Abstract
Respiratory infections are common causes of acute exacerbation of chronic obstructive lung disease (AECOPD). We explored whether the pathogens causing AECOPD and clinical features changed from before to after the coronavirus disease 2019 (COVID-19) outbreak. We reviewed the medical records of patients hospitalized with AECOPD at four university hospitals between January 2017 and December 2018 and between January 2021 and December. We evaluated 1180 patients with AECOPD for whom medication histories were available. After the outbreak, the number of patients hospitalized with AECOPD was almost 44% lower compared with before the outbreak. Patients hospitalized with AECOPD after the outbreak were younger (75 vs. 77 years, p = 0.003) and more often stayed at home (96.6% vs. 88.6%, p < 0.001) than patients of AECOPD before the outbreak. Hospital stay was longer after the outbreak than before the outbreak (10 vs. 8 days. p < 0.001). After the COVID-19 outbreak, the identification rates of S. pneumoniae (15.3 vs. 6.2%, p < 0.001) and Hemophilus influenzae (6.4 vs. 2.4%, p = 0.002) decreased, whereas the identification rates of P. aeruginosa (9.4 vs. 13.7%, p = 0.023), Klebsiella pneumoniae (5.3 vs. 9.8%, p = 0.004), and methicillin-resistant Staphylococcus aureus (1.0 vs. 2.8%, p = 0.023) increased. After the outbreak, the identification rate of influenza A decreased (10.4 vs. 1.0%, p = 0.023). After the outbreak, the number of patients hospitalized with AECOPD was lower and the identification rates of community-transmitted pathogens tended to decrease, whereas the rates of pathogens capable of chronic colonization tended to increase. During the period of large-scale viral outbreaks that require quarantine, patients with AECOPD might be given more consideration for treatment against strains that can colonize chronic respiratory disease rather than community acquired pathogens.
Similar content being viewed by others
Introduction
An acute exacerbation of COPD (AECOPD) is a key event in the natural history of the disease, associated with declining health, worsening lung function, and poor prognosis1,2.{, #9;(GOLD), 2023 #1} Such exacerbations can be triggered by various factors, among which respiratory infections are most common3. Bacteria including Hemophilus influenzae, Moraxella catarrhalis, Streptococcus pneumoniae, and Pseudomonas aeruginosa, as well as various viruses, are strongly associated with exacerbation.
The patterns of infection with respiratory viruses other than severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) changed during and after the global pandemic of coronavirus disease 2019 (COVID-19)4. The incidences of common, seasonal respiratory viral infections dramatically decreased. Influenza infections have been at historically low levels since 2020; the rates of infection by human metapneumovirus, enterovirus, adenovirus, respiratory syncytial virus (RSV), and human rhinovirus (HRV) have also substantially decreased5. Moreover, the rates of certain bacterial infections have fallen since the outbreak. An earlier prospective analysis of surveillance data showed that the transmission rates of S. pneumoniae, H. influenzae, and Neisseria meningitidis decreased in many countries worldwide; these changes were associated with significant reductions in life-threatening invasive diseases6.
Equally impressive decreases in COPD exacerbation rates were reported worldwide during and after the pandemic7, the declines ranged from 44 to 73% globally7,8,9,10. Such findings may be associated with reduction in transmission of respiratory pathogens causing AECOPD3. However, detailed data regarding changes in these pathogens after the COVID-19 outbreak have not been reported. Therefore, this study evaluated whether the pathogens causing AECOPD, and the clinical features of the condition, changed after the COVID-19 outbreak.
Study design and methods
Study design
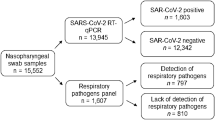
This retrospective multicenter cohort study was performed in four hospitals within the Republic of Korea. We collected the medical records of patients with AECOPD admitted between January 2017 and December 2018 (before the outbreak of COVID-19) and between January 2021 and December 2022 (after the outbreak). The inclusion criteria were: a history of COPD diagnosed via post-bronchodilator forced expiratory volume in 1 s (FEV1)/forced vital capacity (FVC) < 0.7; admission with AECOPD; age > 40 years; and availability of all conventional test data for pathogens causing AECOPD.
AECOPD was defined by a need for additional medication or hospitalization because of worsening clinical symptoms such as cough, sputum production, and/or dyspnea based on the definition within the Global Initiative for Obstructive Lung Disease (GOLD) guidelines1.
Variables
We collected demographic and clinicopathological information including age, sex, all comorbidities, lung function test results, smoking history, body mass index, exacerbation history, and medications used before AECOPD development. Clinical courses were evaluated in terms of intensive care unit admission and hospital mortality rates, inability to be discharged to home (i.e., transfer to a nursing hospital), and total hospitalization period.
Oral medications taken before AECOPD included xanthine derivatives, phosphodiesterase-4 inhibitors, and mucolytic agents. Inhaled treatments included long-acting beta 2-agonists (LABAs), long-acting muscarinic antagonists (LAMAs), inhaled corticosteroids, and combinations of these over ≥ 6 months before AECOPD development.
The microbiologic examination to identify the causative pathogen in patients with AECOPD was based on the first test performed upon hospitalization. Microbiological examinations included cultures of sputum or endotracheal aspirates; sputum polymerase chain reaction (PCR) tests for Chlamydophila pneumoniae, H. influenzae, S. pneumoniae, Mycoplasma pneumoniae, Legionella pneumophila, and viruses; serum tests for immunoglobulin M against C. pneumoniae and M. pneumoniae; urinary antigen tests for L. pneumophila and S. pneumoniae; and nasal swab tests for influenza A and B virus antigens. C. pneumoniae and M. pneumoniae were detected in respiratory specimens via PCR or (indirectly) in serum through immunoglobulin M measurements. L. pneumophila was detected in respiratory specimens via PCR and in urine using an antigen test. H. influenzae was detected in respiratory specimens by either PCR or culture. S. pneumoniae was detected in respiratory specimens by either PCR or culture, and in urine using an antigen test. All single or multiple pathogens identified were recorded.
Statistical analysis
Absent pathogen test data were treated as missing values. Frequencies were expressed as numbers (%); descriptive data were expressed as medians with interquartile ranges. The chi-squared test or Fisher’s exact test was used to compare categorical variables; continuous variables were compared with the Mann–Whitney U test. Factors significantly associated with survival were subjected to Cox proportional hazards modeling after adjustment for age and tested by the log-rank test. The duration of in-hospital survival was defined as the time between admission and hospital discharge, as noted in medical records. Hazard ratios with 95% confidence intervals were calculated. The threshold for statistical significance was set to p < 0.05.
Ethical approval
This study protocol was approved by the Institutional Review Board of Hallym University Kangnam (HKS 2023-11-008) Sacred Heart Hospital. All patient information was anonymized before analysis. Our institutional review boards (the Ethics Committee of Hallym University Kangnam Sacred Heart Hospital, Ethics Committee of Hallym University Sacred Heart Hospital, Ethics Committee of Hallym University Chuncheon Sacred Heart Hospital and the Ethics Committee of Ethics Committee of Hallym University Dontan Sacred Heart Hospital) approved this retrospective study and waived the requirement for informed consent from the patients. This study adhered to all relevant tenets of the 2013 revision of the Declaration of Helsinki.
Results
Patient characteristics
During the study period, 1186 patients with AECOPD were admitted to four hospitals. Among them, 418 patients were hospitalized after the outbreak of COVID-19, a decrease of approximately 44% over the same period compared to before the outbreak of COVID-19 (Table 1). The median age was 77 years, and 84% were men. After the COVID-19 outbreak, the patients were younger and more often men. In terms of comorbidities, congestive heart failure and liver cirrhosis were more common among patients admitted after the outbreak; the frequencies of other comorbidities did not differ between the two periods. The absolute FVC and FEV1 values were higher after the outbreak, but the predicted values did not differ, perhaps because the patients were younger compared with before the COVID-19 outbreak. Before COVID, more patients used LABAs or LAMAs alone than after the outbreak; LABA/LAMA combinations were more commonly utilized after the outbreak. Inhaled corticosteroid use did not differ between the two groups.
After COVID-19, the number of patients coming from home has increased, and the number of patients discharged to other hospitals after treatment has also decreased. Although the patient’s hospitalization period became longer after COVID-19 outbreak, there was no difference in the intensive care unit hospitalization rate or mortality rate between the two groups.
Microbiological analysis: changes after the COVID-19 outbreak
Detection rate of bacteria and virus after COVID-19 outbreak were 29.3% and 5.1%, respectively, which has decreased compared over same period before COVID-19 outbreak (Fig. 1). Before the outbreak, the most frequently identified bacteria were S. pneumoniae (11.7%) and P. aeruginosa (11.1%). However, after the outbreak, the identification rates of S. pneumoniae (15.3 vs. 6.2%, p < 0.001) and Hemophilus influenzae (6.4 vs. 2.4%, p = 0.002) decreased, whereas the identification rates of P. aeruginosa (9.4 vs. 13.7%, p = 0.023), K.pneumoniae (5.3 vs. 9.8%, p = 0.004), and methicillin-resistant Staphylococcus aureus (MRSA) (1.0 vs. 2.8%, p = 0.023) increased. The detection rates of all viruses except COVID-19 considerably decreased after the outbreak. In particular, the influenza A detection rate decreased from 10.4% to 1% (Fig. 2, Table 2). There were 44 cases (5%) in total with mixed bacterial and viral infections, 41 cases (6.6%) before the COVID-19 outbreak and 3 cases (0.6%) after the COVID-19 outbreak (Table 2).
Bacterial and viral detection rates during AECOPD before and after the COVID-19 outbreak: (A) Detection rates of all bacterial species. (B) Detection rates of all viral species. *p < 0.05. MRSA, methicillin-resistant Staphylococcus aureus. RSV, respiratory syncytial virus. COVID-19, coronavirus disease 2019.
Factors prognostic of mortality
Before the outbreak, histories of admission to another hospital and/or COPD exacerbation, and the presence of K. pneumoniae and/or MRSA, were prognostic of mortality. After the outbreak, only MRSA detection was prognostic of mortality (Table 3).
Discussion
The present study revealed changes in the AECOPD hospitalization rates and isolation rates of corresponding respiratory pathogens after the COVID-19 outbreak. The number of patients hospitalized with AECOPD decreased by approximately 44% compared with the number over the same period before the outbreak. Similar phenomena have been reported globally. In the United States, an analysis of the Veterans Affairs Corporate Data Warehouse, a national repository of electronic health records created during visits to all Veterans Affairs facilities, revealed a 48.4% decline in COPD admissions to Veterans Affairs hospitals after the outbreak8. In the United Kingdom, analysis of data from Public Health Scotland and the Secure Anonymized Information Linkage Databank of Wales revealed a 48% pooled reduction in AECOPD requiring hospital admission9. In Singapore, the monthly rate of acute COPD admissions decreased by more than 50% in the first 5 months (February–July 2020) after the outbreak10.
Decreases in AECOPD may be associated with reduced transmission of respiratory-associated pathogens. Most COPD exacerbations are caused by bacterial or viral infections3. Globally, the incidence of AECOPD and transmission levels of respiratory viruses simultaneously decreased11. The present study also showed that the overall detection rates of bacteria and viruses decreased after the outbreak, explaining the observed AECOPD reduction.
H. influenzae, M. catarrhalis, S. pneumoniae, and P. aeruginosa were commonly isolated from AECOPD patients in previous studies12. The most commonly detected viruses in such patients were rhinovirus, influenza A, and RSV13. As in a previous study12,13, S. pneumonia and P. aeruginosa were the frequently identified bacteria, whereas influenza A and rhinovirus were the most common viruses, in the present study. H. influenzae, M. catarrhalis, and RSV were also detected in the present work.
Notably, we found that pathogen detection rates changed after the COVID-19 outbreak. The incidences of S. pneumoniae, H. influenzae, and all viruses except COVID-19 significantly decreased after the outbreak. Such changes have been reported worldwide. A previous study demonstrated significant and sustained reductions in invasive diseases caused by S. pneumoniae, H. influenzae, and N. meningitidis, beginning in early 20206. All viral detection rates have declined worldwide. The Centers for Disease Control and Prevention reported a 98% decrease in influenza activity, from a median of 19.34% to 0.33%, in the United States14. Southern Hemisphere countries (Australia, Chile, and South Africa) have also reported minimal influenza activity. Furthermore, the detection rates of RSV, rhinovirus, metapneumovirus, and parainfluenza virus have decreased4. Consistent with previous international reports, we found that the influenza detection rate substantially decreased from 10.4 to 1.0%.
The changes may be partly explained by the widespread introduction of COVID-19 lockdown policies. S. pneumoniae and H. influenzae are typically transmitted person-to-person via the respiratory route15. Respiratory viruses are transmitted in respiratory droplets and aerosols16. Therefore, widespread adoption of COVID-19 containment policies, such as social distancing and the use of face masks in public spaces, may have reduced the transmission rates of respiratory-related pathogens and COVID-19. The COVID-19 containment policies and relevant public information campaigns slowed the transmission of respiratory-related pathogens, thereby reducing AECOPD rates. Therefore, COVID-19 containment policies are effective in lowering AECOPD levels. Although long-term implementation of strict COVID-19-like containment policies is impossible considering the socioeconomic costs, the COVID-19 experience may aid the establishment of strategies to prevent AECOPD.
The S. pneumoniae and respiratory virus detection rates significantly declined, but the detection rates of P. aeruginosa and MRSA significantly increased, after the COVID-19 outbreak. Both P. aeruginosa and MRSA are associated with poor AECOPD outcomes17,18. In other studies, K.pneumoniae, P.aerusinosa, and MRSA were reported as strains causing colonization in COPD patients19,20,21,22. Additionally, approximately 20% of hospitalized COPD patients is colonized with MRSA18. These pathogens have the capacity to engage in chronic colonization; the corresponding numbers may be less affected by COVID-19 quarantine policies, compared with the numbers of other pathogens. Colonization precedes obvious clinical infection. Murphy et al.23 reported that exacerbations caused by P. aeruginosa were more common in patients with advanced COPD compared with early COPD. The median predicted FEV1 of patients in the present study was 45% and almost half of the patients had a history of AECOPD, reflecting the severity of COPD. These results may explain the increased detection rate of P. aeruginosa after the COVID-19 outbreak.
MRSA is associated with adverse outcomes among patients with AECOPD. Narewski et al.18 reported that COPD patients colonized with MRSA had longer hospitalizations, required longer courses of antibiotics, and was more likely to require intensive care. Additionally, persistent infection with MRSA in patients with cystic fibrosis was associated with a more rapid rate of decline in lung function24. Similarly, we found that MRSA infection was prognostic of mortality before and after the COVID-19 outbreak. COPD patients colonized with MRSA may require close attention.
The present work was a multicenter study including a large number of patients and we comprehensively evaluated the changes in pathogens infecting AECOPD patients before and after the COVID-19 outbreak. However, our study had some limitations. First, this was a retrospective work. Second, we could not evaluate the pathogen status of AECOPD patients after COVID-19 containment policies were completely lifted. It remains unclear whether the observed changes have persisted since easing began. In May 2023, the World Health Organization declared that the COVID-19 public health emergency was over. Thus, an additional study is needed. Third, changes in healthcare utilization by patients with non-COVID-19 conditions during and after the outbreak may have affected pathogen detection rates25,26,27. Forth, this study was conducted on patients hospitalized for acute exacerbations of COPD, patients with severe bronchiectasis were not included, but there may have been patients with focal bronchiectasis, and there is a lack of distinction between respiratory structural abnormalities and their impact on microbial colonization. In the present study, the numbers of patients transferred from other hospitals and discharged to other hospitals after treatment both decreased during and after the outbreak. These findings may be related to difficult transfer between hospitals during lockdown, which could have impacted the pathogen detection rates.
Conclusion
After the COVID-19 outbreak, the number of hospitalized AECOPD patients decreased by almost 44% compared with the number during the same period before the pandemic. After the outbreak, the incidences of community-transmitted AECOPD pathogens tended to decrease, whereas the incidences of pathogens capable of chronic colonization tended to increase. The widespread introduction of COVID-19 containment policies, such as social distancing, may have lowered the transmission of respiratory-associated pathogens, thereby lowering the incidence of AECOPD. When MRSA was identified in AECOPD, patients had a high mortality rate both before and after the COVID-19 outbreak. Regardless of the viral outbreak situation, it is very important to consider treatment for strains associated with chronic colonization or drug resistance in AECOPD patients. However, since strains related to chronic colonization are detected at a higher frequency in AECOPD patients in large-scale viral outbreak situation, chronic colonization strains might need to be given more consideration in the treatment of AECOPD patients in outbreak.
Data availability
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
References
GOLD. Global strategy for prevention, diagnosis and management of COPD, 2023 report. (2023).
Park, Y. B. et al. Revised (2018) COPD clinical practice guideline of the Korean Academy of tuberculosis and respiratory disease: A summary. Tuberc. Respir. Dis. (Seoul) 81, 261–273 (2018).
Sethi, S. & Murphy, T. F. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N. Engl. J. Med. 359, 2355–2365 (2008).
Gomez, G. B., Mahe, C. & Chaves, S. S. Uncertain effects of the pandemic on respiratory viruses. Science 372, 1043–1044 (2021).
Huang, Q. S. et al. Impact of the COVID-19 nonpharmaceutical interventions on influenza and other respiratory viral infections in New Zealand. Nat. Commun. 12, 1001 (2021).
Brueggemann, A. B. et al. Changes in the incidence of invasive disease due to Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria meningitidis during the COVID-19 pandemic in 26 countries and territories in the Invasive respiratory infection surveillance initiative: A prospective analysis of surveillance data. Lancet Digit. Health 3, e360–e370 (2021).
Trujillo, C., Garnet, B., Zadeh, A. V., Urdaneta, G. & Campos, M. Decrease in exacerbations during the coronavirus disease 2019 pandemic in a cohort of veterans with COPD. Chronic Obstr. Pulm. Dis. 8, 572–579 (2021).
Baum, A. & Schwartz, M. D. Admissions to veterans affairs hospitals for emergency conditions during the COVID-19 pandemic. JAMA 324, 96–99 (2020).
Alsallakh, M. A. et al. Impact of COVID-19 lockdown on the incidence and mortality of acute exacerbations of chronic obstructive pulmonary disease: National interrupted time series analyses for Scotland and Wales. BMC Med. 19, 124 (2021).
Tan, J. Y., Conceicao, E. P., Wee, L. E., Sim, X. Y. J. & Venkatachalam, I. COVID-19 public health measures: A reduction in hospital admissions for COPD exacerbations. Thorax 76, 512–513 (2021).
Cookson, W., Moffatt, M., Rapeport, G. & Quint, J. A pandemic lesson for global lung diseases: Exacerbations are preventable. Am. J. Respir. Crit. Care Med. 205, 1271–1280 (2022).
Leung, J. M. et al. The role of acute and chronic respiratory colonization and infections in the pathogenesis of COPD. Respirology 22, 634–650 (2017).
Jafarinejad, H. et al. Worldwide prevalence of viral infection in AECOPD patients: A meta-analysis. Microb. Pathog. 113, 190–196 (2017).
Olsen, S. J. et al. Decreased influenza activity during the COVID-19 pandemic-United States, Australia, Chile, and South Africa, 2020. Am. J. Transpl. 20, 3681–3685 (2020).
Weiser, J. N., Ferreira, D. M. & Paton, J. C. Streptococcus pneumoniae: Transmission, colonization and invasion. Nat. Rev. Microbiol. 16, 355–367 (2018).
Wang, C. C. et al. Airborne transmission of respiratory viruses. Science https://doi.org/10.1126/science.abd9149 (2021).
Almagro, P. et al. Pseudomonas aeruginosa and mortality after hospital admission for chronic obstructive pulmonary disease. Respiration 84, 36–43 (2012).
Narewski, E. R., Kim, V., Marchetti, N., Jacobs, M. R. & Criner, G. J. Is Methicillin-resistant staphylococcus aureus colonization associated with worse outcomes in COPD hospitalizations?. Chronic Obstr. Pulm. Dis. 2, 252–258 (2015).
Jacobs, D. M. et al. Impact of pseudomonas aeruginosa isolation on mortality and outcomes in an outpatient chronic obstructive pulmonary disease cohort. Open Forum Infect. Dis. 7, 5fz546 (2020).
Sethi, S., Maloney, J., Grove, L., Wrona, C. & Berenson, C. S. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 173, 991–998 (2006).
Rodrigo-Troyano, A. et al. Pseudomonas aeruginosa in chronic obstructive pulmonary disease patients with frequent hospitalized exacerbations: A prospective multicentre study. Respiration 96, 417–424 (2018).
Ahmed, F. H. The bacterial colonization in patients with chronic obstructive pulmonary disease (COPD). Int. J. Med. Res. Health Sci. 10(1), 69–74 (2021).
Murphy, T. F. Pseudomonas aeruginosa in adults with chronic obstructive pulmonary disease. Curr. Opin. Pulm. Med. 15, 138–142 (2009).
Dasenbrook, E. C., Merlo, C. A., Diener-West, M., Lechtzin, N. & Boyle, M. P. Persistent methicillin-resistant Staphylococcus aureus and rate of FEV1 decline in cystic fibrosis. Am. J. Respir. Crit. Care Med. 178, 814–821 (2008).
Cassell, K., Zipfel, C. M., Bansal, S. & Weinberger, D. M. Trends in non-COVID-19 hospitalizations prior to and during the COVID-19 pandemic period, United States, 2017–2021. Nat. Commun. 13, 5930 (2022).
Becker, N. V., Karmakar, M., Tipirneni, R. & Ayanian, J. Z. Trends in hospitalizations for ambulatory care-sensitive conditions during the COVID-19 pandemic. JAMA Netw. Open 5, e222933 (2022).
Nguyen, J. L. et al. Pandemic-related declines in hospitalization for non-COVID-19-related illness in the United States from January through July 2020. PLoS One 17, e0262347 (2022).
Author information
Authors and Affiliations
Contributions
Y.S.S., S.J.K., T.K., and H.C.: Conceptualization and Methodology, writing original draft. T.R.S., H.I.K., S.H.J., and J.I.H.: data curation and validation. C.Y.L., S.C., and J.C.: formal analysis. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, S.J., Kim, T., Choi, H. et al. Respiratory pathogen and clinical features of hospitalized patients in acute exacerbation of chronic obstructive pulmonary disease after COVID 19 pandemic. Sci Rep 14, 10462 (2024). https://doi.org/10.1038/s41598-024-61360-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-61360-4
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.