Abstract
Knowing about the antibiotic resistance, serotypes, and virulence-associated genes of Group B Streptococcus for epidemiological and vaccine development is very important. We have determined antimicrobial susceptibility patterns, serotype, and virulence profiles. The antibiotic susceptibility was assessed for a total of 421 Streptococcus agalactiae strains, isolated from pregnant women and neonates. Then, 89 erythromycin and/or clindamycin-resistant strains (82 isolates obtained from pregnant women and seven isolates derived from neonates) were assessed in detail. PCR techniques were used to identify the studied strains, perform serotyping, and assess genes encoding selected virulence factors. Phenotypic and genotypic methods determined the mechanisms of resistance. All tested strains were sensitive to penicillin and levofloxacin. The constitutive MLSB mechanism (78.2%), inducible MLSB mechanism (14.9%), and M phenotype (6.9%) were identified in the macrolide-resistant strains. It was found that macrolide resistance is strongly associated with the presence of the ermB gene and serotype V. FbsA, fbsB, fbsC, scpB, and lmb formed the most recurring pattern of genes among the nine surface proteins whose genes were analysed. A minority (7.9%) of the GBS isolates exhibited resistance to lincosamides and macrolides, or either, including those that comprised the hypervirulent clone ST-17. The representative antibiotic resistance pattern consisted of erythromycin, clindamycin, and tetracycline resistance (71.9%). An increase in the fraction of strains resistant to macrolides and lincosamides indicates the need for monitoring both the susceptibility of these strains and the presence of the ST-17 clone.
Similar content being viewed by others
Introduction
Streptococcus agalactiae (Group B Streptococcus, GBS) inhabits the gastrointestinal and genital tracts as a component of the average human microbiota. However, this species is also associated with various infections affecting newborns, pregnant or postpartum women, and people over the age of 60. The most common diseases in newborns and neonates caused by GBS are pneumonia, sepsis, and meningitis1. Diagnosing skin and soft tissue infections, bacteremia, pneumonia, and osteoarthritis in adults is frequent. GBS is also a known etiologic agent of urinary tract infections, including pyelonephritis and prostatitis2,3,4.
GBS strains are universally sensitive to penicillin, with rare isolates with minimum inhibitory concentrations (MIC) at or above the susceptible breakpoint of 0.12 µg/mL5,6,7. Clindamycin is the recommended antibiotic alternative for patients with a history of allergy to β-lactams. However, there has been a recent increase in the identification of clinical GBS strains resistant to these antibiotics8. In GBS, resistance to macrolides arises mainly from an active drug efflux controlled by the mefA gene or modification of the drug target on the rRNA through methylases encoded by erm genes. Resistance can be expressed as a cross-resistance to macrolides, lincosamides, and streptogramin B (inducible-iMLSB and constitutive-cMLSB) or a resistance to 14- and 15-membered ring macrolides only, while lincosamides, streptogramins, and 16-membered ring macrolides remain active (phenotype M). In addition, the clindamycin-resistant erythromycin-susceptible phenotype (L phenotype) has been reported9,10,11. According to the latest Centers for Disease Control and Prevention (CDC)’ report, clindamycin-resistant GBS belongs to the category of “Concerning Threats”, based on the hazard to human health12. In pregnant women who are allergic to penicillin, it is crucial to consider the sensitivity of GBS to erythromycin and clindamycin. In such cases the perinatal prophylaxis, which includes ampicillin, penicillin, or cefazolin, cannot be administered.
GBS strains exhibit some virulence factors that are important during adhesion, invasion, and avoidance responses from the host's immune system. Surface proteins, including laminin-binding protein (Lmb), serine repeat proteins (Srr), fibrinogen-binding proteins (FbsA, FbsB, and FbsC), Rib, and alpha C, allow bacteria to adhere to host cells, which is crucial for further colonization or invasion of the host organism13,14. Capsular polysaccharides are one of the most essential factors of GBS virulence. Ten serotypes (Ia, Ib, II-IX) have been distinguished based on differences in the structure of capsular polysaccharides. The occurrence of particular serotypes varies, depending on the geographic region of the world and ethnicity, and also varies over time for a given region4,15,16,17,18. Previously, 98% of worldwide identified GBS isolates were found as serotypes Ia, Ib, II, III, IV, and V15. In the US, these six predominant serotypes were accountable for over 99% of early-onset and late-onset GBS-related diseases8. The systematic review demonstrated that serotype III, associated with invasive disease, accounts for 25% of observations but is less common in some South America and Asia countries, where serotypes VI-IX are more frequently identified15. In particular, serotype III isolates belonging to the sequence type (ST) 17 lineage (ST-17) have been defined as hypervirulent. This strain possesses numerous virulence factors, including a surface-anchored protein called hypervirulent GBS adhesin (HvgA). The Gbs2018/hvgA gene encoding this adhesin is being efficiently used to identify the ST-17 strains19,20. Estimating the prevalence of particular serotypes and surface proteins in different geographic regions is crucial as they will become targets for future vaccination, particularly for vaccines under development4,16.
The aim of this study was to determine antimicrobial susceptibility patterns, serotype, and virulence gene profiles of GBS isolates with macrolide and/or lincosamide resistance to highlight the increasing resistance to these groups of antibiotics in pregnant women.
Results
cMLSB found as a dominant phenotype
Of the evaluated 421 strains, 89 (21.1%) isolates were resistant to erythromycin and/or clindamycin and were subjected to further characterization. Among them, 82 were isolated from vaginal or rectal swabs from pregnant women and seven were derived from newborns [blood (n = 4), urine (n = 2), and pharyngeal swabs (n = 1)]. All the erythromycin- and/or clindamycin-resistant strains were sensitive to penicillin and levofloxacin. Resistance to tigecycline was identified in one strain obtained from pregnant women. This strain was also resistant to tetracycline, macrolide, lincosamide, and streptogramin B (cMLSB phenotype). All but three isolates from pregnant women showed resistance to tetracycline (96.6%). 71.9% (n = 64/89) of strains were simultaneously resistant to erythromycin, clindamycin, and tetracycline. Antibiotic resistance profiles are presented in Table 1.
Comparing the occurrence of macrolide, lincosamide, and streptogramin B resistance mechanisms among the tested strains, the iMLSB was identified in 6.74% of strains (n = 6/89; all from pregnant women), cMLSB was found in 76.4% of strains (n = 68/89; 64 from pregnant women, four from newborns), the M phenotype was found in 14.6% of strains (n = 13/89; 10 from pregnant women, three from newborns), whereas the L phenotype was identified in 2.25% of GBS strains (n = 2/89; all from pregnant women) (Fig. 1).
The presence of the ermB gene is a major determinant of erythromycin resistance
In all strains belonging to serotype Ia, the M phenotype (efflux mechanism), determined by the presence of the mefA gene, was found. In contrast, all strains belonging to serotypes Ib and IV showed the cMLSB phenotype, with resistance to clindamycin and erythromycin. The cMLSB phenotype predominated also in strains belonging to serotypes III and V [71.4% (n = 10/14) and 90.4% (n = 47/52) of strains, respectively], followed by the iMLSB [14.3% (n = 2/14) and 5.8% (n = 3/52)], and the M phenotype [14.3% (n = 2/14) and 1.9% (n = 1/52)]. The observed differences in prevalence of resistance mechanisms among different serotypes were statistically significant (p = 5.508e−16). Among 68 isolates with the cMLSB phenotype, 94.1% of samples (n = 64/68) contained the ermB gene, 4.4% of samples (n = 3/68) had the ermA gene, while the remaining 1.5% of samples (n = 1/68) contained both the ermB and mefA genes. All isolates with the iMLSB phenotype contained the ermA gene alone (n = 6/6), while isolates with the M phenotype contained the mefA gene (n = 13/13). In the two strains with the L resistance phenotype, the presence of the lnuB and lsaE genes was confirmed. These strains, one with serotype V and the other with serotype VI, were derived from pregnant women. The profiles of virulence genes, fbsA-fbsB-fbsC-lmb-scpB-rib and fbsB-fbsC-lmb-scpB-bca, were identified in isolates with serotypes V and VI, respectively.
The resistance genotypes and phenotypes identified among strains from pregnant women and newborns are presented in Table 2.
Serotype V was recognized to be predominant
Analyzing the frequencies of particular serotypes among erythromycin- and/or clindamycin-resistant strains, it was found that more than half of these strains (58.4%, n = 52/89; 49 from pregnant women, three from newborns) belonged to serotype V, followed by serotypes III and Ia (15.7%, n = 14/89; 13 from pregnant women, one from newborn) and 11.2%,n = 10/89; eight from pregnant women, two from newborns, respectively) (Table 2).
The most common virulence gene pattern, including the hvgA gene
Distribution of each virulence-associated gene among the individual serotypes is shown in Table 3. Statistically significant differences were found only in the occurrence of the rib gene (p = 9.183e−08) among individual serotypes. The fbsA, fbsB, and fbsC genes were detected in 98.9% (n = 88/89; 81 from pregnant women, seven from newborns) of isolates. Most isolates carried the lmb and scpB genes (84.3% (n = 75/89; 71 from pregnant women, four from newborns) and 95.5% (n = 85/89; 79 from pregnant women, six from newborns), respectively), while rib, bca, alp2/3, and epsilon were present in 30.3% (n = 27/89; 25 from pregnant women, two from newborns), 11.2% (n = 10/89; all from pregnant women), 47.2% (n = 42/89; 40 from pregnant women, two from newborns), and 11.2% (n = 10/89; seven from pregnant women, three from newborns) of isolates, respectively. Virulence gene profiles are presented in Table 1. The most common gene pattern, including the fbsA, fbsB, fbsC, scpB and lmb genes, was recognized in 73 (82.0%) strains.
The occurrence of the ST-17 clone was found in 7.9% of the tested strains (n = 7/89). Five of the ST-17 clones had the cMLSB phenotype, one had iMLSB phenotype, and one had M phenotype. All of them were simultaneously resistant to tetracycline. Strains with the hvgA gene were isolated from pregnant women. No ST-17 clone was found in any of the seven neonatal strains.
Discussion
Resistance of GBS strains to macrolides and lincosamides is discussed worldwide6,7,8. In this study, resistance to erythromycin and clindamycin was found in 20.7% (n = 87/421) and 16.6% (n = 70/421) strains, respectively. Previously, the fraction of strains resistant to erythromycin and clindamycin was recorded as variable and amounted to, respectively, 43.7% and 32.2% in Italy21, 30.0% and 28.0% in Switzerland22, and 14.5% and 14.0% in Sweden23. There is also a worrying trend toward increasing bacterial resistance to these groups of antibiotics seen in other populations24,25,26,27,28.
In line with currently reported data6, the cMLSB resistance phenotype dominated (76.4%) among the evaluated erythromycin and clindamycin-resistant strains, or either. Other resistance phenotypes identified in our study, iMLSB (6.7%) and M (14.6%), were also observed in 2.6% and 19.7% of isolates in an Italian population, respectively6. iMLSB phenotype occurs when resistance to erythromycin induces resistance to clindamycin. This phenotype is detected in clinical laboratories, with a double-disc diffusion test (D-zone test) to prevent clinical treatment failure with clindamycin and the emergence of cMLSB phenotype. The EUCAST guidelines recommend considering the iMLSB phenotype of GBS to be clindamycin-resistant29. We detected the L phenotype in only two strains from pregnant women. These strains were susceptible to erythromycin but resistant to clindamycin. This resistance phenotype is rare in clinical GBS strains but has been previously reported in isolates from the United States11, Spain30, Argentina31, Korea9, and other countries10,32,33.
The study led by the CDC investigators in the US showed that all tested isolates (n = 1727) were susceptible to penicillin, ampicillin, and vancomycin, 44.8% had erythromycin resistance, and 20.8% had constitutive clindamycin resistance8. From 2006 to 2015, the proportion of erythromycin resistance increased significantly from 34.7 to 49.1%, whereas the constitutive clindamycin resistance increased from 14.7 to 26.0%8.
Most of the erythromycin and clindamycin-resistant strains, or either, evaluated in this study were found to be serotype V (58.4%). Previously, in the US, the proportion of isolates with erythromycin resistance was recorded as the highest for serotype II (erythromycin resistance 66.5%) and serotype V (erythromycin resistance 61.1%)8.
In this study, common genetic determinants of erythromycin resistance were the presence of the ermB (73.0%), mefA (16.1%), and ermA [subclass ermTR] (10.1%) genes. All strains presenting with the M phenotype exhibited the presence of the mefA gene, which supports the existing literature findings that indicate the mefA gene exclusively confers resistance to macrolides32. Moreover, 64 isolates showing the cMLB phenotype had only the ermB gene (94.1%). In comparison, three isolates with this phenotype contained only the ermA gene (4.4%). The mefA and ermB genes were simultaneously detected in one isolate with the cMLSB phenotype (1.5%), suggesting the participation of two different genes in the appearance of macrolide and lincosamide resistance. Similar results have been found in the literature10,32,34. We observed a significant correlation between the presence of the ermA gene (100%) and the inducible resistance phenotype in our study. The available literature shows that the inductive phenotype is predominantly linked to a solitary ermA gene, although the existence of the ermB gene can also influence it10,32. Most of the clindamycin-resistant isolates showed cMLSB phenotype determined by the ermB gene. However, two isolates had the L phenotype and were positive for the lnuB and lsaE genes, consistent with previous findings9,10,11.
Recent epidemiological reports indicate the spread of the multidrug-resistant (MDR) subclone ST-17 GBS characterized by the loss of Pl-1 [CC (clonal complex) 17/Pl-2b], simultaneously resistant to macrolides, lincosamides, and tetracycline and other antibiotics20,35,36,37,38,39. Teatero et al. identified 11 CC17 isolates in Canadian patients lacking Pl-1 and acquiring a mobile genetic element encoding resistance to tetracycline, macrolides, and other antibiotics at the same site35. Campisi et al. conducted a whole-genome sequencing analysis and antimicrobial susceptibility testing on 14 isolates with serotype III that are part of the hypervirulent CC17 in China. Thirteen of 14 isolates contained only the sequences encoding the Pl-2b, and in place of the Pl-1 operon, the multi-drug resistance gene clusters harbored in two new versions of integrative and conjugative elements were present36.
Similarly, European studies have reported the emergence of hypervirulent strains belonging to a sublineage characterised by the absence of Pl-1 (CC17/Pl-2b) and exhibiting multidrug resistance20,37,38. In Italy, GBS serotype III was predominant in early-onset (56%) and late-onset (95%) diseases. The resistance to clindamycine reached 29%, and most of the strains (76%) were serotype III-ST17 and contained the genetic markers of the MDR CC17 clone20. Studies conducted on 218 strains isolated from invasive infections in newborns in Portugal yielded comparable findings, in which 50% of the isolates were the CC17. The presence of the clone CC17/Pl-2b simultaneously resistant to macrolides, lincosamides, and tetracycline and exhibiting high-level resistance to streptomycin and kanamycin among the tested strains was confirmed38.
A small fraction of the GBS isolates resistant to macrolides and lincosamides, or either, consisted of the hypervirulent clone ST-17, (7.9%, n = 7/89). The presence of this clone was confirmed only among pregnant women (7/82). Thus, the increase in resistance to erythromycin and clindamycin in Poland is not due to the spread of the hypervirulent MDR ST-17 clone. However, the presence of strains belonging to this clone among pregnant women and the results cited above indicate the need to monitor the drug susceptibility of GBS strains and the frequency of the ST-17 clone.
As in other studies, also in this work, serotype V was dominant among the erythromycin and clindamycin-resistant, or either, strains40,41,42. Subsequently, the tested isolates were identified as serotypes III, Ia, II, Ib, IV, and VI. Two isolates with L phenotype belonged to serotypes V and VI. A similar virulence gene profile was found in these isolates, comprising the fbsA, fbsB, fbsC, lmb, and scpB genes. The isolates with the serotypes V and VI also contained the rib and bca genes, respectively. We detected the fbsA, fbsB, fbsC, and scpB genes in nearly all characterized isolates, and these genes exhibit an even distribution across various serotypes. These results are consistent with previous report, in which the most frequently detected genes were scpB, fbsA, fbsB, cylB, and lmb43,44,45,46. Also, studies utilizing the whole-genome sequencing showed that fibrinogen-binding protein, laminin-binding protein, and serine peptidase coding sequences were detected in ≥ 98% of the assessed samples. In comparison, the rib gene was identified in 27.8% of isolates47.
Healthcare providers utilise maternal screening and intrapartum antibiotic therapy as preventive measures against GBS infections in newborns. However, such early exposure to antibiotics alters the gut microbiota of infants and may have health consequences later in life.
As the frequency of strains resistant to macrolides, lincosamides, and streptogramin B is increasing worldwide, it is essential to characterize these isolates in detail. Epidemiological monitoring of GBS serotypes and their surface proteins is vital in a given geographic region. It plays a substantial role in vaccine design processes and future vaccination strategy implementation.
Methods
GBS strains collection and identification
A total of 421 GBS clinical strains isolated from vaginal or rectal swabs from pregnant women (n = 401) and newborns [(n = 20), including invasive isolates from blood (n = 7), urine (n = 4), ear swabs (n = 2), and pharyngeal swabs (n = 7)] were evaluated. Then, a subset of 89 of erythromycin- and/or clindamycin-resistant strains was assessed in detail, including 82 isolates derived from pregnant women and seven isolates obtained from newborns [blood (n = 4), urine (n = 2), and pharyngeal swabs (n = 1)]. The strains were isolated in the Laboratory of Bacteriology of Obstetrics and Gynecology Clinical Hospital at Poznan University of Medical Sciences (PUMS), from January 2016 to December 2020. The reference strains stored at the Chair and Department of Genetics and Pharmaceutical Microbiology at PUMS were also applied in the assessments.
Specimen collection was performed in the Laboratory of Bacteriology at Gynecological and Obstetric Clinical Hospital PUMS in accordance with recommendations of Centers for Disease Control and Prevention48 and the Polish Gynecological Society49. Briefly, after incubating the vaginal-rectal swabs in Todd-Hewitt broth with colistin (10 µg/ml) and nalidixic acid (15 µg/ml) (OXOID Deutschland Gmbh, Germany) aerobically at 37 °C for 18–24 h, 10 µl of broth was subcultured on Columbia agar with 5% sheep blood (OXOID Deutschland Gmbh, Germany). Then, selected β- or γ-hemolytic colonies were identified to the species level using a latex agglutination test (OXOID) in accordance with the manufacturer's instructions and based on PCR techniques previously described by Kong et al.50. Only one isolate per patient was included in the analysis. Then, the strains were stored for further tests at temperature of − 80 ± 10 °C, in microbanks (Pro-Lab Diagnostics).
GBS strains DNA extraction
DNA extraction from the isolated GBS strains was carried out using the in house developed thermal extraction method7. The DNA sample concentrations were measured using the NanoDrop spectrometer (Thermo Scientific) and the DNA samples were stored for further analyses at − 20 ± 2 °C.
Evaluation of antibiotics susceptibility and mechanisms of resistance to macrolide, lincosamide and streptogramin B
Assessment of antibiotics susceptibility of GBS strains was carried out using the disc diffusion method in accordance with the EUCAST recommendations, applying penicillin (1 IU), erythromycin (15 μg), clindamycin (2 μg), levofloxacin (5 μg), tigecycline (15 μg), tetracycline (30 μg), and levofloxacin (5 μg)29. Interpretation of the results of susceptibility testing was performed in accordance with the EUCAST recommendations29. Isolates which were phenotypically resistant or intermediate resistant to studied antibiotics were reported as resistant strains. Only strains resistant to erythromycin and/or clindamycin were further investigated.
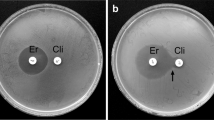
Resistance to macrolides, lincosamides and streptogramin B was evaluated using the double-disc method in accordance with the EUCAST guidelines29. Phenotypes were assigned as follows: (I) inducible mechanism of resistance to macrolide, lincosamide, and streptogramin B (iMLSB), when the zone of growth inhibition around the disc with erythromycin indicating the intermediate sensitivity or resistance and flattening of the zone next to the clindamycin halo on the side of erythromycin (so-called D zone) were visible; (II) constitutive mechanism of resistance to macrolide, lincosamide, and streptogramin B (cMLSB), when the growth inhibition zone around the erythromycin disc pointing to resistance and the zone of inhibition around the clindamycin disc indicating resistance or intermediate sensitivity were observable; (III) M phenotype (M), when the zone of growth inhibition around the disc with erythromycin indicating the intermediate sensitivity or resistance, and the zone indicating sensitivity around the disc with clindamycin in the absence of flattening zone of growth inhibition were apparent; (IV) L phenotype (L), when the zone of growth inhibition around the disc with clindamycin indicating the intermediate sensitivity or resistance, and the zone indicating sensitivity around the disc with erythromycin were visible.
Identification of genes related to antibiotic resistance
The presence of the genes associated with antibiotic resistance, erythromycin and/or clindamycin (ermA, ermB, mefA, lnuB, and lsaE) and tetracycline (tetM), was assessed using a duplex or multiplex PCR technique, described in Supplementary Table S1.
Serotyping and identification of virulence-related genes in macrolide- and/or lincosamide-resistant GBS strains
The use of primers for serotypes Ia, Ib, and II–VII allowed assigning all isolates to single serotypes. Serotypes Ia, Ib, III, and V were identified simultaneously in PCR multiplex reactions, whereas serotypes II, IV, VI, and VII were determined in separate PCR reactions. Supplementary Table S1 presents primers and PCR conditions applied in the serotyping of GBS strains and detection of the fbsA, fbsB, fbsC (protein FbsA, FbsB, FbsC), lmb (protein Lmb), scpB (C5a peptidase), bca, and rib (components of protein C), alp2/3 (protein Alp 2/3) and epsilon (protein Epsilon) virulence-related genes. Identification of hypervirulent ST-17 lineage was performed using a PCR assay based on the detection of the hvgA gene19. To confirm the specificity of the obtained amplicons, the PCR products were purified using a ExoSAP-IT®for PCR product Clean-Up (Affymetrix) and Sanger sequenced (Genomed, Warsaw, Poland).
Statistical analysis
Statistical analysis was performed using χ2 and Fisher exact tests. P values less than 0.05 were considered significant.
Data availability
The authors confirm that all the data supporting the findings of this study are available within the Article and its Supplementary materials S1.
References
Jamrozy, D. et al. Increasing incidence of group B streptococcus neonatal infections in the Netherlands is associated with clonal expansion of CC17 and CC23. Sci. Rep. 10, 9539. https://doi.org/10.1038/s41598-020-66214-3 (2020).
Lim, S., Rajagopal, S., Jeong, Y. R., Nzegwu, D. & Wright, M. L. Group B Streptococcus and the vaginal microbiome among pregnant women: a systematic review. Peer J. 9, e11437. https://doi.org/10.7717/peerj.11437 (2021).
Maeda, T. et al. Comparison between Invasive and Non-Invasive Streptococcus agalactiae isolates from human adults, Based on virulence gene profiles, capsular genotypes, sequence types, and antimicrobial resistance patterns. Jpn J. Infect. Dis. 74, 316–324 (2021).
Bianchi-Jassir, F. et al. Systematic review of Group B Streptococcal capsular types, sequence types and surface proteins as potential vaccine candidates. Vaccine. 38, 6682–6694 (2020).
Humphries, R. M. Group B Streptococcus dynamics in the United States. Clin. Infect. Dis. 72, 1014–1015 (2021).
Genovese, C., D’Angeli, F., Di Salvatore, V., Tempera, G. & Nicolosi, D. Streptococcus agalactiae in pregnant women: serotype and antimicrobial susceptibility patterns over five years in Eastern Sicily (Italy). Eur. J. Clin. Microbiol. Infect. Dis. 39, 2387–2396 (2020).
Kaminska, D. et al. Increasing resistance and changes in distribution of serotypes of Streptococcus agalactiae in Poland. Pathogens. 9, 526 (2020).
Nanduri, S. A. et al. Epidemiology of invasive early-onset and late-onset group B Streptococcal disease in the United States, 2006 to 2015: Multistate laboratory and population-based surveillance. JAMA Pediatr. 173, 224–233 (2019).
Takahashi, T., Maeda, T., Lee, S., Lee, D. H. & Kim, S. Clonal distribution of clindamycin-resistant erythromycin-susceptible (CRES) Streptococcus agalactiae in Korea based on whole genome sequences. Ann. Lab. Med. 40, 370–381 (2020).
Motallebirad, T. et al. Prevalence, population structure, distribution of serotypes, pilus islands and resistance genes among erythromycin-resistant colonizing and invasive Streptococcus agalactiae isolates recovered from pregnant and non-pregnant women in Isfahan Iran. BMC Microbiol. 21, 139 (2021).
Hawkins, P. A. et al. Cross-resistance to lincosamides, streptogramins A and pleuromutilins in Streptococcus agalactiae isolates from the USA. J. Antimicrob. Chemother. 72, 1886–1892 (2017).
Center for Disease Control and Prevention. Antibiotic Resistance Threat in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA (2019).
Creti, R., Fabretti, F., Orefici, G. & von Hunolstein, C. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. J. Clin. Microbiol. 42, 1326–1329 (2004).
Chan, J. M., Gori, A., Nobbs, A. H. & Heyderman, R. S. Streptococcal serine-rich repeat proteins in colonization and disease. Front. Microbiol. 11, 593356 (2020).
Russell, N.J. et al. GBS Maternal Colonization Investigator Group. Maternal colonization with group B Streptococcus and serotype distribution worldwide: Systematic review and meta-analyses. Clin. Infect. Dis. 65, 100–111 (2017).
Berner, R. Group B streptococcus vaccines: One step further. Lancet Infect. Dis. 21, 158–160 (2021).
Perme, T. et al. Genomic and phenotypic characterisation of invasive neonatal and colonising group B Streptococcus isolates from Slovenia, 2001–2018. BMC Infect. Dis. 20, 958 (2020).
Chaguza, C. et al. Population genomics of Group B Streptococcus reveals the genetics of neonatal disease onset and meningeal invasion. Nat. Commun. 13, 4215. https://doi.org/10.1038/s41467-022-31858-4 (2022).
Lamy, M. C. et al. Rapid detection of the “highly virulent” group B Streptococcus ST-17 clone. Microbes Infect. 8, 1714–1722 (2006).
Creti, R., Imperi, M., Berardi, A., Lindh, E., Alfarone, G., Pataracchia, M. & Recchia, S. The Italian Network On Neonatal And Infant Gbs Infections. Invasive Group B Streptococcal Disease in Neonates and Infants, Italy, Years 2015–2019. Microorganisms. 9, 2579. https://doi.org/10.3390/microorganisms9122579 (2021).
Matani, C., Trezzi, M., Matteini, A., Messeri, D. & Catalani, C. Streptococcus agalactiae: prevalence of antimicrobial resistance in vaginal and rectal swabs in Italian pregnant women. Infez. Med. 24, 217–221 (2016).
Capanna, F. et al. Antibiotic resistance patterns among group B Streptococcus isolates: implications for antibiotic prophylaxis for early-onset neonatal sepsis. Swiss. Med. Wkly. 143, w13778 (2013).
Fröhlicher, S. et al. Serotype distribution and antimicrobial susceptibility of group B streptococci in pregnant women: results from a Swiss tertiary centre. Swiss Med. Wkly. 144, w13935 (2014).
Dogan, B., Schukken, Y. H., Santisteban, C. & Boor, K. J. Distribution of serotypes and antimicrobial resistance genes among Streptococcus agalactiae isolates from bovine and human hosts. J. Clin. Microbiol. 43, 5899–5906 (2005).
Phares, C. R. et al. Epidemiology of invasive group B streptococcal disease in the United States, 1999–2005. JAMA. 299, 2056–2065 (2008).
Wang, P. et al. Serotypes, antibiotic susceptibilities, and multilocus sequence type profiles of Streptococcus agalactiae isolates circulating in Beijing China. PLoS One. 10, e0120035 (2015).
Wang, P. et al. Serotype distribution, antimicrobial resistance, and molecular characterization of invasive group B Streptococcus isolates recovered from Chinese neonates. Int. J. Infect. Dis. 37, 115–118 (2015).
Kekic, D. et al. Trends in molecular characteristics and antimicrobial resistance of group B streptococci: A multicenter study in Serbia, 2015–2020. Sci. Rep. 11, 540. https://doi.org/10.1038/s41598-020-79354-3 (2021).
European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters Version 11.0, Valid from 1 January 2021. Available online: https://www.eucast.org (accessed on 12 April 2021).
Arana, D., Rojo-Bezares, B., Torres, C. & Alós, J. I. First clinical isolate in Europe of clindamycin-resistant group B Streptococcus mediated by the lnu(B) gene. Rev. Esp. Quimioter. 27, 106–109 (2014).
Faccone, D. et al. Multiple-clones of Streptococcus agalactiae harbouringlnu B gene. J. Infect. Dev. Ctries. 4, 580–582 (2010).
Compain, F. et al. Molecular characterization of Streptococcus agalactiae isolates harboring small erm(T)-carrying plasmids. Antimicrob. Agents Chemother. 58, 6928–6930 (2014).
De Azavedo, J., McGavin, M., Duncan, C. & Low, D. E. McGeer, APrevelence and mechanisms of macrolide resistance in invasive and noninvasive Group B Streptococcus isolates from Ontario Canada. Antimicrob. Agents Chemother. 45, 3504–3508 (2001).
Santana, F. A. F. et al. Streptococcus agalactiae: Identification methods, antimicrobial susceptibility, and resistance genes in pregnant women. World J. Clin. Cases. 8, 3988–3998 (2020).
Teatero, S. et al. Clonal Complex 17 Group B Streptococcus strains causing invasive disease in neonates and adults originate from the same genetic pool. Sci. Rep. 6, 20047. https://doi.org/10.1038/srep20047 (2016).
Campisi, E. et al. Genomic analysis reveals multi-drug resistance clusters in group B Streptococcus CC17 hypervirulent isolates causing neonatal invasive disease in Southern Mainland China. Front. Microbiol. 7, 1265. https://doi.org/10.3389/fmicb.2016.01265 (2016).
Plainvert, C. et al. Multidrug-resistant hypervirulent group B Streptococcus in neonatal invasive infections, France, 2007–2019. Emerg. Infect. Dis. 26, 2721–2724 (2020).
Martins, E.R., Pedroso-Roussado, C., Melo-Cristino, J. & Ramirez, M. Portuguese Group for the Study of Streptococcal Infections. Streptococcus agalactiae Causing Neonatal Infections in Portugal (2005–2015): Diversification and Emergence of a CC17/PI-2b Multidrug Resistant Sublineage. Front Microbiol. 8, 499. https://doi.org/10.3389/fmicb.2017.00499 (2017).
Martins, E. R., Nascimento, Ó. D., Marques Costa, A. L., Melo-Cristino, J. & Ramirez, M. Characteristics of Streptococcus agalactiae colonizing nonpregnant adults support the opportunistic nature of invasive infections. Microbiol. Spectr. 10, e0108222. https://doi.org/10.1128/spectrum.01082-22 (2022).
Uh, Y., Kim, H. Y., Jang, I. H., Hwang, G. Y. & Yoon, K. J. Correlation of serotypes and genotype of macrolide-resistant Streptococcus agalactiae. Yonsei Med. J. 46, 480–483 (2005).
Gherardi, G. et al. Molecular epidemiology and distribution of serotypes, surface proteins and antibiotic resistance among group B streptococci in Italy. J. Clin. Microbiol. 45, 2909–2916 (2007).
Brzychczy-Włoch, M. et al. Genetic characterization and diversity of Streptococcus agalactiae isolates with macrolide resistance. J. Med. Microbiol. 59, 780–786 (2010).
Bobadilla, F. J., Novosak, M. G., Cortese, I. J., Delgado, O. D. & Laczeski, M. E. Prevalence, serotypes and virulence genes of Streptococcus agalactiae isolated from pregnant women with 35–37 weeks of gestation. BMC Infect. Dis. 21, 73 (2021).
Eskandarian, N. et al. Antimicrobial susceptibility profiles, serotype distribution and virulence determinants among invasive, non-invasive and colonizing Streptococcus agalactiae (group B streptococcus) from Malaysian patients. Eur. J. Clin. Microbiol. Infect. Dis. 34, 579–584 (2014).
Beigverdi, R. et al. Virulence factors, antimicrobial susceptibility and molecular characterization of Streptococcus agalactiae isolated from pregnant women. Acta Microbiol. Immunol. Hung. 61, 425–434 (2014).
Rosenau, A. et al. Evaluation of the ability of Streptococcus agalactiae strains isolated from genital and neonatal specimens to bind to human fibrinogen and correlation with characteristics of the fbsA and fbsB genes. Infect. Immun. 75, 1310–1317 (2007).
Furfaro, L. L., Chang, B. J., Kahler, C. M. & Payne, M. S. Genomic characterisation of perinatal Western Australian Streptococcus agalactiae isolates. PLoS One. 14, e0223256. https://doi.org/10.1371/journal.pone.0223256 (2019).
Verani, J.R., McGee, L., & Schrag, S.J. Prevention of Perinatal Group B Streptococcal Disease. Revised Guidelines from CDC, 2010. MMWR Recomm Rep. 59, No. RR-10 (2010).
Heczko, P. B. et al. Rekomendations for the detection of group B streptococcus (GBS) carriage in pregnant women and for prevention of neonatal infections caused by this pathogen. Zakazenia. 2, 87–96 (2008).
Kong, F., Ma, L. & Gilbert, G. L. Simultaneous detection and serotype identification of Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization. J. Med. Microbiol. 54, 1133–1138 (2005).
Acknowledgements
Thank you for the technical work performed by Magdalena Kopis and Maria Marcysiak.
Author information
Authors and Affiliations
Contributions
Conceptualization, D.K, M.R. and M.G.; methodology, M.R., D.K. and J.D.; formal analysis, M.R., D.K., J.D. and A.Sz-K.; investigation, D.K, M.R., M.K., J.A.K., and J.D.; statistical analysis, D.M.N-M.; interpretation of the results D.K., M.R., J.D., M.K., and A.Sz-K,; writing—original draft preparation, D.K. and M.R.; writing—review and editing, M.G., J.D. and J.A.K.; all authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kamińska, D., Ratajczak, M., Nowak-Malczewska, D.M. et al. Macrolide and lincosamide resistance of Streptococcus agalactiae in pregnant women in Poland. Sci Rep 14, 3877 (2024). https://doi.org/10.1038/s41598-024-54521-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-54521-y
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.