Abstract
Stevia (Stevia rebaudiana Bertoni) is a natural sweetener plant that accumulates highly sweet steviol glycosides (SGs) especially in leaves. Stevia is native to humid areas and does not have a high tolerance to drought which is the most serious abiotic stress restricting its production worldwide. C2H2 zinc finger proteins (C2H2-ZFPs) are a group of well-known transcription factors that involves in various developmental, physiological and biochemical activities as well as in response to abiotic stresses. Here we analyzed C2H2-ZFP gene family in stevia and identified a total of 185 putative SrC2H2-ZF proteins from the genome sequence of S. rebaudiana. We further characterized the identified C2H2-ZF domains and their organization, additional domains and motifs and analyzed their physicochemical properties, localization and gene expression patterns. The cis-element analysis suggested multiple roles of SrC2H2-ZFPs in response to light, phytohormone, and abiotic stresses. In silico analysis revealed that the stevia C2H2-ZFP genes are interactively expressed in different tissues and developmental stages and some C2H2-ZFP genes are involved in response to drought stress. This study provides a background for future exploration of the functional, and regulatory aspects of the C2H2-ZFP gene family in S. rebaudiana.
Similar content being viewed by others
Introduction
Stevia (Stevia rebaudiana Bertoni) is a perennial short-day and medicinal herb of the Asteraceae family that is native to Brazil and Paraguay. It grows near water sources (high relative humidity) and prefers sandy, slightly acidic, and loamy soil with high humus content and good drainage1,2. Drought is the most serious abiotic stress restricting stevia production worldwide. Although stevia cannot tolerate drought, it grows in wide temperatures from 15 to 38 °C3. It is well known for the accumulation of low-calorie content and highly sweet steviol glycosides (SGs) especially in leaves. Despite the surging popularity of SGs, limitations in S. rebaudiana cultivation hinder widespread adoption. To bridge this gap and secure product quality, probing insights into stevia's growth, environmental resilience, and abiotic stress response is of great importance.
Transcription factors control the expression of their target genes by binding to specific DNA motifs. In plants, transcription factors are key regulators of various developmental processes and responding to environmental stresses4. In S. rebaudiana, zinc finger (ZF) transcription factors may modify the expression of genes that might be involved in stevioside biosynthesis5. Additionally, C2H2-ZFPs may contribute to stevia's resilience, helping it adapt to environmental stresses like drought and salinity6. ZF proteins represent a diverse family of transcription factors characterized by the presence of one or more zinc-stabilized structural motifs known as zinc fingers. These finger-like domains fold around a single zinc ion (Zn2+) coordinated by conserved cysteine (Cys) and histidine (His) residues, generating distinct subclasses based on their specific amino acid composition. Zinc finger proteins (ZFPs) play a pivotal role in regulating gene expression by binding to specific DNA sequences within the major groove of the double helix. Each zinc finger domain, functioning as an independent DNA recognition module, recognizes a short, three-nucleotide sequence with high specificity7,8. This modularity allows for combinatorial binding of multiple zinc fingers, enabling ZFPs to target a wide array of DNA sequences with exquisite precision. Hence, ZFPs mediate regulating gene expression and are involved in a wide range of biological processes, including development, differentiation, and stress response. The ZFP family encompasses nine distinct subclasses classified based on the number and arrangement of cysteine and histidine residues, as well as their secondary structure including Cys2/His2 (C2H2), C3H, C3HC4, C2HC5, C4HC3, C2HC, C4, C6 and C89.
The most prevalent subclass, C2H2-ZF proteins (also known as TFIIIA-type ZFPs), are implicated in various critical plant processes, including growth, development, stress resistance, and hormone signaling10,11. The role of C2H2 zinc fingers in gene regulation is to act as transcriptional activators or repressors, depending on the specific protein and the context in which it is functioning. The discovery of the first plant C2H2-ZFP gene, EPF1 from petunia, encoding two typical C2H2 zinc finger motifs, serves as a landmark in understanding the role of these versatile proteins in plant biology12. Many C2H2-type zinc finger protein genes have since been characterized in model and crop plants10. C2H2-ZF proteins with a common pattern of C-X(2–4)-C-X3-F-X5-L-X2-H–X(3–5)-H have 25–30 amino acids with two conserved Cys and two His that coordinate a Zn2 + atom on four sides8. Plant C2H2-ZFPs have a longer and more variable linker between their fingers and the QALGGH conserved sequence in the DNA-contacting surface of each finger13 compared to animals where the distance between two consecutive fingers is often short, seven or eight amino acids, as exemplified in fruit flies and yeast14.
Researchers categorized the plant C2H2-ZF proteins based on the number and arrangement of their C2H2 domains, and the architecture of these domains: Tandem C2H2-ZFPs have closely-spaced C2H2 domains, linked by less than 12 amino acids while dispersed C2H2-ZFPs have C2H2 domains further apart, separated by more than 11 amino acids15. Based on domain architecture, each C2H2 domain has been classified into Q-type (X2-C-X2-C-X7-QALGGH-X3-H) harboring a conserved QALGGH sequence), M-type (further divided into M1 to M5 subtypes based on 1 to 5 degraded amino acids in the QALGGH sequence), Z-type (spacing between the second cysteine and first histidine falls within specific ranges for Z1 and Z2 subtypes), D-type (missing the second histidine compared to other types)16,17. By using these criteria, the researchers created a detailed classification system for plant C2H2-ZFPs, providing a clearer understanding of their diverse structure and potential functions.
Here, we conducted a structural classification and expression analysis of C2H2 zinc finger protein (C2H2-ZFP) gene family in stevia and analyzed the expression of some C2H2-ZFP genes under drought stress.
Materials and methods
Identification of SrC2H2-ZFP genes
A flowchart of the analysis steps is presented in Supplementary Figure S1. The S. rebaudiana genomeic files were received from Xu et al.18, and the multiple sequence alignment (MSA) for C2H2 family was downloaded from the plant transcription factor database (http://planttfdb.gao-lab.org/) which belong to more than 160 species including model plants. The MSA was used it to make a Hidden Markov Model (HMM) profile by hmmbuild option of HMMER package19. This HMM profile was used to identify C2H2-ZFPs proteins of S. rebaudiana. The identified SrC2H2-ZF proteins were grouped based on the structure of each C2H2-ZFP domain, sequence length between the C2H2 zinc-finger domains, and the availability of additional domains in the protein15. SrC2H2-ZFPs were uploaded to the SMART database to distinguish the available domains. The proteins were also inspected manually to confirm the number of C2H2-ZF domains, the position of C2H2-ZF domain, and the space length between two adjacent C2H2-ZFs. The identified SrC2H2-ZF proteins were also assigned to iTAK (http://itak.feilab.net/cgi-bin/itak/online_itak.cgi) to further analyze via C2H2-ZFP domain search20. The longest C2H2-ZF protein variant from each gene that was confirmed to have the C2H2-ZFP domain(s) was considered as stevia C2H2-ZF protein and defined as SrC2H2.[type-description]. The confirmed SrC2H2-ZFPs were grouped into Q-, M-, and or D-type according to Alam et al.16. Briefly, SrC2H2-ZFPs having the conserved QALGGH motif and the conserved X2-C-X2-C-X7-QALGGH-X3-H pattern in the SrC2H2-ZF domains were classified as Q-type; while proteins with one to five degraded amino acids in the QALGGH motif and specific changes in the spacing between two cysteines (C2) and two histidines (H2) were grouped as M1 to M5. The D-type does not have the second histidine in the SrC2H2-ZF domain compared with the Q and M types. This classification placed, SrC2H2-ZFPs containing a single C2H2-ZF domain into 1i (e.g. 1i-Q, 1i-M). C2H2-ZFPs containing two C2H2-ZF domains were defined as 2i (e.g. 2i-Q and 2i-M). The SrC2H2-ZFPs containing at least two types of n non-tandemly repeated C2H2-ZF domains were classified as ni-Mx. Proteins with three or more than three ZF domains with at least two tandemly repeated ZF domains were presented as t1 and t2, respectively. Two ZF domains separated by up to 11 amino acids were considered tandem duplication15.
Physicochemical characterization of C2H2-ZFPs
Physicochemical properties of each of the identified SrC2H2-ZF proteins including amino acid sequence length (AA), molecular weight (MW), isoelectric point (pI), instability index, aliphatic index, and grand average of hydropathicity were obtained from the ProtParam website (https://web.expasy.org/protparam; Gasteiger et al., 2005). The structure of SrC2H2-ZFP genes was extracted from the S. rebaudiana GFF file and the gene structures were presented using the Gene Structure Display Server at http://gsds.cbi.pku.edu.cn21. The conserved domains of the SrC2H2-ZF proteins were searched from Pfam22 and MEME23 web servers and the output tables were visualized in TBtools24. Subcellular localization of the SrC2H2-ZFPs was predicted using the Plant-mPLoc webpage25.
Phylogenetic analysis
The identified SrC2H2-ZF protein sequences were aligned using the ClustalOmega argument of msa function from “msa” package26 in R (The R Project for Statistical Computing, Vienna, Austria). Then, the alignment was used to obtain a neighbor-joining tree by applying 500 bootstrap replicates using the “ape” package27. The resulting newick tree was visualized in R by using the “ggtree” package28.
Evolution of SrC2H2-ZFP gene family
Based on the phylogenetic analysis, the gene pairs encoding the most similar proteins were considered as the recently duplicated genes. The coding sequences (CDS) of these gene pairs were extracted and their Ka/Ks (non-synonymous/synonymous substitution) ratios were obtained using the KaKs_Calculator29. The divergence time for the duplicated genes was predicted by T = Ks/2λ × 10–6, where Ks is the rate of the synonymous nucleotide substitution and λ is the number of substitutions per synonymous site per generation. The λ of 6.5 × 10−9 was used according to Yang, Zhang, Yue, Tian and Chen30.
Expression analysis of C2H2-ZFP genes
RNAseq data for different developmental stages and tissues (NCBI, BioProject ID PRJNA705537)31 and leaf responses to nitrogen deficiency (NCBI, BioProject: PRJNA681456)18 of S. rebaudiana were downloaded from the SRA database of NCBI. After quality control using FastQC and trimming using Trimmomatic32, the low-quality section of reads, the reads of each sample were aligned to the stevia reference genome using the HISAT2 aligner and the aligned reads were assembled using StringTie under its default settings33. Extraction of abundance estimates as FPKM (fragments per kilobase of transcript per million mapped reads) values, for the SrC2H2-ZFP genes and differential expression analysis were done using the ballgown package34. Heatmaps were generated from log2(FPKM + 1) transformed expression values of SrC2H2-ZFP genes using R package “pheatmap”. We also extracted the log2 fold changes (log2FC) of the differentially expressed SrC2H2 genes from the RNAseq results of S. rebaudiana roots and leaves under drought (50% of field capacity, one-day hold) and salinity (120 mM) stresses6 and presented the barplots of those values using the R package “ggplot2”35.
Cis-regulatory elements of C2H2-ZFP genes
The upstream sequences (2-Kb) of S. rebaudiana SrC2H2-ZFP genes were extracted from the stevia genome using TBtools36. The cis-acting elements of the extracted sequences were predicted using the PlantCARE tool37.
Semi-quantitative reverse transcription PCR (sqRT-PCR)
One-month-old seedlings of S. rebaudiana were provided by the Agricultural Research Center of Guilan Province, Iran. Seedlings were transplanted to pots (20 cm diameter and 25 cm height) filled with garden soil and kept in a greenhouse with 22 °C temperature and 16 h of daily light. After two months, we applied drought stress to half of the plants by stopping watering until the leaves wilted while the control plants retained under normal irrigation. We then harvested the second and third leaves from the second lateral stem of both control and stressed plants. The tissues were frozen immediately in liquid nitrogen, and kept at − 80 °C until RNA extraction. Three biological replicates were collected for each of the control and drought treated conditions.
From each frozen stevia leaf sample, total RNA was extracted using acid guanidinium thiocyanate–phenol–chloroform method38. The quality of the isolated RNA was evaluated by both agarose gel electrophoresis and NanoDrop spectrophotometry (Supplementary File S1). To prepare for further analysis, 500 ng of RNA per sample was reverse transcribed into cDNA using a TaKaRa kit (Dalian, China). These cDNA products were diluted tenfold and kept frozen at − 20 °C until needed. Three stevia C2H2-ZFP genes with reasonable expression were selected based on the results of the analysis of publicly available RNAseq data as described above and their expression was analyzed by sqRT-PCR in three technical replicates. Specific primers for the selected genes were designed using the online Primer3Plus tool. The forward and reverse primers of each selected SrC2H2-ZFP gene were designed from two adjacent exons of their cDNA sequences spanning the interval intron. This would allow to identify undesired PCR products if amplified from genomic DNA contamination. The stevia Actin (AF548026.1) was served as an internal reference gene. Band intensities were quantified using ImageJ39, and mean expression between control and drought conditions for each gene was compared using a t-test.
Protein–protein interaction (PPI) network prediction
The Arabidopsis genes orthologous to the stevia C2H2 genes were identified using OrthoVenn340. By referring to the differentially expressed genes in RNAseq results from a stevia drought experiment6, the Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/) was retrieved for predicting protein–protein interactions (default confidence score ≥ 0.40) using the Arabidopsis orthologs of the stevia C2H2-ZFPs that overexpressed under drought stress.
Results
Identification of C2H2-ZFP genes and properties of their proteins
A total of 185 SrC2H2-ZFP genes were identified. In total, 283 transcripts (1–7 transcripts per gene) and on average 1.54 transcripts per gene are encoded. SrC2H2-ZF gene IDs, longest transcripts and protein IDs, assigned gene names, chromosomal positions, general domain architecture and C2H2 domain sequences are presented in Supplementary Dataset S1 and the longest protein sequences of each od SrC2H2-ZF genes are available in Supplementary File S2. The molecular weights of the longest proteins of the SrC2H2-ZFP gene transcripts range from 12,448.27 to 158,960.08 Daltons (Da) and the isoelectric point (PIs) ranged from 4.88 to 10.05. All of the SrC2H2-ZF proteins had a negative grand average of hydropathicity indicating the hydrophilic nature of StC2H2-ZF proteins. The range of the instability indices of the SrC2H2-ZF proteins was between 32.52 and 75.72. For the majority of the proteins, the instability index was higher than 40.00, suggesting that most SrC2H2-ZF proteins are unstable (Supplementary Dataset S2).
We generally classified the stevia SrC2H2-ZF proteins into non-tandem and tandem groups. Each of these two groups was further divided into subgroups based on the number and pattern of ZFs and the conservation rate of the QALGGH domain (Fig. 1). For example, 1i subgroup contains a single C2H2-ZFPs domain (e.g. 1i-Q, 1i-M), 2i harbours two C2H2-ZF domains (e.g. 2i-Q and 2i-M), and ni-Mx contains at least two types of n non-tandemly repeated C2H2-ZF domains. C2H2-ZFPs containing 3 or 4 non-tandem ZF domains were defined as 3i and 4i, respectively. Proteins with three or more than three ZF domains with at least two tandemly repeated ZF domains were presented as t1 and t2, respectively. For instance, t1-M has three M-type ZF domains and t2-Mx has at least four ZF domains composed of at least two different ZF types (Supplementary Dataset S1). Conserved motifs of SrC2H2-ZF proteins in S. rebaudiana were also identified by MEME. Ten conserved motifs were determined as shown in Fig. 2 from which motifs 1, 3, and 9 are C2H2 zinc finger proteins.
Grouping of 185 SrC2H2-ZF proteins of S. rebaudiana based on the number and pattern of C2H2 zinc finger (ZF) domains. Each group has further been divided into subsets based on the variation in C2H2 domain sequences. Numbers of SrC2H2-ZFPs in the 10 different subsets containing 1–4 non-tandem C2H2-ZF domains and number of proteins with up to three (t1) or more than three (t2) C2H2-ZF domains with at least two tandemly repeated domains are presented. Detailed information on corresponding genes is available in Supplementary Dataset S1.
Additional domains outside of the C2H2-ZF domains
To improve the classification of the 185 S. rebaudiana C2H2-ZFPs, the complete protein sequence of each SrC2H2-ZFP was examined using the SMART tool to identify any other domains besides the C2H2-ZF domains. In total, 14 other known domains outside the C2H2-ZF domains were recognized on 21 different SrC2H2-ZF proteins (Supplementary Dataset S1). These domains include the transmembrane domain, Zinc ion binding (ZnF_U1, ZnF_AN1, ZnF_Rad18), Histone binding/phosphorylation/methylation domains (PreSET, SET, PostSET, JmjN, JmjC), coiled coil, EXOIII, DnaJ, JmjN, JmjC, PUG, UBX. The first group consists of 164 proteins which solely harbored C2H2-ZF domains, without any other additional detected domains. Seven SrC2H2-ZF proteins contained coiled-coil domain. Each of ZnF_U1 and EXOIII were present in three different SrC2H2-ZFPs. Each of transmembrane and ZnF_AN1 were present in two SrC2H2-ZF proteins and 4 other SrC2H2-ZFPs proteins contained a combination of two or three other domains. Based on the subcellular localization predictions, 179 (97%) members of S. rebaudiana C2H2-ZFPs are localized in the nucleus, 5 (3%) C2H2-ZFs are localized in the chloroplast and nucleus, and 1 is localized in Golgi apparatus and nucleus (Supplementary Dataset S1).
Phylogeny of the Stevia C2H2-ZF proteins
We assessed the phylogenetic relationships of the stevia SrC2H2-ZF proteins. The result is presented in Fig. 3, and the detailed gene IDs is available in Supplementary Dataset S1. The phylogenetic tree shows that the bootstrap supports for some main clades are considerably low, however, most of the SrC2H2-ZFPs in the same subgroup were clustered together with a high bootstrap support indicating that a high structural variation exists among different types of SrC2H2-ZFPs. Generally, the peptide sequences were relatively conserved among different members of SrC2H2-ZFPs of each subgroup, suggesting that the phylogenetic classification led to a reasonable tree.
Phylogenetic classification of S. rebaudiana C2H2-ZF proteins. The proteins were aligned by Clustal-omega and the phylogenetic tree was determined by using the ape package in R with the Neighbor–Joining (NJ) algorithm and a bootstrap analysis of 1,000 replicates. The tree represents 10 S. rebaudiana C2H2-ZFP clades as shown by numbers 1 to 10 outside the circle. The color of the protein names is based on C2H2-ZFP types. Brown circles outside of the tree indicate the mean expression of the corresponding genes based on RNAseq analysis of seven different tissues31, where small and big circles indicate means less or more than 1 FPKM, respectively.
Structural properties and evolutionary divergence of the SrC2H2-ZFP gene family
Gene structure analysis (Supplementary Figure S2) revealed that out of the 185 identified C2H2-ZFP genes, 92 (50%) lack intron. The remaining genes contained 1 to 11 introns with length ranging from 31 (in Streb.Contig00126G000030) to 19,496 (in Streb.1G026630) bases. Only three genes had intron longer than 9000 bases (Supplementary Dataset S3). No correlation between the number of C2H2-ZF domains and the number of exons in corresponding gene (Supplementary Dataset S1) was observed for stevia C2H2-ZFPs (R2 = 9.0%). We analyzed the evolutionary time divergence between coding sequences of 15 most recently duplicated pairs of the SrC2H2-ZFP genes (Supplementary Dataset S4). The result revealed that these genes have been duplicated and diverged 0.08–14.2 million years ago (MYA). However, for most (73%, 11 out of 15) of the analyzed genes, the divergent time ranged from 0.08 to 2.9 MYA, suggesting a recent duplication event for SrC2H2-ZFP genes. All the studied gene pairs had a Ka/Ks value less than 1, indicating that they have been subjected to purifying selection.
Analysis of chromosomal locations of SrC2H2-ZFP genes
A physical map of the location of the SrC2H2-ZFP genes on the S. rebaudiana chromosomes is illustrated in Fig. 4. The SrC2H2-ZFP genes were mapped to 11 stevia chromosomes plus the unassembled part of the genome. The SrC2H2-ZFP genes showed uneven distribution across the chromosomes with an average of 16 genes on each chromosome and a higher density on chromosome 1 which harbored 32 genes. SrC2H2-ZFP genes were mainly localized toward the distal ends of the chromosome arms. The genes with similar ZF domain architecture are mostly clustered together on the chromosomes.
Physical map of C2H2-ZFP genes on the Stevia rebaudiana chromosomes. The density of C2H2-ZFPs along the chromosomes is highly accompanied by general gene density in the S. rebaudiana genome31. The genes are colored according to the Fig. 1 classification. Ten C2H2-ZFP genes were located on unassembled contigs and were not presented on this map.
Cis-regulatory elements and miRNAs
To illuminate how S. rebaudiana SrC2H2-ZFP genes respond to external cues, the 2 kb promoter region of 185 SrC2H2-ZFP genes was analyzed using the PlantCARE database. In total, 5319 potential regulatory elements possibly responding with light, hormonal signals, defense and stress-related, etc. (Fig. 5 and Table 1) were identified. Considering only the factor presented in Table 1, each SrC2H2-ZFP gene promoter harbored 2 (for Streb.2G014040 promoter) to 61 (for Streb.2G005970 promoter) cis-acting elements. While light-responsive elements predominated, binding sites for hormonal, drought, and low-temperature signals were also identified. Although cis-elements responding to light was most widely distributed, we only presented the number of identified cis-acting elements related to hormone and environmental stresses in Fig. 5. Streb.5G008640 and Streb.2G005860 had the most abundant cis-acting elements related to hormone (22) and low-temperature (7). The maximum drought related element (MBS binding element) was 3 which was observed for the promoters of Streb.9G010930, Streb.11G011770, Streb.11G018970, and Streb.7G004510 genes (Table 1). A considerable number of cis-acting elements related to abscisic acid (ABA) compared to other stimuli was observed in SrC2H2-ZFP promoter regions (Fig. 5). Different types of promoter cis-components were observed, such as ABRE (abscisic acid response element), AuxRR and TGA (auxin response element), CGTCA (methyl jasmonate response element), GARE, P-box and TATC-Box (gibberellin response element), TCA and SARE (salicylic acid response element), DRE (dehydration, low temperature and salt stress responsive element), MBS (drought response element), LTR (low temperature response element), and TC rich repeats that may be involved in the defense and stress response.
Heatmap of possible cis-acting elements in the 2-kb upstream of the SrC2H2-ZFP genes of S. rebaudiana. The elements were predicted using the PlantCARE website37. The number of the identified elements (shown on the right) for each gene is presented inside the cells and the stimuli are indicated on the left.
We found miRNA targets in intronic sequences for five genes including Streb.1G035890 (ath-miR5658), Streb.2G029620 (pta-miR171), Streb.11G004260 (bdi-miR845), Streb.6G002350 (gma-miR5035-3p) and Streb.5G019230 (ath-miR5658), all at the expectation value of 1.5 for Streb.1G035890 which was 1. At the cDNA level, 6 SrC2H2-ZFP genes were the target for ath-miR5658 and one was the target for osa-miR2055 with cut-off expectation threshold of 0, 1 or 1.5 (Supplementary Dataset S3).
SrC2H2-ZFPs expression patterns
To get insight into the SrC2H2-ZFPs expression patterns, we used two independent set of publicly available RNAseq reads, one from seven different developmental stages of S. rebaudiana including root at seedling stage (RS), stem at seedling stage (SS) leaves at seedling stage (LS), leaf at vegetative (LV), leaf at bud (LB), leaf at initial flowering (LIF) and leaf at peak flowering stage (LPF)31 and the other set from leaves under control and nitrogen deficiency18. From 185 identified C2H2-ZF proteins, 91 and 109 had a mean expression of less than 1 FPKM (fragments per kilobase of transcript per million fragments mapped) in different developmental stages and under nitrogen deficiency conditions, respectively. These low transcriptionally active genes were discarded from the expression analysis. The expression heatmap of the remaining genes generated from RNAseq data of developmental stages (Fig. 6A) indicates that two groups of C2H2-ZFP genes exist based on their expression patterns. Group 1 genes are highly expressed in all S. rebaudiana tissues while group 2 genes are expressed specifically at seedling or flowering stages. Among the latter, some genes expressed specifically at the RS (root at seedling) stage and switched off in aerial parts of S. rebaudiana while a few genes (e.g. SrC2H2.1i-Q.43) showed a reverse expression pattern. Some genes such as SrC2H2.t1-M.03 (Streb.4G011560) expressed evenly in all stages. Some genes such as SrC2H2.2i-Q.26 and SrC2H2.2i-Q.11 showed extremely high expression at the RS (root at seedling) stage. We further intended to analyze RNAseq data from experiments conducted under various stress treatments to elucidate the expression patterns on C2H2-ZFP genes under stress conditions as well. Based on the RNAseq data from the nitrogen deficiency investigation, none of the members of the C2H2-ZFP family was significantly affected by nitrogen deficiency in the leaves of stevia under nitrogen-deficient conditions (Fig. 6B), although three members of the C3H-ZF family were among the upregulated genes18.
SrC2H2-ZFP gene expression patterns. a) Expression patterns of SrC2H2-ZFP genes in different tissues of S. rebaudiana. RS, SS, and LS: root, stem and leaf at the seedling stage, respectively. LV, LB, LIF, and LPF: leaf at vegetative, bud, initial flowering and peak flowering stages, respectively. b) Expression patterns of C2H2-ZFP genes in stevia leaves under Control and nitrogen deficiency (ND). The expression rates are based on Log2(FPKM + 1) and C2H2-ZFP genes with a mean expression of less than 1 FPKM were excluded.
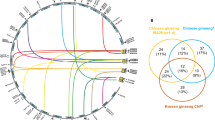
We further investigated the expression changes of SrC2H2-ZFP genes under drought and salinity conditions from the RNAseq results of a recently published study6. From the 185 SrC2H2-ZFP genes, 25 were up- or down-regulated in stevia leaves or roots by drought or salt stress (Fig. 7, Supplementary Dataset S5). Of these, six SrC2H2-ZFP genes were up- and eight were down-regulated in stevia lives followed by drought stress. No significantly overexpressed SrC2H2-ZFP gene was detected in leaves or roots under salinity conditions, although 14 were down-regulated in roots. Arabidopsis orthologs of the stevia SrC2H2-ZFP genes which were differentially expressed under drought or salt stresses were determined using BlastP (Supplementary Dataset S6) and protein interaction networks for the Arabidopsis orthologs of four significantly overexpressed stevia genes under drought was constructed using STRING database (Fig. 8). The corresponding proteins are ZAT12, ZAT9, ZAT8 and T18C15_3. ZAT12 or RHL41 (AT5G59820) is orthologous to SrC2H2.2i-Q.20 (Fig. 8A), ZAT9 (AT3G60580) is orthologous to SrC2H2.3i-Mx.06 (Fig. 8B), ZAT8 (AT3G46080) is orthologous to SrC2H2.2i-Q.23 (Fig. 8C) and T18C15_3 (AT1G49900) is orthologous to SrC2H2.2i-Q.19 (Fig. 8D). The predicted functional partners for the above network clusters are presented in Table 2, showing the score and number of nodes and edges in each cluster, and, localization of the partner proteins. Most of the partner proteins were transcription factors and predicted to be localized in nucleus, although a few proteins showed cytoplasm, endoplasmic reticulum (ER) or chloroplast localization (Table 2).
Barplots analysis of differentially expressed SrC2H2-ZFP genes (FDR < 0.05) in leaves (a and b) and roots (c and d) of S. rebaudiana under drought and salinity stresses based on published RNAseq data6. Up- and down-regulation are indicated by cyan and orange colors, respectively. Under salinity conditions, no significantly up- or downregulated C2H2 genes were found for leaf (c) but many of them have been down-regulated in root (b).
The protein–protein interaction networks for the Arabidopsis orthologs of four significantly overexpressed stevia genes under drought stress: (a) ZAT12 or RHL41 (AT5G59820) which is orthologous to SrC2H2.2i-Q.20; (b) ZAT9 (AT3G60580) is orthologous to: SrC2H2.3i-Mx.06; (c) ZAT8 (AT3G46080) which is orthologous to SrC2H2.2i-Q.23; (d) T18C15_3 (AT1G49900) is orthologous to SrC2H2.2i-Q.19. Nodes are proteins, and edges are interactions, colored by evidence type.
Relative expression of SrC2H2-ZFP genes under drought stress
To get insight into the expression of SrC2H2-ZFP genes, three C2H2 genes (including SrC2H2.t1-Mx.26, SrC2H2.1i-M.01, and SrC2H2.3i-Mx.01), were selected for expression analysis. Supplementary Dataset S7 details the sqRT-PCR primers and their expected amplicon sizes. SqRT-PCR was performed with the cDNA samples prepared from leaves total RNA. Although low-level variations were observed, none of the analyzed genes showed significant expression levels between the control and drought-stressed conditions as revealed by statistical comparisons after quantifying the RT-PCR band intensities (Supplementary Figure S3).
Discussion
Classification of the SrC2H2-ZF proteins
The C2H2-ZFP gene family is one of the well-studied gene families and has been studied in many plants such as Arabidopsis41, petunia42, wheat43,44, cotton45,46, soybeans15, rice47 and sorghum17. These proteins are well-known for their involvement in various developmental processes48,49 as well as in response to abiotic stresses such as drought and salinity10,11, 50,51,52. We identified a total of 185 putative SrC2H2-ZF proteins from the genome sequence of S. rebaudiana that in total encode 283 transcripts. All the SrC2H2-ZF proteins were predicted to localize in the nucleus supporting their role as a transcription factor in the nucleus. 5 C2H2-ZF proteins were also predicted to be localized in chloroplast suggesting that they may be also involved in chloroplast gene expression.
We followed a generally accepted method for the classification of stevia C2H2-ZF proteins based on the number and patterns (tandem or dispersed) of zinc fingers distribution41, and the plant-specific QALGGH conserved motif13. Based on this method, the C2H2-ZFP family in stevia is divided into 5 main groups and 10 subgroups (Fig. 1 and Supplementary Dataset S1). Alternatively, the C2H2-ZFP gene family in Arabidopsis has been divided into A, B, and C groups where A1 (containing tandemly arranged ZFs) and C1 (one or two to five dispersed ZFs and with a QALGGH motif in the ZF domain) compose the two largest and evolutionarily youngest and active C2H2-ZFPs41.
Most of the stevia C2H2-ZF proteins contain one or more C2H2 zinc fingers with the QALGGH motif and were classified as Q-type. The Q-type C2H2-ZFPs are plant-specific and their abundance suggests their involvement in plant developmental and physiological processes as well as responses to biotic and abiotic stresses11,12,13,53.
Phylogeny, physical distribution, and gene duplication
Phylogeny, physical mapping and domain and motif analysis of the stevia C2H2-ZF proteins indicate that the genes with similar ZF domain architecture are mostly clustered together in the genome suggesting tandem gene duplication and possibly their similar biological functions. Gene duplication is a major drive of evolving the paralog members within gene families54, commonly undergoing new functionalization or pseudogenizations55,56,57.
Analysis of the evolutionary time divergence between coding sequences of 15 most recently duplicated pairs of the SrC2H2-ZFP genes (Supplementary Dataset S3) revealed a duplication time divergence of 0.08–14.2 MYA. However, for most (11 out of 15, 73%) of the analyzed gene pairs, the divergent time ranged from 0.08 to 2.9 MYA, suggesting a recent duplication event for SrC2H2-ZFP genes. All the studied gene pairs had a Ka/Ks value less than 1, indicating that they have been subjected to purifying selection, thus, they are conserved and expected to retain conserved functionality, while pairs with a high divergence time value may acquire new functions58.
Evolution of SrC2H2-ZFP gene family
Cis-regulatory elements are non-coding regions of DNA that govern the transcription of nearby genes. Cis elements including enhances and silencers are present in variable distances from the gene and can be several kilobase away from the gene59. A reasonably higher number of cis-acting elements for abscisic acid compared to other stimuli was observed in SrC2H2-ZFP promoter regions emphasizing a general involvement in abscisic acid pathways related to abiotic stress responses. In fact, C2H2-ZF proteins can induce abiotic stress tolerance through ABA-mediated signal pathways10,11.
SrC2H2-ZFP gene expression
The analysis of the publicly available RNAseq data revealed that the stevia C2H2-ZFP genes are interactively expressed in different tissues and developmental stages. C2H2-type zinc finger proteins play widespread functions in plant tolerance to different abiotic stresses, such as drought, salinity, cold, osmotic and oxidative stresses10,11. Genes of the C2H2 family have shown differential expression under salt and drought stresses in Brassica rapa16. Responsive C2H2 genes were also identified under cold and drought in Sorghum bicolor17 and Chinese cabbage60, and under salt and drought stresses in Solanum tuberosum61 and Larix kaempferi62.
We also looked for the expression changes of SrC2H2-ZFP genes under drought and salinity stresses6. Out of the 185 SrC2H2-ZFP genes, 25 genes (~ 13%) were up- or down-regulated in stevia leaves or roots by drought or salt stress. The analysis of the Arabidopsis orthologous of these genes (Supplementary Dataset S5) showed that they are involved in abiotic stress tolerances possibly via gibberellin (GAF1-2, AT3G50700)63 and abscisic acid signaling10. SrC2H2.2i-Q.23 is one of the genes up-regulated in leaves (Fig. 7A) but down-regulated in roots under drought stress (Fig. 7C). It has been reported that overexpression of the Arabidopsis ortholog of SrC2H2.2i-Q.23 (i.e. ZAT18, AT3G53600) improves drought tolerance as revealed by higher leaf water content and antioxidant enzyme activities, but lower content of reactive oxygen species after drought treatment compared to control plants64. ZAT18 also is connected with RAS1 (Fig. 8A) which is a negative regulator of salt tolerance probably by enhancing abscisic acid sensitivity65.
We further analyzed the expression of SrC2H2.t1-Mx.26, SrC2H2.1i-M.01, and SrC2H2.3i-Mx.01 genes by sqRT-PCR. The sqRT-PCR reactions were performed with the cDNA samples belonging to the total RNA isolated from the leaves. Although low-level variations were observed, the expression of none of the analyzed genes significantly changed by the drought stress (Supplementary Figure S2). So, the analyzed genes were not among those that are affected by drought. We selected these genes because they showed expression in leaves based on the results of the analysis of publicly available RNAseq data for different tissues and developmental stages which was not under any stress condition31.
Cis-regulatory elements and miRNA targets
The analysis of cis-acting elements in SrC2H2-ZFP promoter regions suggests that the stevia C2H2-ZFP family members generally responds to light and phytohormones indicating that they could play a role in abiotic stresses via hormone responses such as abscisic acid (ABA), methyl jasmonate (MeJA), salicylic acid (SA), and auxin. Some SrC2H2-ZFP genes harbours considerable number of ABA responsive elements and some genes contain both ABA and drought response cis-elements in their promoter regions suggesting that they may respond to drought stress through the ABA signaling pathway66. At the maximum expectation value of 1.5, we found miRNA targets in intronic sequences for five genes and at the cDNA level, 5 SrC2H2-ZFP genes were the target for ath-miR5658 (Supplementary Dataset S3). In Arabidopsis, miR5658 is mainly seen in the nucleus and activate the expression of AT3G25290 (an auxin-responsive family protein) directly by binding to its promoter67. Hence, possibly there are other regulation layers for SrC2H2-ZFP genes involving auxin pathway and miR5658 miRNA.
Conclusion
A comprehensive genome-wide analysis of C2H2-ZFPs in S. rebaudiana led to the identification of 185 SrC2H2-ZFP genes distributed over all the chromosomes. SrC2H2-ZF proteins were grouped into 5 groups and 12 subgroups based on the number and pattern of C2H2 zinc finger domains. All the SrC2H2-ZF proteins were predicted to localize in the nucleus. The Q-type C2H2-ZFPs were the most abundant C2H2-ZF proteins. As supported by physical mapping and phylogeny, we argued that gene duplication events have significantly expanded and evolved the stevia C2H2-ZFP genes. The SrC2H2-ZFP Expression patterns in different tissues and developmental stages as well as under drought and salinity conditions confirmed that Q-type C2H2-ZFPs along with the M and Mx types are interactively involved in development and drought stress responses. Some genes such as SrC2H2.2i-Q.19 were upregulated in both roots and leaves under drought condition. The study identified some target gene for further genetic improvement of stevia. The protein–protein interaction networks can further clarify the potential functions of C2H2-ZF proteins. In conclusion, our results provide a root for in-detailed characterization of C2H2-ZFP functions in growth and development and abiotic stress response in stevia.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Lemus-Mondaca, R., Vega-Gálvez, A., Zura-Bravo, L. & Ah-Hen, K. Stevia rebaudiana Bertoni, source of a high-potency natural sweetener: A comprehensive review on the biochemical, nutritional and functional aspects. Food Chem. 132, 1121–1132 (2012).
Soejarto, D., Compadre, C. M., Medon, P., Kamath, S. & Kinghorn, A. D. Potential sweetening agents of plant origin. II. Field search for sweet-tasting Stevia species. Econ. Bot. 37, 71–79 (1983).
Doherty, J. (University of Hawai'i at Manoa, A PhD Dissertation, 2023).
Riechmann, J. L. & Ratcliffe, O. J. A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 3, 423–434 (2000).
Zhang, T. et al. The SrWRKY71 transcription factor negatively regulates SrUGT76G1 expression in Stevia rebaudiana. Plant Physiol. Biochem. 148, 26–34 (2020).
Pal, P. et al. Genome-wide transcriptional profiling and physiological investigation elucidating the molecular mechanism of multiple abiotic stress response in Stevia rebaudiana Bertoni. Sci. Rep. 13, 19853 (2023).
Elrod-Erickson, M. & Pabo, C. O. Binding Studies with Mutants of Zif268: Contribution of individual side chains to binding affinity and specificity in the Zif268 zinc finger-DNA complex. J. Biol. Chem. 274, 19281–19285 (1999).
Pavletich, N. P. & Pabo, C. O. JJo. B. C. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 Å. Science 252, 809–817 (1991).
Kosarev, P., Mayer, K. F. & Hardtke, C. S. Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. 3, 1–12 (2002).
Han, G. et al. C2H2 zinc finger proteins: Master regulators of abiotic stress responses in plants. Front. Plant Sci. 11, 115 (2020).
Liu, Y., Khan, A. R. & Gan, Y. C2H2 zinc finger proteins response to abiotic stress in plants. Int. J. Mol. Sci. 23, 2730 (2022).
Takatsuji, H., Mori, M., Benfey, P. N., Ren, L. & Chua, N. H. Characterization of a zinc finger DNA-binding protein expressed specifically in Petunia petals and seedlings. EMBO J. 11, 241–249 (1992).
Takatsuji, H. Zinc-finger proteins: The classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39, 1073–1078 (1999).
Schuh, R. et al. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Krüppel, a Drosophila segmentation gene. Cell 47, 1025–1032 (1986).
Yuan, S. et al. Genome-wide identification and classification of soybean C2H2 zinc finger proteins and their expression analysis in legume-rhizobium symbiosis. Front. Microbiol. 9, 126 (2018).
Alam, I., Batool, K., Cui, D. L., Yang, Y. Q. & Lu, Y. H. Comprehensive genomic survey, structural classification and expression analysis of C2H2 zinc finger protein gene family in Brassica rapa L.. PloS One 14, e0216071 (2019).
Cui, H. et al. Genome-wide analysis of C2H2 zinc Finger gene family and Its response to cold and drought stress in sorghum [Sorghum bicolor (L.) Moench]. Int. J. Mol. Sci. 23, 5571 (2022).
Sun, Y. et al. Comparative transcriptome analysis provides insights into steviol glycoside synthesis in stevia (Stevia rebaudiana Bertoni) leaves under nitrogen deficiency. Plant Cell Rep. 40, 1709–1722 (2021).
Potter, S. C. et al. HMMER web server: 2018 Update. Nucleic Acids Res. 46, W200–W204 (2018).
Zheng, Y. et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 9, 1667–1670 (2016).
Hu, B. et al. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31, 1296–1297 (2015).
El-Gebali, S. et al. The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 (2019).
Bailey, T. L. et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Chen, C., Chen, H., He, Y. & Xia, R. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv 289660 (2018).
Chou, K.-C. & Shen, H.-B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PloS One 5, e11335 (2010).
Bodenhofer, U., Bonatesta, E., Horejš-Kainrath, C. & Hochreiter, S. msa: An R package for multiple sequence alignment. Bioinformatics 31, 3997–3999 (2015).
Paradis, E. & Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019).
Yu, G. et al. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. 8, 28–36 (2017).
Zhang, Z. et al. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 4, 259–263 (2006).
Yang, S., Zhang, X., Yue, J.-X., Tian, D. & Chen, J.-Q. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genom. 280, 187–198 (2008).
Xu, X. et al. The chromosome-level Stevia genome provides insights into steviol glycoside biosynthesis. Hortic. Res. 8, 129 (2021).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650 (2016).
Frazee, A. C. et al. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 33, 243 (2015).
Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 3, 180–185 (2011).
Chen, C. et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202 (2020).
Lescot, M. et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327 (2002).
Chomczynski, P. & Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: Twenty-something years on. Nat. Protoc. 1, 581–585 (2006).
Abràmoff, M. D., Magalhães, P. J. & Ram, S. J. Image processing with ImageJ. Biophotonics Int. 11, 36–42 (2004).
Sun, J. et al. OrthoVenn3: An integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res. 51, W397–W403 (2023).
Englbrecht, C. C., Schoof, H. & Böhm, S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genom. 5, 1–17 (2004).
Kobayashi, A. et al. Seven zinc-finger transcription factors are expressed sequentially during the development of anthers in petunia. Plant J. 13, 571–576 (1998).
Faraji, S., Rasouli, S. H. & Kazemitabar, S. K. Genome-wide exploration of C2H2 zinc finger family in durum wheat (Triticum turgidum ssp. durum): Insights into the roles in biological processes especially stress response. Biometals 31, 1019–1042 (2018).
Li, Y. et al. Comprehensive genomic survey, structural classification and expression analysis of C2H2-type zinc finger factor in wheat (Triticum aestivum L.). BMC Plant Biol. 21, 1–18 (2021).
Salih, H., Odongo, M. R., Gong, W., He, S. & Du, X. Genome-wide analysis of cotton C2H2-zinc finger transcription factor family and their expression analysis during fiber development. BMC Plant Biol. 19, 1–17 (2019).
Rehman, A. et al. Identification of C2H2 subfamily ZAT genes in Gossypium species reveals GhZAT34 and GhZAT79 enhanced salt tolerance in Arabidopsis and cotton. Int. J. Biol. Macromol. 184, 967–980 (2021).
Agarwal, P. et al. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 65, 467–485 (2007).
Lyu, T. & Cao, J. Cys2/His2 zinc-finger proteins in transcriptional regulation of flower development. Int. J. Mol. Sci. 19, 2589 (2018).
Li, Y. et al. Fine mapping and genetic analysis identified a C2H2-type zinc finger as a candidate gene for heading date regulation in wheat. Theor. Appl. Genet. 136, 140 (2023).
Liu, K. et al. Genome-wide analysis of C2H2 zinc finger family and their response to abiotic stresses in apple. Gene 148164 (2024).
Feng, Q. et al. The C2H2 family protein ZAT17 engages in the cadmium stress response by interacting with PRL1 in Arabidopsis. J. Hazard. Mater. 465, 133528 (2024).
Huang, R. et al. Zinc finger transcription factor MtZPT2-2 negatively regulates salt tolerance in Medicago truncatula. Plant Physiol. 194, 564–577 (2024).
Kubo, K.-I. et al. Cys2/His2 zinc-finger protein family of petunia: Evolution and general mechanism of target-sequence recognition. Nucl. Acids Res. 26, 608–615 (1998).
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D. & May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 1–21 (2004).
Flagel, L. E. & Wendel, J. F. Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564 (2009).
Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 18, 292–298 (2003).
Goldtzvik, Y., Sen, N., Lam, S. D. & Orengo, C. Protein diversification through post-translational modifications, alternative splicing, and gene duplication. Curr. Opin. Struct. Biol. 81, 102640 (2023).
Hurst, L. D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 18, 486–487 (2002).
Biłas, R., Szafran, K., Hnatuszko-Konka, K. & Kononowicz, A. K. Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cult. (PCTOC) 127, 269–287 (2016).
Chen, L., Jiang, C., Ye, L., Gao, Y. & Hou, X. Genome-wide identification of C2H2-ZFPs and functional analysis of BcZAT12 and BcZAT10 under cold stress in non-heading Chinese cabbage. Environ. Exp. Bot. 217, 105574 (2024).
Liu, Z. et al. Genome-wide identification and analysis of the Q-type C2H2 gene family in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 153, 327–340 (2020).
Shao, L. et al. Identification of C2H2 zinc finger genes through genome-wide association study and functional analyses of LkZFPs in response to stresses in Larix kaempferi. BMC Plant Biol. 23, 298 (2023).
Fukazawa, J. et al. DELLAs Function as Coactivators of GAI-ASSOCIATED FACTOR1 in Regulation of Gibberellin Homeostasis and Signaling in Arabidopsis. Plant Cell 26, 2920–2938 (2014).
Yin, M. et al. The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J. Exp. Bot. 68, 2991–3005 (2017).
Ren, Z. et al. RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. 107, 5669–5674 (2010).
Lim, C. W., Baek, W., Jung, J., Kim, J.-H. & Lee, S. C. Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 16, 15251–15270 (2015).
Yang, G. et al. MicroRNAs transcriptionally regulate promoter activity in Arabidopsis thaliana. J. Integr. Plant Biol. 61, 1128–1133 (2019).
Acknowledgements
The University of Kurdistan financially supported this work.
Author information
Authors and Affiliations
Contributions
S.N. conducted experiments. R.E. conducted experiments and revised the manuscript. M.M. helped with research designation and revised the manuscript. G.M. conceived and designed research, conducted data analysis, and wrote the manuscript. All the authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nikraftar, S., Ebrahimzadegan, R., Majdi, M. et al. Genome-wide analysis of the C2H2-ZFP gene family in Stevia rebaudiana reveals involvement in abiotic stress response. Sci Rep 14, 6164 (2024). https://doi.org/10.1038/s41598-024-56624-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-56624-y
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.