-grade next generation sequencing (NGS) as per standard of care, which identified a somatic pathogenic NBN mutation and an in-frame SEL1L::NTRK1 fusion (Table 1). The patient then received palliative chemotherapy with the modified FOLFIRINOX regimen for 13 months. The patient achieved an initial response with a marked decrease in cancer antigen (CA)19-9 and a reduction in hepatic tumor burden, but treatment was eventually discontinued due to the development of a new liver lesion and an increase in the size of prior liver lesions (Fig. 1a). Given the presence of an NTRK1 fusion in the primary resection specimen, the patient received targeted treatment with the NTRK inhibitor larotrectinib. However, the patient rapidly progressed with multiple new liver lesions and an increase in the size of prior liver lesions, resulting in treatment discontinuation 2 months later.
A new liver biopsy (herein referred to as the metastatic liver biopsy) was taken at this time and sent for clinical-grade NGS (Table 1). Pending the results and based on the detection of a pathogenic NBN mutation in the primary resection specimen, the patient was enrolled on TRESR (NCT04497116) and received a therapeutic dose level of the ataxia telangiectasia and Rad3-related (ATR) inhibitor camonsertib (RP-3500, Fig. 1a). The patient had stable disease at 6 weeks, but treatment was discontinued after 12 weeks due to progression of the target liver lesions (32% increase). The patient was then treated with gemcitabine and paclitaxel (nab-paclitaxel was not available due to a nationwide shortage), on which he also experienced progression of disease after 2 months due to worsening peritoneal carcinomatosis. Subsequently, he was briefly treated on a clinical trial with TST001, an anti-claudin 18.2 monoclonal antibody, before discontinuing the study due to clinical progression after which the patient was enrolled in a hospice and passed away 1 month later.
Genomic analysis
Genomic analyses of the tissue specimens obtained during the course of treatment were performed with clinical-grade NGS assays, as well as with research-use-only NGS assays performed as part of TRESR (NCT04497116; Table 1). This included the use of the Synthetic Lethal Interactions for Precision Diagnostics (SNiPDx™) panel, a novel NGS-based diagnostic panel specifically designed to detect and distinguish between monoallelic and biallelic LOF alterations in genes determined to be synthetic lethal with camonsertib15. The primary resection specimen collected 3 months after diagnosis was initially evaluated by clinical-grade NGS and retrospectively by SNiPDx™, and anchored multiplex polymerase chain reaction (PCR) (AMP)-based RNA sequencing (Table 1)15,16,17,18. The metastatic liver biopsy specimen, collected after treatment with FOLFIRINOX and larotrectinib and just prior to initiation of camonsertib, was initially evaluated by clinical-grade NGS, and retrospectively by SNiPDx™ and whole genome sequencing (WGS)15,19,20,21.
DNA analysis of both tissue specimens and peripheral blood mononuclear cells (PBMCs) by multiple methodologies identified a somatic (absent in PBMCs) pathogenic 1-nucleotide deletion in exon 10 of NBN (c.1146del; p.E383Kfs*21; ClinVar Variation ID: 822251; Table 1 and Fig. 1b). This deletion results in a premature stop codon 21 codons downstream of p.E383 and is predicted to truncate the protein leading to nonsense-mediated decay. Analysis of the allele-specific copy number by SNiPDx™ and WGS revealed copy-neutral loss of heterozygosity of the NBN wild-type allele with subsequent duplication of the mutant allele, consistent with a clonal somatic biallelic inactivation of NBN (Fig. 1c and Supplementary Fig. 1). In addition, a subclonal RB1 (c.649 C > T; p.Q217*) pathogenic mutation was detected in the primary tumor resection. No germline or somatic pathogenic alterations affecting TP53, ATM, BRCA1, BRCA2, PALB2, or other DDR/HR-related genes included in the assays were detected. No RET, BRAF, or RAF1 fusions were detected by WGS, targeted DNA sequencing or targeted RNA sequencing; however, we detected a complex structural rearrangement involving exon 1 of SEL1L and exon 8 of NTRK1 resulting in an expressed, in-frame, likely oncogenic SEL1L::NTRK1 fusion containing the NTRK1 kinase domain (Table 1 and Fig. 1d, e).
Novel NBN reversion mutation in the metastatic biopsy
A genomic comparison of the biopsies from the liver metastasis obtained upon progression on FOLFIRINOX and the primary tumor resection revealed a novel SMAD4 homozygous deletion, an MCL1 amplification, and a second frameshift mutation in NBN. This NBN mutation, which was detected only in the liver metastasis specimen, but absent in the primary resection specimen, resulted in an additional 2-nucleotide deletion (c.1154_1155del; p.K385fs*27) 8 nucleotides downstream of the initial somatic mutation (Table 1 and Fig. 1b). Visual inspection of the genomic sequencing reads revealed that the NBN mutations were in cis (data not shown) and predicted to re-establish the NBN coding frame.
Multimodal detection of NTRK1 rearrangement and fusion
Following progression on FOLRIFINOX, the patient received larotrectinib due to the presence of an NTRK1 fusion. The patient, however, rapidly progressed within 2 months of initiating therapy. Given the lack of response to NTRK-targeted therapy, we aimed to validate the presence of the NTRK1 fusion. Targeted RNA sequencing of the primary resection confirmed the presence of a SEL1L::NTRK1 fusion and further delineated the breakpoints for the fusion transcript to be at exon 1 of SEL1L and exon 8 of NTRK1, resulting in an in-frame fusion containing the NTRK kinase domain. To understand the genomic mechanisms underlying this rare molecular anomaly, we examined the genomic rearrangements identified by WGS of the metastatic liver biopsy. No direct interaction was identified between SEL1L and NTRK1, rather, both genes rearranged with the same intron of CENPC (intron 17) resulting in a “bridged fusion” between SEL1L intron 1 and NTRK1 intron 6 (Fig. 1d, e)22. These data demonstrate that the skipping of NTRK1 exon 7 and usage of the next proximal splice site, exon 8, led to productive expression of the fusion transcript. Thus, the combined analysis of targeted RNA sequencing and WGS elucidated both the genomic and post-transcriptional details of this rare molecular event. The lack of larotrectinib antitumor activity in this case cannot be explained by the absence of an in-frame NTRK1 fusion, suggesting that another mechanism of primary resistance may be involved.
Characterization of genomic signatures
To identify additional genomic features of the tumor, we performed WGS on the metastatic liver biopsy specimen only, as the available primary resection specimen had insufficient tumor fraction and residual DNA. This analysis revealed the presence of 8789 single nucleotide variants (SNVs), 319 insertions and deletions (indels; 161 insertions and 158 deletions), and 37 structural variants (16 translocations, 8 deletions, 8 inversions, and 5 tandem duplications; Fig. 1e). The tumor specimen was found to be microsatellite-stable with a tumor mutational burden of 3.0 mutations per megabase, and an estimated tumor ploidy of 1.7. The dominant mutational signatures were related to aging (signatures 1 [12%] + 5 [29%]), followed by HRD (signatures 3 [27%] + 8 [11%]; Fig. 1f). The tumor, however, displayed only a partial set of the genomic features consistent with HRD. Despite the dominant HRD-related single nucleotide substitution signature and genome-wide loss-of-heterozygosity patterns, the enrichment for deletions with microhomology was modest, and rearrangement signatures associated with HRD were not observed. Consistent with these findings, the HRDetect score was 0.24, well below the cutoff used to classify a tumor as HRD (0.7). These results are consistent with genomic scars present in cancers with LOF alterations in genes upstream in the DDR pathway23,24,25.
Discussion
PACCs have been shown to harbor DDR/HR DNA repair defects, but somatic loss of NBN has not been linked to the development of pancreatic cancers in general, or specifically to PACC. In this case study, we performed detailed multimodal genomic profiling of multiple biopsies from the same patient with PACC revealing a comprehensive genomic characterization of the tumor evolution during multiple lines of therapy.
Our findings provide circumstantial evidence to suggest that the somatic NBN biallelic mutation likely played a role in the development and/or progression of this PACC. First, the biallelic inactivation of NBN through somatic mutation and copy-neutral loss of heterozygosity was linked to genomic signatures consistent with a partial HRD profile, but lacking long indels with (micro)homology, the main driver of HRDetect scores. This is expected, given that HRDetect was trained on genomes from patients with BRCA1/BRCA2 LOF in breast cancer, whereas NBN plays a role further upstream of BRCA1/BRCA2 in the repair of DNA double-strand breaks12,23. Although the genomic “scars” present in canonical HR-related genes (eg, BRCA1, BRCA2, PALB2, RAD51C, RAD51D, and RAD51B) and their respective absence in cancers with alterations affecting DDR-related genes (eg, ATM and CHEK2) have been described, the spectrum of genomic signatures in tumors with LOF of less common HR/DDR genes, including NBN, has yet to be fully characterized. Our findings emphasize the importance of further studies characterizing the genomic scars stemming from LOF of specific HR/DDR genes, which may not only provide mechanistic insights about back-up DNA repair mechanisms operating in absence of these genes, but also novel therapeutic opportunities based on synthetic lethality approaches26.
Second, an additional alteration predicted to restore the NBN reading frame was detected following initial treatment and therapeutic vulnerability to FOLFIRINOX. We interpreted this to be a novel reversion mutation based on (1) the potential selective pressure of the platinum-containing regimen FOLFIRINOX; (2) the biallelic nature of the primary NBN mutation in the tumor; (3) the proximity; and (4) the cis nature of the second NBN mutation to the first; and (5) the potential to re-establish the coding frame in the absence of an intervening stop codon. Identification of an emergent NBN reversion mutation in the post-therapy specimen might suggest an adaptive resistance mechanism mediated by restoration of NBN function. Reversion alterations in BRCA1/2, PALB2, RAD1C, and RAD51D have been documented as mechanisms of resistance to platinum-containing therapies and PARP inhibitors27. To our knowledge, this is the first report of a reversion in NBN following treatment with platinum or PARP inhibitor therapy in any tumor type.
Third, the patient did not respond to a selective NTRK inhibitor despite demonstrated expression of an in-frame NTRK1 fusion containing the kinase domain. This was unexpected, given the high response rates for NTRK inhibitors across tumor types including pancreatic cancers9,10. Most notably, a recent case report described a patient with PACC with the same SEL1L::NTRK1 fusion reported in this study, but lacking any other likely driver alterations, who had an exceptional response to the NTRK inhibitor, larotrectinib7. In contrast, one can posit that the lack of response to NTRK inhibition in our case might be explained by the presence of the NBN LOF mutation and genomic instability that has been associated with resistance to targeted therapies20,28.
Taken together, our findings are consistent with NBN LOF having played a role in the progression of this PACC, and provide evidence that LOF of DDR/HR-related genes may have a causative role in PACC more broadly. This observation warrants the consideration of other HR/DDR genes in ongoing studies of PARP inhibitors in pancreatic cancers, including PACC (NCT04858334 and NCT05286827). Finally, our report highlights the unique insights and the clinical impact stemming from comprehensive genomic analysis of rare tumors, such as PACC.
Methods
The patient reported in this case study was enrolled in a monotherapy arm of TRESR (NCT04497116), an ongoing, modular, phase 1/2a, first-in-human, multicenter, open-label, non-randomized, dose-escalation, dose-expansion study of camonsertib administered orally as a single agent, or in combination with talazoparib or gemcitabine in patients with advanced solid tumors. The study was conducted in accordance with the Declaration of Helsinki and Council for International Organizations of Medical Sciences International Ethical Guidelines, applicable International Conference on Harmonization Good Clinical Practice Guidelines, and applicable laws and regulations. The patient provided written informed consent which allowed for in-depth genomic investigation of the tumor and normal tissue specimens provided. The protocol was approved by the Institutional Review Board or ethics committee at the treating institution, Sarah Cannon Research Institute/Tennessee Oncology.
Tissue specimen review
Formalin-fixed paraffin-embedded (FFPE) tissue specimens corresponding to the primary resection (November 2018) and metastatic liver biopsy (July 2021) were retrieved. After biopsy, tissue specimens were immediately fixed in 10% neutral buffered formalin for 12–24 h. Genomic DNA and total RNA were extracted (Invitae; San Francisco, CA) using the AllPrep DNA/RNA FFPE kit (Qiagen, cat# 80234) and quantified using the Quant-iT dsDNA Assay Kit, broad range (ThermoFisher, cat# Q33120) at NeoGenomics using documented standard operating procedures as part of conduct of the trial (Supplementary Table 1). Corresponding hematoxylin and eosin sections were reviewed by a board-certified pathologist to review tumor cellularity and histology to confirm diagnoses. BCL-10 staining of the screening tissue to confirm the diagnosis was performed at NeoGenomics.
Biomarker monitoring
Serum CA19-9 levels were monitored routinely via commercial laboratory testing.
Clinical-grade NGS
Prior to enrollment into TRESR (NCT04497116), the patient’s tumor tissue specimens were analyzed by Tempus xT and Foundation Medicine FoundationOne CDx to guide therapy selection as part of standard of care at the treating institution16,19.
Retrospective central NGS testing
SNiPDx™ targeted sequencing
SNiPDx™ is a novel targeted sequencing panel capable of distinguishing monoallelic and biallelic LOF alterations in select DNA damage repair genes15. DNA (minimum of 30 ng) was extracted from 10 × 5 µm FFPE slides. DNA was analyzed on a custom AMP panel comprising 26 genes, referred to as SNiPDx™ 15. Libraries were quantitated using quantitative PCR (qPCR; Kapa Biosystems, Woburn, MA, USA) per manufacturer protocol. Amplicon sequencing was performed on the NovaSeq platform (Illumina, San Diego, CA, USA) according to the manufacturer’s standard protocol. Paired-end sequence data were processed using methods developed for AMP to align error-corrected reads18. AMP libraries were processed using the VariantPlex Pipeline from Archer Analysis Platform v6.2.8.
Genome-wide major and minor copy numbers were inferred by FACETS29. Briefly, copy number alterations and allelic imbalances in the 26 SNiPDx™ target genes and other genomic regions were calculated on the basis of the Log2R (ie, the Log2 ratio of single nucleotide polymorphism coverage in a tumor specimen to coverage in a matched panels of normal), and Log2 odds ratio (Log2OR; calculated from the number of reads reporting the alternative allele:number of reads reporting the reference allele), adjusted by tumor purity and ploidy.
Whole genome sequencing
All FFPE-isolated DNA was quantified by KAPA SYBR FAST qPCR quantification and quality control (QC) kit (Kapa Biosystems, Woburn, MA, USA). QC criteria required a minimum DNA concentration of >1 ng/µL and minimum amplifiable ratio of >0.1. WGS was performed on the NovaSeq targeting 60x coverage for tumor and 30x coverage for PBMCs. Genome sequencing covered a minimum of 95% of the genome with 20x coverage or higher. Nucleic acid from the submitted specimen was isolated and the library products were sequenced with 2 × 150 base pair reads using the Illumina NovaSeq sequencing instrument (Illumina, San Diego, CA). After alignment to the reference genome (GRCh37/hg19), off-target, low quality, and duplicate reads were removed from the analysis. The targeted regions were assessed for average depth of coverage and other data quality thresholds.
WGS analysis was performed as previously described20,21. Raw sequencing reads from FASTQ were aligned using the Burrow-Wheelers Aligner30 to the GRCh37/hg19 reference. SNVs were called using MuTect31. Short indels were called using VarScan232, Strelka33, Platypus34, and Scalpel35. Allele-specific copy number analysis was performed using FACETS29. Mutational signatures were decomposed using SIG NAL36. Structural variants were called using Manta37 and GRIDSS38. HRD scores were calculated using HRDetect23.
Targeted RNA sequencing
Targeted RNA sequencing was performed utilizing the Archer® FusionPlex® Custom Solid Panel with Anchored Multiplex PCR (AMP™; Invitae) at the Memorial Sloan Kettering Integrated Genomics Operation as previously described17,39. Results were analyzed utilizing the FusionPlex® standard analytical suite as previously described17,40.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
DNA and RNA sequencing data from this study are not publicly available because they contain potentially identifiable information on the patient, and there is no consent to deposit the data into a repository. For eligible studies, qualified researchers may request access to individual patient-level clinical data through a data request platform. At the time of writing, this request platform is Vivli (https://vivli.org/ourmember/roche/). Datasets may be requested 18 months after a clinical study report has been completed and, as appropriate, once the regulatory review of the indication or drug has completed. Access to patient-level data from this trial may be requested and will be assessed by an independent review panel, which decides whether the data will be provided, taking the risk of patient re-identification into consideration. Once approved, the data are available for up to 24 months. Anonymized records for individual patients across more than one data source external to Roche cannot, and should not, be linked owing to a potential increase in risk of patient re-identification. For up-to-date details on Roche’s Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see https://go.roche.com/data_sharing.
Code availability
The analysis of SNiPDx™ data is based on R package FACETS, where version 0.6.1 was used, downloaded from URL: https://github.com/mskcc/facets.
References
Calimano-Ramirez, L. F. et al. Pancreatic acinar cell carcinoma: A comprehensive review. World J. Gastroenterol. 28, 5827–5844 (2022).
Jiao, Y. et al. Whole-exome sequencing of pancreatic neoplasms with acinar differentiation. J. Pathol. 232, 428–435 (2014).
Furukawa, T. et al. Whole exome sequencing reveals recurrent mutations in BRCA2 and FAT genes in acinar cell carcinomas of the pancreas. Sci. Rep. 5, 8829 (2015).
Chmielecki, J. et al. Comprehensive genomic profiling of pancreatic acinar cell carcinomas identifies recurrent RAF fusions and frequent inactivation of DNA repair genes. Cancer Discov. 4, 1398–1405 (2014).
Chou, A. et al. RET gene rearrangements occur in a subset of pancreatic acinar cell carcinomas. Mod. Pathol. 33, 657–664 (2020).
Prall, O. W. J. et al. RAF1 rearrangements are common in pancreatic acinar cell carcinomas. Mod. Pathol. 33, 1811–1821 (2020).
Gupta, M. et al. Targeting the NTRK fusion gene in pancreatic acinar cell carcinoma: A case report and review of the literature. J. Natl. Compr. Canc Netw. 19, 10–15 (2021).
Campoverde, L., Vyas, M. & Bullock, A. Pancreatic acinar cell carcinoma in the setting of a BRCA1 germline mutation. Ann. Intern. Med. Clin. Cases 1, e220596 (2022).
Drilon, A. et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N. Engl. J. Med. 378, 731–739 (2018).
Drilon, A. et al. Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor entrectinib: Combined results from two phase I trials (ALKA-372-001 and STARTRK-1). Cancer Discov. 7, 400–409 (2017).
Subbiah, V. et al. Tumour-agnostic efficacy and safety of selpercatinib in patients with RET fusion-positive solid tumours other than lung or thyroid tumours (LIBRETTO-001): a phase 1/2, open-label, basket trial. Lancet Oncol. 23, 1261–1273 (2022).
Yamamoto, H. & Hirasawa, A. Homologous recombination deficiencies and hereditary tumors. Int. J. Mol. Sci. 23, 348 (2021).
Belhadj, S. et al. NBN pathogenic germline variants are associated with pan-cancer susceptibility and in vitro DNA damage response defects. Clin. Cancer Res. 29, 422–431 (2023).
Amin, M. B. et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 67, 93–99 (2017).
Glodzik, D. et al. Detection of biallelic loss of DNA repair genes in formalin-fixed, paraffin-embedded tumor samples using a novel tumor-only sequencing panel. J. Mol. Diagn. 25, 295–310 (2023).
Beaubier, N. et al. Clinical validation of the tempus xT next-generation targeted oncology sequencing assay. Oncotarget 10, 2384–2396 (2019).
Solomon, J. P. et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod. Pathol. 33, 38–46 (2020).
Zheng, Z. et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat. Med. 20, 1479–1484 (2014).
Milbury, C. A. et al. Clinical and analytical validation of FoundationOne(R)CDx, a comprehensive genomic profiling assay for solid tumors. PLoS One 17, e0264138 (2022).
Selenica, P. et al. APOBEC mutagenesis, kataegis, chromothripsis in EGFR-mutant osimertinib-resistant lung adenocarcinomas. Ann. Oncol. 33, 1284–1295 (2022).
Riaz, N. et al. Precision radiotherapy: Reduction in radiation for oropharyngeal cancer in the 30 ROC trial. J. Natl. Cancer Inst. 113, 742–751 (2021).
PCAWG Transcriptome Core Group. et al. Genomic basis for RNA alterations in cancer. Nature 578, 129–136 (2020).
Davies, H. et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med. 23, 517–525 (2017).
Polak, P. et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat. Genet 49, 1476–1486 (2017).
Riaz, N. et al. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat. Commun. 8, 857 (2017).
Setton, J. et al. Synthetic lethality in cancer therapeutics: the next generation. Cancer Discov. 11, 1626–1635 (2021).
Pettitt, S. J. et al. Clinical BRCA1/2 reversion analysis identifies hotspot mutations and predicted neoantigens associated with therapy resistance. Cancer Discov. 10, 1475–1488 (2020).
Lukow, D. A. et al. Chromosomal instability accelerates the evolution of resistance to anti-cancer therapies. Dev. Cell 56, 2427–2439.e2424 (2021).
Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Koboldt, D. C. et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Saunders, C. T. et al. Strelka: Accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012).
Rimmer, A. et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet 46, 912–918 (2014).
Fang, H. et al. Indel variant analysis of short-read sequencing data with Scalpel. Nat. Protoc. 11, 2529–2548 (2016).
Degasperi, A. et al. A practical framework and online tool for mutational signature analyses show inter-tissue variation and driver dependencies. Nat. Cancer 1, 249–263 (2020).
Chen, X. et al. Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32, 1220–1222 (2016).
Cameron, D. L. et al. GRIDSS: Sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res. 27, 2050–2060 (2017).
Yang, S. R. et al. A performance comparison of commonly used assays to detect RET fusions. Clin. Cancer Res. 27, 1316–1328 (2021).
Benayed, R. et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin. Cancer Res. 25, 4712–4722 (2019).
Acknowledgements
We would like to thank the patient, his family, and all investigators and staff involved in the study. We thank Parham Nejad for leading the biomarker operations for TRESR (NCT04497116) and facilitating the analyses presented in this case report study. Editorial support was provided by Allison TerBush, PhD of Onyx (a division of Prime, London, UK), supported by Repare Therapeutics according to Good Publication Practice guidelines. All authors accept final responsibility for the content of the manuscript and for the decision to submit the manuscript for publication. The patient was treated as part of TRESR (NCT04497116), which was funded by Repare Therapeutics Inc. J.S.R.F. is funded in part by the Breast Cancer Research Foundation, a Susan G Komen Leadership Grant, and the NIH/NCI P50 CA247749 01 and P30 CA08748 grants.
Author information
Authors and Affiliations
Contributions
M.S.P. treated the patient. D.U., I.M.S., V.R., and J.S.R.F. designed the study, which was supervised by I.M.S., V.R., and J.S.R.F. D.U., I.M.S., J.D.S., A.J., and P.S. analyzed the data. All authors contributed to writing the report and approved the final version.
Corresponding author
Ethics declarations
Competing interests
I.M.S., J.D.S., A.J., D.U., and V.R. are employees and own stock in Repare Therapeutics. J.S.R.F. declares personal/consultancy fees from and membership of the scientific advisory board of Repare Therapeutics. J.S.R.F. also reports personal/consultancy fees from Bain Capital, LP, Goldman Sachs, Paige.AI, Inc., SAGA Diagnostics, and Swarm, membership of the scientific advisory boards for Daiichi Sankyo, Merck, Paige.AI, Inc., Personalis, and Roche Tissue Diagnostics, and membership of the Board of Directors of Grupo Oncoclínicas, outside the scope of this study. M.S.P. reports consulting fees paid to their institution from AstraZeneca, Bayer, CytomX Therapeutics, Daiichi Sankyo, Ipsen, Novartis, Pfizer Inc., and Seagen Inc., and research support to their institution from Arcus Biosciences, Astellas Pharma Inc., BeiGene, BioNTech, Bristol Myers Squibb, Codiak BioSciences, CytomX Therapeutics, Eisai, Gilead Sciences, Inc., Gritstone Oncology, HiberCell, Immune-Onc Therapeutics, Inc., Leap Therapeutics, OncXerna Therapeutics, Panbela Therapeutics, Inc., Revolution Medicines, Surface Oncology, SQZ Biotechnologies, Translational Genomics, TransThera Sciences, ZielBio, and 1200 Pharma, outside the scope of this study. P.S. has no competing interests to disclose.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pelster, M.S., Silverman, I.M., Schonhoft, J.D. et al. Post-therapy emergence of an NBN reversion mutation in a patient with pancreatic acinar cell carcinoma. npj Precis. Onc. 8, 82 (2024). https://doi.org/10.1038/s41698-024-00497-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-024-00497-x