Abstract
Molecular profiling of childhood CNS tumors is critical for diagnosis and clinical management, yet tissue access is restricted due to the sensitive tumor location. We developed a targeted deep sequencing platform to detect tumor driver mutations, copy number variations, and heterogeneity in the liquid biome. Here, we present the sensitivity, specificity, and clinical relevance of our minimally invasive platform for tumor mutation profiling in children diagnosed with CNS cancer.
Similar content being viewed by others
Introduction
Central nervous system (CNS) tumors are the leading cause of childhood cancer-related death1. Molecular classification of tumor subtype is increasingly important for clinical management1,2, yet remains a challenge due to sensitive neuroanatomical locations and restricted tissue access. CNS tumors, including diffuse midline glioma (DMG), exhibit mutational heterogeneity3,4 which may not be captured by diagnostic surgical biopsy5. Moreover, these tumors are not amenable to repeat surgical biopsies, thus tumor genome evolution at disease progression5 remains largely unexplored.
To address the lack of ‘tumor visibility’, we previously developed a digital droplet PCR (ddPCR)-based approach to detect DMG driver mutations in circulating tumor DNA (ctDNA) from patient biofluids6,7,8. However, this approach is suitable only for monitoring a limited number of single nucleotide variants (e.g., up to 4 multiplexed mutations), and is not sufficient to detect larger-scale alterations. To achieve comprehensive tumor mutation profiling in the liquid biome, we validated a commercially available, targeted next generation sequencing (NGS) platform covering a panel of 523 cancer-associated genes (TSO500ctDNATM), encompassing all major prognostic and driver mutations associated with DMG5,9 and adapted this platform to detect tumorigenic alterations in ctDNA. In this proof-of-concept study, we evaluated platform performance in a cohort of paired plasma (1-2 mL) and CSF (500 µL-1 mL) specimens from children diagnosed with DMG (n = 10, Table 1). Paired tumor tissue was analyzed by whole exome (WES) or whole genome sequencing (WGS).
Results and discussion
Benchmarking ctDNA sequencing library preparation
Sequencing libraries were generated from cell free DNA between 90–250 base pairs (bp) in length to capture tumor-derived fragments10,11. We first benchmarked three metrics of ctDNA library preparation: (1) DNA input, (2) percentage of target exons with ≥500X coverage (‘Exons 500X’), and (3) median exon coverage (MEC). We sequenced multiple libraries from CSF (ID 846) ranging from 30–75 ng input (Fig. 1a, Supplementary Data 1). High assay reproducibility was observed when sequencing two replicates of 30 ng input DNA (Fig. 1a). All DNA inputs tested generated high quality libraries, with optimal QC results obtained at 60 ng. Specifically, 30 ng input resulted in Exons 500X of 73–74%, whereas increasing to 60 ng input improved Exons 500X to >90% (Fig. 1a, left). DMG driver mutations were detected at all inputs (Fig. 1b, right). Thus, subsequent CSF specimens were analyzed at ≥60 ng input where attainable. Plasma/serum specimens were analyzed at ≥50 ng input, given lower DNA yields (Table 1). Most (n = 7/10) CSF specimens achieved target yield (≥60 ng), and all liquid specimens successfully generated sequencing libraries.
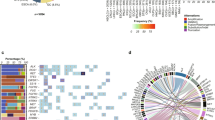
a Left: MEC (left y-axis) and Exons 500X (%; right y-axis) of sequencing libraries generated from 30–75 ng DNA input (ID 846CSF). ‘30ng-1’ and ‘30ng-2’ indicate technical replicates of 30 ng inputs. Right: Detection of DMG-associated mutations in genes H3-3A, PPM1D, and PIK3CA from libraries generated at 30, 50, 60, and 75 ng input. b Concordance of DMG-associated mutations detected in paired tumor tissue (T), CSF (C), and plasma (P) or serum (S). ‘X’: variant detected in tumor but not in paired liquid specimen. VAF variant allele frequency. c Representative overlap between tumor-associated mutations (SNVs, insertion/deletions) identified in paired specimens from two patients (IDs 933 and 1446). d Left: Comparison of H3K27M detection by ctDNA sequencing (black) and ddPCR (grey) in paired specimens (n = 8). Right: Detection of H3-3A K27M (10% VAF) by deep sequencing (ID 933CSF), with zero false positive reads of H3C2 K27M. e CNV plots showing chromosomes (chr) 4 and 12 (ID 1549). KIT and PDGFRA gains were detected in tumor and CSF, while KRAS gain was detected exclusively in CSF. X-axis = chromosome position; y-axis = fold-change (FC) calculated based on pre-established baseline; y = 1.3, CNV reporting threshold. f Detection of KIT (n = 1), PDGFRA (n = 1), KRAS (n = 1) and MDM4 (n = 2) gains in tumor and biofluid pairs. y = 1.3: CNV reporting threshold. g DNA methylation array-derived CNV plots of chr1 from pre-treatment (PT) and postmortem (PM) tumor tissue (ID 760), showing gain of chr1q containing MDM4, as validation of MDM4 gain. Y-axis: log-2 CN ratio. T tumor, C CSF, P plasma, S serum, CN copy number.
ctDNA deep sequencing reveals clinically relevant mutations
Deep sequencing achieved concordant mutation detection in paired tumor tissue and biofluids (Fig. 1b). In general, tumor and CSF pairs displayed higher concordance than tumor and plasma/serum pairs (Fig. 1b, c). Given that our cohort was comprised of DMGs, we evaluated detection of histone H3K27M mutation, present in 80% of DMGs and most commonly affecting H3-3A (60%) or H3C2 (20%) genes12,13,14,15. Most patients in our cohort harbored H3K27M (n = 9/10), confirmed by tumor DNA sequencing (Fig. 1b). Of the nine H3K27M mutations detected in tumor, eight were present in CSF and three in plasma/serum, for a positive percent agreement of 89% and 33%, respectively, with tumor results. We compared these results to our ddPCR-based H3K27M assay6,7 using eight paired liquid specimens, and detected similar H3K27M variant allele frequencies (VAF) using both approaches (Fig. 1d). In one case (ID 933CSF), ddPCR results were inconclusive, with one false positive H3C2 K27M-mutant allele detected (Supplementary Data 2). In contrast, deep sequencing resolved histone mutation status, detecting H3-3A K27M (9.8% VAF) with zero false positive reads at H3C2 locus, in agreement with tumor sequencing results (Fig. 1d). In one patient wildtype for H3 (ID 760), zero false positive reads of H3K27M were detected. Collectively, our results demonstrated high platform sensitivity and specificity for CSF mutation detection.
We next profiled clinically relevant histone partner mutations9,15 and detected alterations in genes including TP53, PPM1D, and PTEN in tumor and ctDNA pairs (Fig. 1b). To evaluate the clinical utility of our platform, we focused on CSF collected at upfront diagnosis or during the course of treatment (IDs 933, 1549). In these live CSF specimens, we identified several tumor driver mutations including H3-3A, TP53, PPM1D, ATRX, and KRAS mutations, which were confirmed present in paired tumor (Fig. 1b). Strikingly, although 1549 CSF yielded immeasurably low DNA (Table 1), oncogenic mutations were detected including H3K27M, KRAS and TP53 gain-of-function mutations (44-53% VAF) (Fig. 1b), supporting the utility of our platform for early detection of clinically relevant mutations even in cases with extremely low starting DNA input.
Capturing tumor genomic heterogeneity in liquid biome
We next assessed whether ctDNA was informative of tumor heterogeneity when compared to tissue. In one case (ID 1291CSF) we identified mutations including PTEN deletion, BCOR frameshift, and ACVR1 activating mutation that were absent from tumor WES. In a second case (ID 760 P), we detected a low frequency PIK3CA activating mutation, also absent from paired tumor WES. We inspected WES data and found that these variants were indeed present in raw sequencing files at subthreshold VAF (<10%), thus filtered and not reported. In line with our previous work5,9, mutations uncovered by liquid biome analyses may represent low frequency/sub-clonal events, further emphasizing the temporal and spatial heterogeneity and sub-clonal evolution of the DMG genome. Our observation indicates a potential advantage of ctDNA analyses to uncover tumor tissue genomic heterogeneity, including detection of sub-clonal mutations activating key DMG-associated growth factor signaling pathways.
Detection of tumor copy number variation in ctDNA
Finally, we assessed the feasibility of tumor copy number variation (CNV) detection in ctDNA. Interestingly, our platform proved more informative for CNV detection when compared to tumor WES. In patient 1549, CSF and tumor specimens showed concordance on KIT and PDGFRA gains (Fig. 1e, f). However, KRAS gain was detected exclusively in CSF, albeit at a relatively modest amplification (<2-fold change), representing a putative sub-clonal event not captured by tumor WES. Notably, this patient also harbored KRAS activating mutation, suggesting that KRAS gain may be associated with oncogene activation in this tumor, as reported in other cancers16,17,18. Additionally, MDM4 gains were detected in ctDNA (IDs 760, 1291) (Fig. 1f). In patient 760, tumor WES indicated a sub-threshold, but similar, gain of MDM4 (fold change 1.76 and 1.78 in tumor and plasma, respectively). This copy number gain, detected at a similar level by tumor and ctDNA sequencing, was subsequently confirmed by tumor DNA methylation analyses revealing amplification of chromosome 1q harboring MDM4 locus (Fig. 1g). Further studies are warranted to optimize the appropriate thresholds for CNV detection in ctDNA using large cohorts of CNS cancer and healthy pediatric control CSF and plasma.
Discussion
Our collective data present a minimally invasive, targeted deep sequencing platform capable of detecting and monitoring clinically relevant mutations in the liquid biome of children diagnosed with DMG and other CNS tumors, with remarkable sensitivity and specificity in CSF specimens. While CSF collection for molecular analyses is routine for children and young adults with certain CNS cancers (e.g., medulloblastoma and other embryonal tumors19), the procedure is currently not part of standard-of-care for children diagnosed with DMG. Our findings provide an opportunity for prospective clinical trials to incorporate CSF collection and deep sequencing for DMG molecular profiling as a less invasive alternative to surgical tissue biopsies. Though technology is becoming increasingly sensitive, future studies are needed to improve somatic mutation detection in plasma/serum. Additionally, one limitation of our study was the inclusion of retrospectively collected and postmortem specimens. Though the time from death to autopsy processing (Table 1) did not correlate to the total cell free DNA concentration, nor to the maximum somatic VAF detected in CSF (Supplementary Fig. 1), further studies are warranted using larger cohorts of live CSF specimens. These prospective studies will require standardization of collection time and processing protocols for uniformity across specimens. Importantly, while monitoring changes in the frequency of single hotspot mutations in ctDNA provides only limited insight into treatment response in DMGs7,20, the use of a large gene panel provides a comprehensive profile of the evolving tumor genomic landscape. Our results establish a foundation for comprehensive tumor mutation profiling in ctDNA, improving the ability to assess tumor molecular heterogeneity and genomic evolution in children diagnosed with CNS cancer.
Methods
Biological specimens
All patient specimens were collected after written informed consent was obtained from each patient or patient’s guardian for participation in a clinical trial, or biorepository as approved by the respective Institutional Review Boards of Children’s National Hospital (IRB #1339, #747) and University of California San Francisco (San Francisco, CA; IRB #14-13895). Patient specimens were collected during treatment (PNOC003, NCT02274987) or upon autopsy. All patient identifiers were removed with de-identified numerical identifiers were assigned. CSF from children with DMG (n = 10, Table 1) was collected during treatment or at autopsy. CSF specimens were centrifuged for 5000 × g for 10 min at 4 °C, supernatant was collected, aliquoted and stored at −80 °C. Plasma (n = 8) from children with DMG was collected as whole blood in purple top potassium EDTA tubes, inverted, and centrifuged at 2000 × g for 15 min at 4 °C. Serum (n = 2) from children with DMG was collected in gel-barrier tubes with clot activator and gel, incubated for 30 min at room temperature, and centrifuged at 2000 × g for 15 min at 4 °C. Plasma/serum supernatants were aliquoted into cryovials and stored at −80 °C. Tumor tissue (n = 10) was obtained from DMG patients during treatment or at autopsy and stored at −80 °C.
DNA isolation and quantification
Tumor tissue genomic DNA was isolated from frozen tissue specimens using Gentra Puregene Tissue Kit (QIAGEN) per the manufacturer’s instructions. Genomic DNA was quantified using Qubit dsDNA Broad Range Assay Kit. Cell free DNA was isolated from 500 µL-1 mL of CSF and 1-2 mL of plasma or serum (Table 1) using QIAamp Circulating Nucleic Acid Kit (QIAGEN) (ID 933CSF) or MagMax Cell-Free DNA Isolation Kit (ThermoFisher) (all other liquid specimens) according to the manufacturers’ instructions. DNA quantity was assessed using Qubit dsDNA High Sensitivity Assay kit.
Tape Station analysis of cell free DNA
Cell free DNA size was evaluated using the Agilent Technologies TapeStation 4200 High Sensitivity D5000 assay according to the manufacturer’s instructions.
Digital droplet PCR
ddPCR detection and quantification of H3-3A and H3C2 K27M variant allelic frequency (VAF) were performed as previously described6,7. Briefly, cell free DNA isolated from 500 µL CSF or 1 mL plasma/serum was subjected to pre-amplification using sequence-specific primers, dropletization, and PCR using sequence-specific primers and fluorescent probes. The numbers of mutant and wildtype droplets were quantified using RainDrop Analyst II Software (RainDance Technologies). VAF values were calculated as the number of mutant droplets divided by the sum of mutant and wildtype droplets. H3K27M VAF values represent the average of technical duplicates (CSF) or triplicates (plasma/serum).
Tumor whole exome sequencing
Whole exome libraries from each sample were generated by shearing 50 ng of genomic DNA to 150–180 bp using Covaris LE220 sonicator (Covaris, Woburn, MA). The following library preparation procedure was automated on a SciClone G3 liquid handling workstation using custom scripts. Sheared genomic DNA was processed using Kapa Hyper library construction and dual index kit (Kapa/Roche, Wilmington, MA) through end-repair, dA tailing, ligation with indexed Illumina adaptors on a SciClone G3 liquid handling workstation (Perkin Elmer, Waltham, MA). Adaptor-ligated libraries were purified using AMPure XP beads (Beckman Coulter, Indianapolis, IN) and then amplified by PCR (12 for germline specimens or 14 cycles for tumor FFPE specimens) using KAPA HiFi polymerase (KAPA/Roche). Amplified libraries were purified using AMPure XP beads, and 750 ng of each library was hybridized to a biotinylated RNA bait set (SureSelect XT Human V6 + COSMIC, Agilent Technologies, Santa Clara, CA) at 65 °C for 16 h. The captured genomic DNA fragments were enriched by Dynabeads MyOne Streptavidin T1 beads (Invitrogen/ThermoFisher Scientific) and amplified for 10 cycles of PCR using KAPA HiFi polymerase (KAPA/Roche). Amplified post-hybridization libraries were purified using AMPure XP beads, checked for size distribution (300–400 bp) using Agilent TapeStation 4200 and quantitated using Quant-iT™ High-Sensitivity dsDNA assay kit (ThermoFisher Scientific) on a SpectraMax M2e microplate reader (Molecular Devices, Hampton, NH). Final libraries were pooled at equimolar ratios and quantitated with a ddPCR Library Quantification Kit for Illumina TruSeq (Bio-Rad) on a QX200 digital PCR system (BioRad Laboratories, Hercules, CA). Final pooled libraries were diluted to either 1.2 nM or 2.0 nM depending on NovaSeq run mode: XP or Standard. Then, denatured final pool was clustered on a NovaSeq S4 flowcell (Illumina, San Diego, CA). Sequencing was performed on a NovaSeq 6000 (Illumina) using 2 × 150 bp paired-end sequencing mode.
Analysis of tumor-only WES data
An in-house bioinformatics pipeline was used to process tumor-only WES data. FASTQ data were generated using the bcl2fastq tool (Illumina, v2.18) and run through FASTQC for quality confirmation. Reads were mapped to the human hg19 reference genome using the Burrows-Wheeler alignment tool21. The resulting bam files were processed using GATK best practice workflow22. GATK HaplotypeCaller and Platypus23 were used to call variants. Copy number data was inferred from WES data through use of the CNVKit algorithm24, using a pool of normal Hapmap cell line specimens as references. The variants identified by WES were further annotated by the MoCha Oncogenic MOI Annotator (MOMA: https://github.com/FNL-MoCha/moma), a sequencing platform agnostic tool used to annotate variants as Mutations of Interest (MOIs) or Variants of Unknown Significance (VuS). These classifications are based on data from annotating variants with Annovar25 and mapping variants to OncoKB26.
Targeted cell free DNA NGS
Libraries were prepared using ≤50 ng DNA originating from plasma and serum, and ≤100 ng from CSF, using the Illumina TruSightTM Oncology 500 ctDNA with unique molecular identifiers (UMIs) and duplex barcodes for error correction, then enriched by target capture, pooled, and denatured according to the manufacturer instructions, and as described27. The denatured final pool was clustered on a NovaSeq S4 flowcell (Illumina, San Diego, CA). Sequencing was performed on a NovaSeq 6000 (Illumina) using 2 × 150 bp paired-end sequencing in XP mode.
TSO500ctDNA sequencing data analysis
Cell free DNA sequencing data were analyzed using the TSO500 analytical pipeline zipmocom 0.8.2.10, as described in Zhao et al.27. The TSO500ctDNA analysis pipeline was modified from the original FFPE pipeline27. This pipeline included Burrows-Wheeler Aligner to align reads to the reference genome; Unique Molecular Identifiers (UMIs) for read collapsing; Gemini to stitch together collapsed sequences into consensus fragments; Pisces somatic variant calling; and Nirvana to annotate variants. The variants annotated by this analysis were further annotated by MOMA as described above.
The original FFPE pipeline was modified to allow for lower VAF detection, and used a baseline derived from healthy donor plasma specimens to subtract background signal specific to cell free DNA materials. Alignment, collapsing, utilization of UMIs and post hoc filtering for germline variants were shared between the two pipelines, using thresholds relevant for different sample types. Limits of detection were set as 0.25% VAF for single nucleotide variants and insertion/deletions, 0.5% for translocations, and 1.3-fold change for CNVs.
Analysis of 765T whole genome sequencing data
One tumor tissue sample (765T) failed to generate a WES library. Thus, we queried previously generated, publicly available WGS data from this sample to determine tumor mutation status. Data were available from the Children’s Brain Tumor Network (CBTN) on the Cavatica platform (cavatica.sbgenomics.com).
To estimate VAF of mutations of interest in 765T, the somatic mutation caller Mutect2 in tumor-only mode from GATK28 was used.
DNA methylation analysis
DNA was extracted from frozen tumor tissue (ID 760), bisulphite converted using EZ DNA Methylation-Gold kit (Zymo Research) and hybridized onto Infinium MethylationEPIC BeadChip using Infinium MethylationEPIC BeadChip Kit per the manufacturer’s instructions (Illumina). BeadChip arrays were scanned using iScan Reader (Illumina). Data (IDAT files) were uploaded to the Molecular Neuropathology CNS Tumor Classifier Version v11b4 (https://www.molecularneuropathology.org/mnp) to generate CNV plots29.
Primer sequences
For ddPCR detection of H3K27M mutation, H3-3A and H3C2 primer sequences are listed in Panditharatna et al.7. For DNA sequencing, standard Illumina sequencing primers were used, are sequences are available at: https://support-docs.illumina.com/SHARE/AdapterSeq/Content/SHARE/AdapterSeq/TruSight/Tumor170/TruSightTumor500ctDNA.htm.
Statistical analysis
To test for correlations between the time from death to autopsy processing and (a) CSF cell free DNA concentration or (b) maximum somatic VAF detected in CSF (Supplementary Fig. 1), Spearman correlations were performed and two-tailed p-values were reported.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All sequencing datasets generated during the current study are available from the Blood Profiling Atlas in Cancer (BloodPAC) Data Commons (dataset identifier bpa-NCI). The sequencing data presented in the current publication have also been deposited in and are available from the dbGaP database under dbGaP accession phs003022.v1.p1. DNA methylation array data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE210323 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE210323).
Code availability
Code used to analyze sequencing data are available at: https://github.com/FNL-MoCha/nextgenseq_pipeline.
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019).
Louis, D. N. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 131, 803–820 (2016).
Hoffman, L. M. et al. Spatial genomic heterogeneity in diffuse intrinsic pontine and midline high-grade glioma: implications for diagnostic biopsy and targeted therapeutics. Acta Neuropathol. Commun. 4, 1 (2016).
Vinci, M. et al. Functional diversity and cooperativity between subclonal populations of pediatric glioblastoma and diffuse intrinsic pontine glioma cells. Nat. Med. 24, 1204–1215 (2018).
Mueller, S. et al. A pilot precision medicine trial for children with diffuse intrinsic pontine glioma-PNOC003: A report from the Pacific Pediatric Neuro-Oncology Consortium. Int. J. Cancer 145, 1889–1901 (2019).
Bonner, E. R. et al. Detection and Monitoring of Tumor Associated Circulating DNA in Patient Biofluids. J. Vis. Exp. https://doi.org/10.3791/59721 (2019).
Panditharatna, E. et al. Clinically Relevant and Minimally Invasive Tumor Surveillance of Pediatric Diffuse Midline Gliomas Using Patient-Derived Liquid Biopsy. Clin. Cancer Res. 24, 5850–5859 (2018).
Li, D. et al. Standardization of the liquid biopsy for pediatric diffuse midline glioma using ddPCR. Sci. Rep. 11, 5098 (2021).
Nikbakht, H. et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat. Commun. 7, 11185 (2016).
Underhill, H. R. et al. Fragment Length of Circulating Tumor DNA. PLoS Genet 12, e1006162 (2016).
Mouliere, F. et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 10, https://doi.org/10.1126/scitranslmed.aat4921 (2018).
Chan, K. M. et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 27, 985–990 (2013).
Wu, G. et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet 44, 251–253 (2012).
Castel, D. et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 130, 815–827 (2015).
Mackay, A. et al. Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell 32, 520–537.e525 (2017).
Modrek, B. et al. Oncogenic activating mutations are associated with local copy gain. Mol. Cancer Res. 7, 1244–1252 (2009).
Soh, J. et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One 4, e7464 (2009).
Kerr, E. M., Gaude, E., Turrell, F. K., Frezza, C. & Martins, C. P. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature 531, 110–113 (2016).
Gajjar, A. et al. Comparison of lumbar and shunt cerebrospinal fluid specimens for cytologic detection of leptomeningeal disease in pediatric patients with brain tumors. J. Clin. Oncol. 17, 1825–1828 (1999).
Mueller, S. et al. Mass cytometry detects H3.3K27M-specific vaccine responses in diffuse midline glioma. J. Clin. Investig. 130, 6325–6337 (2020).
Li, H. Aligning sequencing reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997 (2013).
Poplin, R. et al. Scaling accurate genetic variant discovery to tens of thousands of samples. BioRxiv, https://doi.org/10.1101/201178 (2018).
Rimmer, A. et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet 46, 912–918 (2014).
Talevich, E., Shain, A. H., Botton, T. & Bastian, B. C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol. 12, e1004873 (2016).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Chakravarty, D. et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017, https://doi.org/10.1200/PO.17.00011 (2017).
Chen Zhao, T. J., et al. TruSight Oncology 500: Enabling Comprehensive Genomic Profiling and Biomarker Reporting with Targeted Sequencing. BioRxiv, https://doi.org/10.1101/2020.10.21.349100 (2020).
Van der Auwera, G. A. & O’Connor, B. D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. (O’Reilly Media, Inc., 2020).
Hovestadt, V. & Zapatka, M. conumee: Enhanced copy-number variation analysis using Illumina DNA methylation arrays. R package version 1.9.0, https://doi.org/10.18129/B9.bioc.conumee.
Acknowledgements
The authors would like to acknowledge the generosity of all patients and their families. This work was supported by funding from the Isabella Kerr Molina Foundation, the National Cancer Institute and National Institutes of Health under contract HHSN261200800001E, Smashing Walnuts Foundation (Middleburg, VA), the V Foundation (Atlanta, GA), The Gabriella Miller Kids First Data Resource Center, Clinical and Translational Science Institute at Children’s National (5UL1TR001876-03), Matthew Larson Foundation (Franklin Lake, NJ), The Lilabean Foundation for Pediatric Brain Cancer Research (Silver Spring, MD), Kortney Rose Foundation (Oceanport, NJ), The Swifty Foundation (Woodridge, IL), Michael Mosier Defeat DIPG Foundation (Bethesda, MD), The Swiss National Science Fund (SNSF), Pediatric Brain Tumor Foundation (PBTF) (Asheville, NC), Pediatric Cancer Research Foundation (Irvine, CA), Kisses 4 Kayla Foundation, The Brad Kaminsky Foundation, and the We Love You Connie Foundation (Toronto, CA). This work is included in the forthcoming Doctoral Dissertation entitled “Characterizing the Evolving Genomic Landscape of Histone and Partner Gene Mutations in Pediatric High-Grade Gliomas” by E.R.B. of The George Washington University.
Author information
Authors and Affiliations
Contributions
E.R.B., C.K., J.N.: Conceived and designed the project. E.R.B: Wrote the manuscript text and created figures. E.R.B., R.H., A.D., B.D., L.C., R.P., A.P., D.H.: Designed and conducted experiments, contributed to data analysis and interpretation. J.N., R.H., A.E., M.B., C.N.K., H.G.D., P.M.W., C.K., J.D., R.P., S.M.: Contributed to the preparation of manuscript text and figures. E.R.B., A.E., M.B., C.K.: Contributed to the collection and processing of clinical and postmortem specimens. A.E., M.B., C.N.K., L.K., S.M.: Contributed to the acquisition of patient clinical data. J.N.: Provided critical input on the manuscript text and figures, supervised the project, and served as corresponding author.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bonner, E.R., Harrington, R., Eze, A. et al. Circulating tumor DNA sequencing provides comprehensive mutation profiling for pediatric central nervous system tumors. npj Precis. Onc. 6, 63 (2022). https://doi.org/10.1038/s41698-022-00306-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-022-00306-3
This article is cited by
-
Sequencing of cerebrospinal fluid cell-free DNA facilitated early differential diagnosis of intramedullary spinal cord tumors
npj Precision Oncology (2024)
-
Recent Developments in Blood Biomarkers in Neuro-oncology
Current Neurology and Neuroscience Reports (2023)
-
Molecular neuropathology: an essential and evolving toolbox for the diagnosis and clinical management of central nervous system tumors
Virchows Archiv (2023)