Abstract
Extracellular vesicles (EVs) have been implicated in metastasis and proposed as cancer biomarkers. However, heterogeneity and small size makes assessments of EVs challenging. Often, EVs are isolated from biofluids, losing spatial and temporal context and thus lacking the ability to access EVs in situ in their native microenvironment. This work examines the capabilities of label-free nonlinear optical microscopy to extract biochemical optical metrics of EVs in ex vivo tissue and EVs isolated from biofluids in cases of human breast cancer, comparing these metrics within and between EV sources. Before surgery, fresh urine and blood serum samples were obtained from human participants scheduled for breast tumor surgery (24 malignant, 6 benign) or healthy participants scheduled for breast reduction surgery (4 control). EVs were directly imaged both in intact ex vivo tissue that was removed during surgery and in samples isolated from biofluids by differential ultracentrifugation. Isolated EVs and freshly excised ex vivo breast tissue samples were imaged with custom nonlinear optical microscopes to extract single-EV optical metabolic signatures of NAD(P)H and FAD autofluorescence. Optical metrics were significantly altered in cases of malignant breast cancer in biofluid-derived EVs and intact tissue EVs compared to control samples. Specifically, urinary isolated EVs showed elevated NAD(P)H fluorescence lifetime in cases of malignant cancer, serum-derived isolated EVs showed decreased optical redox ratio in stage II cancer, but not earlier stages, and ex vivo breast tissue showed an elevated number of EVs in cases of malignant cancer. Results further indicated significant differences in the measured optical metabolic signature based on EV source (urine, serum and tissue) within individuals.
Similar content being viewed by others
Introduction
Extracellular vesicles (EVs) are nanometer- to micrometer-sized bilipid-membrane bound particles released from all cell types1. EVs play a role in cell-to-cell communication and can transport encapsulated molecules between organs via the circulatory system. In cancer, EVs have been implicated for their role in metastasis2 and in promoting tumorigenesis in the local tumor environment through processes such as immunosuppression3,4 and angiogenesis5,6. Furthermore, breast cancer EVs can reprogram the metabolism of target cells7. Despite these findings, there are still many unknowns regarding the systemic roles that EVs play in cancer and metastasis due to the difficulty of studying them in vivo and in diverse environments.
EVs can be isolated from easily accessible biofluids such as blood serum and urine, and have been suggested as candidates in “liquid biopsy” for screening and diagnostics for a variety of different diseases including cancer8. These biofluids are collected for routine tests in all medical care facilities, which would enable convenient translation of biofluid-derived EV tests into current standard-of-care. One significant barrier to characterizing EVs in liquid biopsy is the lack of single-EV analysis methods with biochemical/functional specificity. Most methods to analyze EVs focus on “bulk” analysis that provides indiscriminatory genomic, metabolomic, or proteomic information for a sample with a large number of EVs, or rely upon structural or physical features such as using nanoparticle tracking analysis (NTA) for EV size and concentration9, dynamic light scattering (DLS) for EV size distribution10, transmission electron microscopy (TEM) for EV size and morphology11, or atomic force microscopy for EV size and physical properties12. Single-EV analysis methods are needed to untangle the heterogeneity of EVs, especially in early cancer detection where only a small proportion of EVs may be indicative of disease13. In recent years, several studies achieved label-free biochemical characterization of EVs using methods such as spontaneous Raman spectroscopy14,15 and surface-enhanced Raman spectroscopy16,17,18. Raman methods can achieve single-EV resolution and have discovered high biochemical heterogeneity in EVs19. Flow cytometry can additionally be used for single-EV level resolution to examine surface markers20. Single-EV proteomic analysis has recently been achieved via droplet barcode sequencing21. However, these methods lack the ability to characterize EVs in situ, either in vivo or ex vivo, which provides essential spatial and dynamic information. In vivo imaging of EVs with exogenous fluorescent markers presents a pathway to EV tracking and visualization in situ22,23, but these methods lack information on the functional state and biochemical makeup of EVs.
Single-EV functional and biochemical characterization of both isolated and in situ EVs was recently achieved with label-free nonlinear optical microscopy using endogenous contrasts from autofluorescent molecules within EVs24,25,26,27,28,29,30. Autofluorescence is primarily collected from reduced nicotinamide adenine dinucleotide (phosphate) (NAD(P)H) and flavin adenine dinucleotide (FAD), key metabolic cofactors that are ubiquitous throughout biological systems. The relative autofluorescence intensities and lifetimes of these cofactors are dependent on the activity of metabolic pathways within cells, and these cofactors are passed on into EVs. Optical metrics quantifying NAD(P)H and FAD can thus be used to make inferences regarding cells and the EVs they produce. The autofluorescence of these cofactors has been studied extensively in cell culture15,31,32 and tissues33,34 in a variety of healthy and disease states including cancer. Importantly, due to the cancer-related shift in energy metabolism toward reliance on glycolysis instead of mitochondrial metabolism, NAD(P)H and FAD autofluorescence show distinct signatures in cancer31,33,34 and in other altered metabolic states15,35. This is attributed to the difference in relative abundance of NAD(P)H and FAD as metabolic pathways shift and the longer fluorescence lifetime of protein-bound NAD(P)H compared to “free” NAD(P)H36,37,38,39.
Label-free nonlinear optical microscopy can quantify and spatially map these shifts in relative abundance via intensity-based imaging and shifts in the protein-bound vs. free states with fluorescence lifetime imaging microscopy (FLIM). One label-free nonlinear optical microscopy method previously used to characterize EVs is simultaneous label-free autofluorescence multiharmonic (SLAM) microscopy40. Briefly, this method uses a spectrally- and temporally-optimized pulsed laser excitation to simultaneously image multiple nonlinear optical contrast mechanisms within biological samples: two-photon excited autofluorescence of FAD, three-photon excited autofluorescence of NAD(P)H, second harmonic generation (SHG) of collagen, and third harmonic generation (THG) of interfaces to visualize sample structure. From SLAM images, the optical redox ratio (ORR), a normalized metric to characterize the redox balance, can be computed from FAD and NAD(P)H intensities. From multiphoton FLIM of NAD(P)H, the mean fluorescence lifetime (τ), and phasor components (g and s) are computed to describe the multidimensional fluorescence decay curve. EVs contain active enzymes41, which suggests that the protein-bound NAD(P)H signature in EVs is due to enzymatic activity of proteins that use NADH or NADPH as cofactors (NADH and NADPH exhibit similar fluorescence excitation and emission, and their optical signature is often referred to as NAD(P)H to account for the contribution from both molecules). Thus, the fluorescence lifetime provides a functional quantification of what is occurring inside the EV, and may provide insight on the parent cell metabolism.
Label-free microscopy has previously been used to explore the optical signatures of isolated EVs from cell culture media24,25, ex vivo EVs in breast cancer tissue24,26,27, in vivo EVs in rat mammary tumor tissue24 and human patient-derived xenografts from pancreatic cancer implanted into mice28, isolated urinary EVs in canine bladder cancer29, and isolated urinary EVs in human colorectal cancer30. These studies have indicated that the optical characteristics of EVs are strongly associated with cancer. Notably, one study showed that in vivo rat tumors exhibited a significantly elevated concentration of NAD(P)H-rich EVs24, a biomarker that is indicative of the metabolic preference of cancer cells for glycolysis, which leads to increased NADH levels. Previous work also showed that EVs isolated from cell culture had significantly different NAD(P)H fluorescence lifetime than parent cells, but with a lifetime profile most similar to that of the parent cell cytosol, and less similar to the parent cell nucleus and mitochondria25, indicating that EVs can be representative markers of parent cell metabolism. These previous studies using label-free nonlinear optical microscopy have primarily focused on large EVs. Likely due to their larger volume which enables larger EVs to carry a higher number of autofluorescent molecules, large EVs produce a stronger autofluorescence signal than small EVs25. In this manuscript “EV” is used as a general term to refer to both isolated large EVs (pelleted by ultracentrifugation at 12,000×g) and non-isolated EVs in tissue.
Here we explore and compare the optical metabolic signatures of isolated EVs from urine and serum with in situ EVs in freshly excised intact ex vivo breast tissue from human study participants. We explore and compare optical signatures between groups of healthy and breast cancer participants, including benign and malignant breast cancer from stages 0 to II. This work presents the first inter- and intra-individual comparison of optical metabolic signatures across multiple EV sources. Furthermore, it presents the first exploration into the capability of using optical metabolic signatures of EVs in liquid biopsy for breast cancer. A limited number of participants (n = 34) were recruited in this preliminary study in order to understand capabilities and establish candidate biomarker signatures for future, larger-scale analyses. Results indicate that several optical metabolic metrics have the potential to indicate the presence of early-stage cancer and that optical metabolic signatures differ greatly across EV sources.
Methods
Ethics statement
This study involved human participants and was approved by Carle Foundation Hospital and University of Illinois Urbana-Champaign (Urbana, IL) Institutional Review Boards (IRB #18CCC1708). All research procedures performed were in accordance with the ethical standards of the Institutional and National research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from all individual participants involved in the study and all eligible participants completed a one-time medical history questionnaire. All confidential data was stored a in REDCap (Research Electronic Data Capture) database.
Participant selection
The study accrued 40 participants in total: 30 malignant breast cancer patients, 6 benign breast tumor patients, and 4 individuals without cancer scheduled for breast reduction surgery as normal controls. Not all samples were able to be processed and imaged due to logistical considerations; only samples able to be processed and successfully imaged are presented, leading to a final cohort of 34 participants: 24 breast cancer-positive, 6 benign breast tumor, and 4 breast-reduction surgery normal controls (Fig. 1a). Serum and urine were collected from all participants, and ex vivo tissue was collected from participants when available (tumor and/or extra non-tumor tissue > 1 cm in size). Participant demographics and relevant health information (including age, race/ethnicity, clinical staging of breast cancer, and more) collected from the electronic medical record (EMR) is provided in Table S1, as well as which biofluid and tissue samples were available and imaged from each participant. Cancer stages of malignant tumors and patient age are graphically represented in Fig. S1.
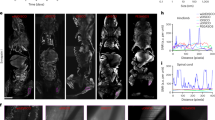
Workflow for characterizing EVs in breast cancer using single-EV capabilities of nonlinear optical microscopy and example dataset from one participant. (a) Study overview including participant cancer status, EV isolation, and tissue extraction, and imaging. (b) Concentration and size of isolated EVs. (c) NAD(P)H single-EV fluorescence lifetime distribution; (d) NAD(P)H single-EV phasor distribution for isolated EVs from urine (yellow) and serum (red) from a single participant with malignant breast cancer. (e) Single-EV FAD autofluorescence intensity distribution. (f) Single-EV NAD(P)H intensity distribution for isolated EVs from urine (yellow), serum (red), ex vivo tumor tissue (green), and normal-appearing ex vivo tissue from the same participant “Tumor adj.” (light blue). Panels (b–f) all show data collected from samples from one single participant with malignant breast cancer, indicating the broad range of different variables that were collected from each participant.
Tissue specimen collection
Human breast tissue specimens were collected during either breast-conserving surgery (lumpectomy) or mastectomy, for malignant or benign tissues, or during breast reduction surgery (normal tissue, no patient history of cancer). The resected specimens were placed in a phosphate buffered saline (PBS) and the cold ischemia start time was documented. After no more than 45 min past cold ischemia time, specimens were bread-loafed to identify the gross tumor (if present). If the tissue was > 1 cm in size, the pathologist or assistant selected a small sample of tissue from the tumor and/or visually healthy tissue and placed the sample(s) in vials with saline. Visually healthy tissue from participants with malignant breast cancer was termed “tumor adjacent tissue” whereas visually healthy tissue from breast reduction surgery participants was considered to be a healthy control. In some cases, only tumor tissue or tumor adjacent tissue was available from a patient with malignant cancer (Table S1). Tumor stage and grade were determined by a board-certified pathologist. Tissue samples were transported to the imaging lab on ice and imaged within 2–3 h of excision.
Biofluid collection
Pre-operative urine samples were collected from the research participants in 120 mL sterile urine specimen cups at a random time of day. Pre-operative whole venous blood was collected from research participants into 10 mL BD Vacutainer polyethylene terephthalate serum separation tubes (SST). Urine and blood samples were transported to the sample processing laboratory on ice within 1 h of collection and processed within 1 h after receiving. Sample volumes are given in Table S2.
EV isolation
The workflow from sample retrieval to EV imaging, including EV isolation and validation, is presented in Fig. 1a. EVs were isolated from biofluids as previously described29. Urine samples were diluted to a final volume of 45 mL using PBS if less than 45 mL was acquired. If more than 45 mL was acquired, only 45 mL was used and the remaining sample was discarded. After clotting for 30 min, the venous whole blood was centrifuged at 1200×g for 10 min to separate serum from red blood cells. A 3–5 mL volume of serum was collected and resuspended in a final 42 mL PBS volume. Both serum and urine samples were centrifuged at 800×g for 10 min at 4 °C. A 40 mL volume of supernatant was transferred into new 50 mL conical Protein LoBind tube (Eppendorf), which was centrifuged at 2000×g for 30 min at 4 °C to remove particulate matter. These first two centrifugation steps were performed in a benchtop centrifuge (Rotina 380 R, Hettich). Supernatant (34 mL) was centrifuged at 12,000×g for 1 h at 4 °C to isolate EVs in an ultracentrifuge (Sorvall WX +, Thermo Fisher) using a Surespin630/36 rotor and associated tubes (PP thinwall tube, 36 mL, Thermo Scientific). The resulting urinary EV or serum EV pellet was dissolved in 55 μL of PBS supplemented with 25 mM D(+)-Trehalose (Fig. 1a). This type of procedure is sometimes referred to as isolating large EVs (LEVs) or microvesicles (MVs); throughout the manuscript we use the general term “EV” and indicate whether that EV sample was isolated from biofluids using this procedure or whether EVs were segmented from images of intact tissue.
EV counting and size distribution analysis
Isolated EV count and size were evaluated by NTA (NanoSight NS300, Malvern Panalytical, Malvern, UK). The EV analytes were diluted in PBS to a final volume of 2 mL. In NTA, the dilution factor was recorded during each run to enable an automated particle concentration upgrade by the NTA v.3.3 (104) software. The data was primarily acquired using a camera sensitivity level set to either 12 or 13 and syringe pump speed set to 75. Valid particle track number was determined at a detection threshold of 3 or 5. Each NTA run lasted for 3 min. Video data capture time was set to either 30 s or 60 s to collect 6 or 3 repeated measurement data points. The main NTA results were EV concentration (expressed as particles/mL normalized to the initial volume of raw sample) and mean diameter (Fig. 1b).
Transmission electron microscopy
The presence of EVs in the samples that were processed by differential centrifugation was confirmed by transmission electron microscopy (Fig. S3). For TEM imaging, 20 μL of EVs were adsorbed to a 200-mesh Formvar-coated copper grid (Ted Pella) for 3 min. Excess sample was drained using filter paper before being stained negatively with a 2% w/v uranyl acetate solution. The samples were examined by TEM at high magnification (FEI Tecnai G2 F20 S-TWIN STEM, 120 kV).
Western blotting
The expression of EV biomarkers was detected by Western blotting, which was performed as described previously42. Proteins from lysed serum or urine EVs were separated by LDS-PAGE under reducing conditions using 4–12% gradient NuPAGE gels. Resolved proteins were transferred onto 0.22 µm pore-size nitrocellulose membrane, which was subdivided into distinct molecular weight zones, blocked with 3% BSA solution for 2 h and individually immunoblotted with a 1:1000 dilution of either unconjugated Anti-ALIX rabbit monoclonal antibody (Boster-Bio, #M01751), anti-TSG101 rabbit polyclonal antibody (Abcam, #ab30871), anti-CD9 rabbit monoclonal antibody (Boster-Bio, #M01202), anti-CD81 rabbit monoclonal antibody (Boster-Bio, #M01281-1), Anti-GM130/GOLGA2 (Golgi organelle contamination indicator) mouse monoclonal 6D4 antibody (Boster-Bio, # M05865-2) or anti-Calreticulin/CALR (ER organelle presence indicator) (Boster-Bio, #A00894-1) antibody overnight followed by four 7 min wash cycles in prechilled 1X TBS-T buffer, probing with 1:10,000 horse anti-mouse IgG Cell Signaling Technology, #7076S) or 1:40,000 goat anti-rabbit IgG (H + L) (Thermo Fisher, #31460) HRP-linked secondary antibody for 1 h at room temperature, four 5 min washes in 1X TBS-T buffer and a final wash in dH2O prior to chemiluminescent signal development by a 5 min-long blot incubation with SuperSignal™ West Pico PLUS chemiluminescent substrate (Thermo Scientific) and automated protein band visualization in Bio-Rad ChemiDoc MP imaging system at optimal non-saturating 24 s (ALIX), 44 s (TSG101), 180 s (CD9, CD81, GM130 and CALR) exposure time. Data is shown in Fig. 1a.
EV verification and validation
The minimal information for studies of extracellular vesicles 2018 (MISEV2018) guidelines provide an extensive set of parameters to report and control in EV-related studies43. Detailed steps for EV isolation via ultracentrifugation are described above and initial sample volumes are given in Table S2, as suggested by MISEV2018. For EV validation, NTA, TEM, and Western blotting were used, as described above; the use of multiple complementary techniques is strongly suggested by MISEV 2018. Using Western blotting, ESCRT components ALIX and TSG101 were confirmed in isolated EVs, along with tetraspanins CD9 and CD81. GM130 and CALR are associated with non-EV intracellular components, were also examined. Accordingly, the term “extracellular vesicle” or “EV” was used as a general term since general EV size, morphology, and markers were confirmed in the isolated particles examined in this study, but no specific subtype of EV was selected with certainty.
Nonlinear optical microscopy
EVs were imaged on two different previously described custom nonlinear optical microscopy systems, both configured in an inverted epi-detection geometry using 1.05 NA water immersion objective lenses (XLPLN25XWMP2, Olympus)24,25. While detailed system diagrams and descriptions are available in previous publications24,25, both imaging systems are depicted in Fig. S2. For imaging with SLAM microscopy, isolated EVs were embedded in 99% molecular biology grade glycerol (Sigma, USA) in a 1:4 EVs:glycerol ratio24,29. Fresh ex vivo breast tissue specimens were also imaged on the SLAM system. Samples were kept on ice until imaging, and then placed on top of a large cover glass (260454, Ted Pella) and imaged at room temperature from below. Briefly, the SLAM system uses a shaped supercontinuum (1110 ± 30 nm, 35 fs) to excite four different types of nonlinear optical contrast within biological specimens: SHG, THG, two-photon excited autofluorescence of FAD, and three-photon excited autofluorescence of NAD(P)H26,40. Thus, each pixel of a SLAM image contains four channels of intensity, each corresponding to one of these four contrasts (Fig. S2a). SHG primarily shows collagen in biological samples, and is not present in EVs. THG derives contrast from changes in refractive index, highlighting biological features such as cell and EV membranes. Samples were imaged using 14 mW of incident power, with a 200 × 200 μm field of view (FOV), with a frame rate of 0.125 Hz, and no frame averaging. For biofluid-derived EVs, 25 total FOV per sample were acquired; for tissue samples, 9–81 total FOV were acquired per sample depending on sample size.
EVs were additionally characterized with two-photon exited FLIM to characterize NAD(P)H-protein interactions25. This custom system creates a time-trace for each pixel by time-tagging collected photons in order to characterize the fluorescence decay curve of each pixel of the image (Fig. S2b). From this curve, fluorescence lifetime and associated metrics can be determined, enabling inferences about the free and protein-bound NAD(P)H within the sample. For FLIM, EVs were embedded in a 0.2% agarose gel in a 1:5 ratio (EVs in PBS:agarose) and imaged with an average power of 25 mW using 750 nm excitation. An average of 20 frames per FOV was acquired, with 16 total FOV per sample, each 180 × 180 μm. The frame rate of the system was 0.2 Hz. The number of EVs imaged per sample is given in Table S3; if 10 or fewer EVs were imaged, the dataset was discarded and excluded due to low number. The mean number of EVs imaged per sample were: 1330 for urinary EV FLIM, 579 for urinary EV SLAM, 134 for serum EV FLIM, 691 for serum EV SLAM, 26,954 for ex vivo tumor SLAM, and 8,228 for ex vivo non-tumor SLAM, with large standard deviations similar to or larger than the means, as listed in Table S3.
Image processing, interpretation, and statistical analysis
Image/data analysis and visualization was performed using custom Matlab scripts. EVs were segmented using a blob-detection algorithm, based on previously demonstrated25,28 and validated methods24,27, but segmentation validation using tagged EVs was not performed in this study due to the limited number of EVs per sample and potential for fluorescence bleedthrough. Mean fluorescence lifetime (τ) was determined using phasor analysis to determine the cosine (g) and sine (s) components at the modulation frequency of the laser (ω)44; ORR was computed the same as in previous EV studies24,29.
The mean fluorescence lifetime (τ) of NAD(P)H increases when it is bound to a protein; an increase is associated with more NAD(P)H-related protein activity in the EVs. Phasor components g and s represent the fluorescence lifetime profile in a two-dimensional space that can be used to better represent and understand combinations of multiple lifetime species within one pixel, such as combinations of free and protein-bound NAD(P)H. Phasor components are useful since they can describe a shift in the fluorescence decay curve that is not evident in just the mean lifetime, since the mean lifetime is made of multiple components of bound and free NAD(P)H. ORR is generally assumed to correspond to an increase in mitochondrial activity, where FADH2 is converted to FAD (leading to an increase in FAD autofluorescence signal) and NADH is converted to NAD + (leading to a decrease in NADH autofluorescence signal). Thus, high ORR is associated with relatively higher mitochondrial activity and lower glycolytic metabolism (in which NAD + is reduced to NADH, increasing NADH autofluorescence signal), and low ORR is associated with relatively lower mitochondrial activity and higher glycolytic metabolism. It is important to note that multiple formulae for ORR and related metrics exist, which should be carefully considered when comparing these results to those of other studies.
To compare matched data from the same participant (ex: mean FAD in urine vs. serum), paired Student’s t-tests were used. To compare data from different participants (ex: mean FAD in malignant vs. benign tumors), non-paired Student’s t-tests were used. Linear correlation coefficient (R) was additionally calculated to compare optical metabolic signatures across participants, and additionally used to examine confounding variables such as participant age.
Results and discussion
Label-free nonlinear optical microscopy enables multidimensional analysis with single-EV resolution
Intact breast tissue and biofluid-derived isolated EVs from urine and serum were imaged with label-free nonlinear optical microscopy to characterize and extract the optical metabolic profiles from single EVs (Fig. 1). Samples were collected from 30 participants with breast tumors (24 malignant, 6 benign) and 4 control participants (Fig. 1a). Single-EVs were segmented, quantified, and visualized in isolated EV samples and in situ EVs in ex vivo tissue, and a variety of metrics were extracted from these images using two-photon FLIM of NAD(P)H and SLAM microscopy. One example EV characterization dataset from a single participant with malignant breast cancer is presented in Fig. 1b–f, with the extracted optical metabolic metrics from SLAM and FLIM (Fig. 1c–f) and EV size and concentration from NTA (Fig. 1b). For optical metabolic metrics in Fig. 1c–f, each point represents a single segmented EV. The example images in Fig. 2 represent just a portion of the imaging data collected for a single participant. Single EVs can be visualized as dots (annotated with arrows) in Fig. 2, including in situ EVs in ex vivo tumor tissue (Fig. 2a,d), and isolated EVs with SLAM (Fig. 2b,e) and FLIM (Fig. 2c,f). EVs within samples from a single participant show optical metabolic heterogeneity, indicated by the broad profiles (Figs. 1d–f, 2).
Example images of EVs in situ in tissue and isolated EVs. (a) Example SLAM image of ex vivo tumor tissue with 4-color map displaying each SLAM modality. (b) Example SLAM image of urinary isolated EVs. (c) Example FLIM image of urinary isolated EVs with rainbow colormap displaying mean NAD(P)H fluorescence lifetime. (d–f) Insets corresponding to smaller field of view from (a–c), respectively, showing single-EVs zoomed-in.
This study presents the first comparison of optical metabolic signatures of single EVs from different human biofluid and tissue sources and examines their potential for use in breast cancer screening and diagnostics. EVs can be imaged in situ in excised breast biopsy tissues (Fig. 2a) and in isolated EV samples from urine and serum (Fig. 2b,c). Single EVs are segmented and quantified via a variety of nonlinear optical metrics that report on metabolic function. Two commonly used and previously validated metrics are ORR and NAD(P)H fluorescence lifetime: NAD(P)H and FAD autofluorescence intensities are used to compute the ORR (Eq. 2) and NAD(P)H fluorescence lifetime is determined (Eq. 1) for each segmented EV. In cells, ORR increases with more electron transport chain activity (FAD produced and/or NADH depleted) and decreases with increased glycolysis (NADH produced). NAD(P)H fluorescence lifetime increases when NAD(P)H is protein-bound, though the lifetime can depend on the protein36,45. Thus, optical metabolic metrics provide information on the metabolic state of EVs and can reflect the metabolism of the parent cells.
Within isolated EVs, seven metrics were examined for control (normal), benign, and malignant breast cancer: mean NAD(P)H fluorescence lifetime (from FLIM, Fig. 3a,b), phasor components g and s (from FLIM, Fig. 3d,e), mean diameter (from NTA, Fig. 3g,h), concentration (from NTA, Fig. 3j,k), mean THG (from SLAM, Fig. 3m,n), and mean ORR (from SLAM, Fig. 3p,q). EV signatures were also examined by clinical stage (Fig. 4, Fig. S5), tumor grade (Fig. S6), and participant age (Fig. S7). Age did not show a significant correlation with any optical metabolic metrics (Fig. S7).
Optical metabolic characterization of control, benign, and malignant EVs across different sources. EV source is indicated by column, and patient cancer status is indicated as: normal control (blue), benign (orange), malignant (yellow), and non-tumor tissue from participants with malignant cancer “tumor adjacent” (light blue). Example SLAM images from (c) normal control, (f) benign, and (i) malignant ex vivo tissues are additionally shown using the same color scale. Significance: *p < 0.05; not significant if unmarked; in (e) the s value for benign and malignant is significantly different.
Optical metabolic characterization of EVs by cancer stage for selected metrics. Three metrics showed significant differences based on participant cancer stage: (a) EVs per mm2 segmented from ex vivo tissue images, (b) mean EV THG in ex vivo tissue images, and (c) mean EV ORR in EVs isolated from serum. Significance: *p < 0.05, **p < 0.01, ***p < 0.001, not significant if unmarked.
Certain EV optical metabolic signatures are dependent on cancer status in isolated EVs
Urinary EVs from participants with malignant cancer exhibited significantly elevated NAD(P)H fluorescence lifetime compared to EVs from control participants (Fig. 3a, p = 0.040), though neither showed a significant difference compared to benign cases. This significantly elevated NAD(P)H fluorescence lifetime in urinary EV samples from participants with malignant cancer compared to non-cancer controls (Fig. 3a), indicates a higher amount of NAD(P)H-related enzymatic activity in EVs from participants with malignant breast cancer. This could either be due to a compositional change: the urinary EVs from participants with malignant cancer contained more NAD(P)H-related proteins, or due to an activity change: the NAD(P)H-related proteins are significantly more active in urinary EVs in the case of malignant cancer. Metabolic reprogramming is a hallmark of cancer, and a variety of NAD(P)H-related enzymes have been implicated in key pathways associated with this reprogramming such as isocitrate dehydrogenase, glucose-6-phosphate dehydrogenase, and lactate dehydrogenase46,47. Urinary EVs are primarily derived from genitourinary endothelial cells48. For this reason, urinary EVs have predominantly been investigated for biomarkers of prostate, bladder, and kidney cancers49. However urinary EVs have also shown promise as biomarkers of other cancers such as non-small cell lung cancer50. Further studies are needed to better understand the source of this increased NAD(P)H fluorescence lifetime signature in EVs and determine if the EVs with high NAD(P)H fluorescence lifetime originate from the tumor, from local or distal immune cells, or from the kidney or bladder. Additional analysis on single-EV clustering of phasor components was performed to examine different subpopulations of EVs based on fluorescence lifetime, but did not improve performance for distinguishing cancer status (Fig. S4). No significant differences were found in the fluorescence lifetime of serum-derived EVs based on cancer presence, grade, or status.
Various studies have provided strong evidence for EVs and related particles as cancer biomarkers (including breast cancer) in blood plasma51,52,53 and serum54. In this study, the mean ORR in serum-derived isolated EVs from participants with stage II breast cancer, but not stage I or 0, was significantly decreased compared to control (p = 0.025, Fig. 4c). However no significant differences were found between optical metabolic metrics for EVs isolated from the serum of participants when all stages of malignant breast cancer were grouped together into the overarching category of “malignant” (Fig. 3). This may suggest that breast cancer-related optical metabolic signatures of EVs in breast cancer may develop over time and increase with disease severity since a significant difference was only visible in stage II and not the earlier stages. Accordingly, tumor-related EVs may not reach circulation until cancer has reached stage II or later stages. Lower ORR and higher NAD(P)H intensity have previously been reported in optical metabolic imaging of breast cancer cell lines, in vivo rat mammary tumors, ex vivo human breast cancer24, and urinary EVs in bladder cancer29. This shift has also been extensively reported in cells and tissues, due to the shift towards glycolytic metabolism often associated with cancer31,33,34. However, a lower ORR in ex vivo tissue EVs was not observed in this study, conflicting with previous results that found a higher proportion of NAD(P)H-rich (low ORR) EVs in in vivo rat mammary tumors compared to control animals24. This may be due to the lower tumor grades in this study or due to time-dependent changes in the optical metabolic characteristics of tissues and after removal from the body. Previous studies showed that optical metabolic signatures of isolated EVs can change after 48 h of storage at − 20 °C29 and that in vivo vs. ex vivo imaging can alter optical metabolic metrics of EVs in intact tissue28. Additionally, previous work on breast cancer found more significant changes in ORR and NAD(P)H in the later stages (II and III) of cancer24, whereas this study primarily recruited earlier stage participants and did not have any participants with stage III breast cancer (Table S1, Fig. S1). Despite the trends of lower ORR in participants with malignant cancer compared to control participants in urinary EVs, no statistical significance was found in urinary EV ORR between normal, malignant, and benign cases (Fig. 3p), between clinical stages (Fig. S5a), or between tumor grade (Fig. S6a), which in part could be due to the small number of samples or the relatively larger contribution of non-tumor related EVs in urinary samples.
EV concentration and THG signature are dependent on cancer status for in situ EVs
In ex vivo tissue samples, three EV metrics were examined from SLAM images: concentration in EVs per mm2 (Fig. 3l), mean THG (Fig. 3o), and mean ORR (Fig. 3r). The segmented EV density was significantly higher in malignant tumor tissue (p = 0.039) and adjacent non-tumor tissue (p = 0.049) from participants with malignant cancer compared to tissue from control participants (Fig. 3l). Furthermore, the EV THG intensity is higher in the malignant tumor images compared to the control images (Fig. 3o, p = 0.022). Similar to Fig. 3l,o, EVs per mm2 and EV THG signature were both significantly elevated in stage 0 and stage II cancer compared to controls (Fig. 4a,b). When grouped by tumor grade, similar trends emerged in ex vivo tissue EVs per mm2 and THG (Fig. S6c). This increased EV concentration in tumors has previously been observed using label-free nonlinear optical microscopy in ex vivo breast cancer biopsy specimens27. It also corresponds well with studies which found that cancer cells produce more EVs, especially in the case of hypoxia55. THG signal increases with particle diameter in index-matched and non-index-matched solvents56. Thus, this increase in THG signature in tumor tissue EVs could be due to the increased size of cancer-related EVs, sometimes termed large oncosomes57. However, no strong positive correlation was found between the size of isolated EVs and their THG signature, which suggest that the size of EVs isolated from urine and serum was not increased (Fig. 3), indicating that this increased THG signature is more relevant in tissue, and not associated with the size of isolated EVs.
All optical metabolic signatures are dependent on EV source
In order to assess how optical metabolic signatures depend on EV source (urine, serum, tissue), paired Student’s t-tests were used to compare the signatures across all metrics for all participants. NAD(P)H fluorescence lifetime and phasor component g showed significant differences between serum and urine, with significantly higher fluorescence lifetimes in serum samples, indicating a larger presence of protein-bound NAD(P)H (Fig. 5a,b, p < 0.001), which has a longer lifetime than free NAD(P)H36,37,38. Furthermore, compared to serum-derived EVs, isolated urinary EVs were significantly larger in mean diameter (Fig. 5c, p = 0.004) and had lower concentration per mL of original biofluid (Fig. 5d, p = 0.020), as determined by NTA. FAD and NAD(P)H intensities were significantly lower in serum-derived isolated EVs compared to urinary isolated EVs and ex vivo tissue EVs measured in situ (Fig. 5e,f, p < 0.001). THG signature was significantly elevated in ex vivo tissue EVs in situ, averaging 105 times higher intensity than the isolated EVs (Fig. 5g, p < 0.001). The ORR was significantly different between all sources, with the lowest ORR in serum-derived EVs and the highest ORR in tissue EVs (Fig. 5h). Thus, clear differences between EVs isolated from urine and serum emerged from almost all variables, as well as between the two biofluid-based isolated EVs and ex vivo tissue EVs (Fig. 5). The origin of the EV populations in biofluids and tissue varies greatly, which likely accounts for the dramatic optical differences.
Optical metabolic characterization of EVs from different sources. The mean value of each metric was computed for measured EVs from all sources across all cancer status groups for: (a) mean NAD(P)H fluorescence lifetime from FLIM, (b) g and s phasor lifetime components from FLIM, (c) mean size from NTA, (d) concentration of isolated EVs per mL of initial biofluid from NTA, (e) mean FAD SLAM intensity, (f) mean NAD(P)H SLAM intensity, (g) mean THG SLAM intensity, and (h) mean ORR, calculated from NAD(P)H and FAD intensities from SLAM. Significance using paired Student’s t-test to examine intra-individual changes: ns not significant; *p < 0.05, **p < 0.01, ***p < 0.001.
Despite these differences, a variety of variables showed high linear correlation across different EV sources from the same participant (Fig. 6, Table 1). The linear correlation coefficient was calculated for all metrics within (Fig. 6a–c) and between (Fig. 6d–f) EV sources. When examining the correlation coefficients, it is important to note that ORR is calculated from FAD and NAD(P)H intensities (Eq. 2) and mean lifetime (τ) is calculated from g and s values (Eq. 1), causing stronger linear relationships between these variables since they are dependent. Selected high absolute value (> 0.7, < − 0.7) correlation coefficients are listed in Table 1. All coefficients are listed in Tables S4–S9. Notably, NAD(P)H intensity is highly positively correlated between urinary EVs and tissue EVs (R = 0.798), pointing to a link between tumor EVs and urinary EVs. Serum EV ORR is highly negatively correlated with tissue EV NAD(P)H intensity (R = -0.737); since ORR decreases with increased NAD(P)H intensity, this further strengthens the link between NAD(P)H signatures across EV sources. Similarly, urinary EV ORR is highly negatively correlated with serum EV NAD(P)H (R = − 0.874). Overall, these results suggest that some optical metabolic metrics may be conserved across different EV sources, which warrants future investigation.
Linear correlation coefficient examining optical metabolic variables within and between EV source. Correlations within a single-source for: (a) isolated urinary EVs, (b) isolated serum EVs, and (c) ex vivo tissue EVs. Correlations comparing different sources for: (d) isolated urinary EVs vs. ex vivo tissue EVs, (e) isolated serum EVs vs. ex vivo tissue EVs, and (f) isolated urinary EVs vs. isolated serum EVs.
Implications for future work on label-free nonlinear optical microscopy of EVs
This preliminary study had some limitations. Primarily, the study population was small, which means that the results may not be universal, and that these small numbers impact the statistical significance of the findings. The significantly elevated NAD(P)H fluorescence lifetime in urinary EVs (Fig. 3a) and lower ORR in serum-derived EVs (Fig. 4c), support the potential for optical metabolic characterization of EVs in liquid biopsy and should be confirmed on a larger scale. Additionally, this study focused only on early-stage breast cancer. While this population is important for screening and diagnostics, only examining early-stage participants means that the results for later stages of breast cancer could show a different outcome. Also, various factors in urine and blood collection may impact results, such as the time of the day, medications or supplements, or the presence of other diseases or conditions.
A limitation for the translation of nonlinear optical microscopy for clinical use is the high cost of equipment and specialized skills needed to build and operate these systems. Future work could examine the possibility to characterize single-photon NAD(P)H and FAD autofluorescence with lower cost laboratory equipment such as confocal and widefield micro-scopes, which are more accessible and can be used for EV analysis, as has been demonstrated with label-based fluorescence techniques for EV imaging58. However, the longer wavelengths used for excitation in nonlinear optics enable better depth penetration into biological tissue, which would be lost when using a single-photon fluorescence imaging system. Also, future work could incorporate NAD(P)H and/or FAD FLIM into a SLAM channel to enable colocalization of all metrics.
Another aspect for future investigation is the relative abundance of cancer-related EVs in biofluids. Future work could involve the use of tumor-specific labels or markers to examine what subpopulations of EVs in urine and serum are from the tumor, and investigate other suspected origins such as blood, immune, and bladder cells. Nonlinear optical microscopy can also provide spatial and temporal information in vivo and could be used to examine EV trafficking throughout the body and in and around the tumor microenvironment. By observing the distribution, transfer, and transport of EVs between tissues and the blood and lymphatic circulatory systems, a better understanding of the relationship between EVs in tissues and different biofluids could be achieved.
Conclusions
Nonlinear optical microscopy is a novel tool with great potential to examine EVs from a variety of different sources, including isolated EVs and in situ EVs in ex vivo tissue samples. It enables direct comparison of the optical metabolic properties from multiple EV sources to evaluate their potential as biomarkers and the relationship between EVs from these different sources. Overall, isolated urinary EV autofluorescence lifetime of NAD(P)H and ex vivo tissue EV density were significantly elevated in cases of malignant tumors, while the ORR of isolated serum-derived EVs showed significant changes in stage II of malignant cancer, but not in earlier stages of malignant cancer. EV optical metabolic properties were highly dependent on whether the EVs were in intact breast tissue or were isolated from serum or urine. Despite that, EV optical signatures showed some correlations in NAD(P)H autofluorescence and ORR values that could indicate some properties of individual EVs are conserved despite differences in source. Further investigations are needed to better understand the relationship between EV source and optical metabolic properties.
Data availability
The data and analysis codes generated in this study will be made publicly available upon publication via Open Science Framework at: https://osf.io/d37ac/?view_only=523ce62c46aa4f3b8524c1b13a2c425559.
References
Van Niel, G., d’Angelo, G. & Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell. Biol. 19, 213–228 (2018).
Becker, A. et al. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell 30, 836–848 (2016).
Cooks, T. et al. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 9, 771 (2018).
Park, J. E. et al. Hypoxia-induced tumor exosomes promote M2-like macrophage polarization of infiltrating myeloid cells and microRNA-mediated metabolic shift. Oncogene 38, 5158–5173 (2019).
Nazarenko, I. et al. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 70, 1668–1678 (2010).
Huang, M. et al. New insights into the regulatory roles of extracellular vesicles in tumor angiogenesis and their clinical implications. Front. Cell. Dev. Biol. 9, 791882 (2021).
Bertolini, I. et al. Small extracellular vesicle regulation of mitochondrial dynamics reprograms a hypoxic tumor microenvironment. Dev. Cell. 55, 163–177 (2020).
Yu, D. et al. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer 21(1), 56 (2022).
Gardiner, C., Ferreira, Y. J., Dragovic, R. A., Redman, C. W. G. & Sargent, I. L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Ves. 2(1), 19671 (2013).
Rikkert, L. G., Nieuwland, R., Terstappen, L. W. M. M. & Coumans, F. A. W. Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J. Extracel. Ves. 8(1), 1555416 (2019).
Palmieri, V. et al. Dynamic light scattering for the characterization and counting of extracellular vesicles: a powerful noninvasive tool. J. Nanoparticle Res. 16, 2583 (2014).
Sajidah, E. S. et al. Spatiotemporal tracking of small extracellular vesicle nanotopology in response to physicochemical stresses revealed by HS-AFM. J. Extracell. Ves. 11, 12275 (2022).
Ferguson, S., Yang, K. S. & Weissleder, R. Single extracellular vesicle analysis for early cancer detection. Trends Mol. Med. 28, 681–692 (2022).
Penders, J. et al. Single particle automated Raman trapping analysis of breast cancer cell-derived extracellular vesicles as cancer biomarkers. ACS Nano 15, 18192–18205 (2021).
Liu, Z., et al. Mapping metabolic changes by noninvasive, multiparametric, high-resolution imaging using endogenous contrast. Sci. Adv. 4, eaap9302 (2018).
Park, J. et al. Exosome classification by pattern analysis of surface-enhanced Raman spectroscopy data for lung cancer diagnosis. Anal. Chem. 89, 6695–6701 (2017).
Kazemzadeh, M. et al. Label-free classification of bacterial extracellular vesicles by combining nanoplasmonic sensors with machine learning. IEEE Sens. J. 22, 1128–1137 (2021).
Jalali, M. et al. MoS2-Plasmonic nanocavities for Raman spectra of single extracellular vesicles reveal molecular progression in glioblastoma. ACS Nano 17, 12052–12071 (2023).
Smith, Z. J. et al. Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. J Extracell. Ves. 4, 28533 (2015).
Nolan, J. P. & Duggan, E. Analysis of individual extracellular vesicles by flow cytometry. Methods Mol. Biol. 1678, 79–92 (2018).
Banijamali, M. et al. Characterizing single extracellular vesicles by droplet barcode sequencing for protein analysis. J. Extracell. Ves. 11, 12277 (2022).
Linnemannstöns, K. et al. Microscopic and biochemical monitoring of endosomal trafficking and extracellular vesicle secretion in an endogenous in vivo model. J. Extracell. Ves. 11, 12263 (2022).
Hyenne, V. et al. Studying the fate of tumor extracellular vesicles at high spatiotemporal resolution using the zebrafish embryo. Dev. Cell. 48, 554–572 (2019).
You, S. et al. Label-free visualization and characterization of extracellular vesicles in breast cancer. Proc. Natl. Acad. Sci. 116, 24012–24018 (2019).
Sorrells, J. E. et al. Label-free characterization of single extracellular vesicles using two-photon fluorescence lifetime imaging microscopy of NAD(P)H. Sci. Rep. 11, 3308 (2021).
Tu, H. et al. Concurrence of extracellular vesicle enrichment and metabolic switch visualized label-free in the tumor microenvironment. Sci. Adv. 3, e1600675 (2017).
Sun, Y., et al. Intraoperative visualization of the tumor microenvironment and quantification of extracellular vesicles by label-free nonlinear imaging. Sci. Adv. 4, eaau5603 (2018).
Park, J. et al. In vivo label-free optical signatures of chemotherapy response in human pancreatic ductal adenocarcinoma patient-derived xenografts. Commun. Biol. 6, 980 (2023).
Park, J. et al. Label-free optical redox ratio from urinary extracellular vesicles as a screening biomarker for bladder cancer. Am. J. Canc. Res. 12, 2068 (2022).
Borisov, A. V. et al. A criterion of colorectal cancer diagnosis using exosome fluorescence-lifetime imaging. Diagnostics 12, 1792 (2022).
Ostrander, J. H. et al. Optical redox ratio differentiates breast cancer cell lines based on estrogen receptor status. Cancer Res. 70, 4759–4766 (2010).
Podsednik, A., Jiang, J., Jacob, A., Li, L. Z. & Xu, H. N. Optical redox imaging of treatment responses to Nampt inhibition and combination therapy in triple-negative breast cancer cells. Int. J. Mol. Sci. 22, 5563 (2021).
Skala, M. C. et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. 104, 19494–19499 (2007).
Xu, H. N., Tchou, J., Feng, M., Zhao, H. & Li, L. Z. Optical redox imaging indices discriminate human breast cancer from normal tissues. J. Biomed. Opt. 21, 114003 (2016).
Yaseen, M. A. et al. Fluorescence lifetime microscopy of NADH distinguishes alterations in cerebral metabolism in vivo. Biomed. Opt. Express 8(5), 2368–2385 (2017).
Blacker, T. S. et al. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat. Commun. 5, 3936 (2014).
Lakowicz, J. R., Szmacinski, H., Nowaczyk, K. & Johnson, M. L. Fluorescence lifetime imaging of free and protein-bound NADH. Proc. Natl. Acad. Sci. 89(4), 1271–1275 (1992).
Ranjit, S., Malacrida, L., Stakic, M. & Gratton, E. Determination of the metabolic index using the fluorescence lifetime of free and bound nicotinamide adenine dinucleotide using the phasor approach. J. Biophotonics 12(11), e201900156 (2019).
Lagarto, J. L. et al. Characterization of NAD(P)H and FAD autofluorescence signatures in a Langendorff isolated-perfused rat heart model. Biomed. Opt. Express 9(10), 4931–4978 (2018).
You, S. et al. Intravital imaging by simultaneous label-free autofluorescence-multiharmonic microscopy. Nat. Commun. 9, 2125 (2018).
Angulo, M. A., Royo, F. & Falcón-Pérez, J. M. Metabolic nano-machines: Extracellular vesicles containing active enzymes and their contribution to liver diseases. Curr. Pathobiol. Rep. 7, 119–127 (2019).
Aksamitiene, E., Hoek, J. B. & Kiyatkin, A. Multistrip Western blotting: a tool for comparative quantitative analysis of multiple proteins. Methods Mol. Biol. 1312, 197–226 (2015).
Théry, C. et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Ves. 7(1), 1535750 (2018).
Digman, M. A., Caiolfa, V. R., Zamai, M. & Gratton, E. The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 94, L14–L16 (2008).
Sharick, J. T. et al. Protein-bound NAD(P)H lifetime is sensitive to multiple fates of glucose carbon. Sci. Rep. 8, 5456 (2018).
Hanahan, D. Hallmarks of cancer: New dimensions. Cancer Discov. 12, 31–46 (2022).
Ward, P. S. & Thompson, C. B. Metabolic reprogramming: A cancer hallmark even Warburg did not anticipate. Cancer Cell 21, 297–308 (2012).
Svenningsen, P., Sabaratnam, R. & Jensen, B. L. Urinary extracellular vesicles: Origin, role as intercellular messengers and biomarkers; efficient sorting and potential treatment options. Acta Physiol. 228, e13346 (2020).
Lourenço, C., Constâncio, V., Henrique, R., Carvalho, Â. & Jerónimo, C. Urinary extracellular vesicles as potential biomarkers for urologic cancers: An overview of current methods and advances. Cancers 13, 1529 (2021).
Li, Y., Zhang, Y., Qiu, F. & Qiu, Z. Proteomic identification of exosomal LRG1: A potential urinary biomarker for detecting NSCLC. Electrophoresis 32, 1976–1983 (2011).
Hoshino, A. et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 182, 1044–1061 (2020).
Fang, S. et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PloS One 12, e0175050 (2017).
Hinestrosa, J. P. et al. Early-stage multi-cancer detection using an extracellular vesicle protein-based blood test. Commun. Med. 2, 29 (2022).
Lee, J. U., Kim, W. H., Lee, H. S., Park, K. H. & Sim, S. J. Quantitative and specific detection of exosomal miRNAs for accurate diagnosis of breast cancer using a surface-enhanced Raman scattering sensor based on plasmonic head-flocked gold nanopillars. Small 15, 1804968 (2019).
King, H. W., Michael, M. Z. & Gleadle, J. M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 12, 421 (2012).
Shcheslavskiy, V. I., Saltiel, S. M., Faustov, A., Petrov, G. I. & Yakovlev, V. V. Third-harmonic Rayleigh scattering: theory and experiment. JOSA B 22, 2402–2408 (2005).
Minciacchi, V. R. et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget 6, 11327 (2015).
Schürz, M. et al. EVAnalyzer: High content imaging for rigorous characterization of single extracellular vesicles using standard laboratory equipment and a new open-source ImageJ/Fiji plugin. J. Extracell. Ves. 11, 12282 (2022).
Sorrells J. Nonlinear optical imaging of breast cancer EVs. Open Science Framework, 2023. https://osf.io/d37ac/?view_only=523ce62c46aa4f3b8524c1b13a2c4255.
Acknowledgements
This work was supported in part by the National Institutes of Health under grants (R01CA213149, R01CA241618) and the NIH/NIBIB P41 Center for Label-free Imaging and Multiscale Biophotonics (CLIMB) (P41EB031772). J.E.S. was supported in part by NIH/NIBIB under award number T32EB019944. The authors thank Darold Spillman for his technical and administrative support, Sixian You, Ronit Barkalifa, and Rishyashring Iyer for their mentorship, and Lingxiao Yang, Prabuddha Mukherjee, and Aneesh Alex for their logistical support throughout this study.
Author information
Authors and Affiliations
Contributions
S.A.B., M.M., E.A., E.J.C., J.E.S., and J.P. designed the study. A.M.H., K.A.C., and Z.G.L. performed breast surgeries and provided pathological determinations. E.J.C., M.M., and E.A. handled sample transport, pathological reports, and participant information. E.A., J.P., and J.E.S. performed EV isolation and characterization. J.E.S., J.P., E.A., E.M.M., and M.M. performed data analysis. J.E.S., J.P., E.A., M.M., and S.A.B. wrote the manuscript with input from all coauthors. S.A.B. obtained funding to conduct the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sorrells, J.E., Park, J., Aksamitiene, E. et al. Label-free nonlinear optical signatures of extracellular vesicles in liquid and tissue biopsies of human breast cancer. Sci Rep 14, 5528 (2024). https://doi.org/10.1038/s41598-024-55781-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-55781-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.