Abstract
Huntington’s disease (HD) is caused by an aberrant expansion of CAG repeats in the HTT gene that mainly affects basal ganglia. Although striatal dysfunction has been widely studied in HD mouse models, other brain areas can also be relevant to the pathology. In this sense, we have special interest on the retina as this is the most exposed part of the central nervous system that enable health monitoring of patients using noninvasive techniques. To establish the retina as an appropriate tissue for HD studies, we need to correlate the retinal alterations with those in the inner brain, i.e., striatum. We confirmed the malfunction of the transgenic R6/1 retinas, which underwent a rearrangement of their transcriptome as extensive as in the striatum. Although tissue-enriched genes were downregulated in both areas, a neuroinflammation signature was only clearly induced in the R6/1 retina in which the observed glial activation was reminiscent of the situation in HD patient’s brains. The retinal neuroinflammation was confirmed in the slow progressive knock-in zQ175 strain. Overall, these results demonstrated the suitability of the mouse retina as a research model for HD and its associated glial activation.
Similar content being viewed by others
Introduction
Huntington’s disease (HD) (OMIM #143100) is a fatal rare disorder without a cure, and its estimated prevalence is 1–15 per 100,000 people worldwide1. HD is caused by an aberrant expansion (fully penetrant with > 39) of CAG repeats at exon 1 of the HTT locus, which triggers a progressive symptomatology, usually starting in mid-adulthood (35–45 years old) that includes cognitive and motor impairments, psychiatric disorders and other symptoms (weight loss, sleep disturbance, etc.) until final death2. Although the most prominent signs are derived from malfunctioning and degeneration of the basal ganglia and the corticostriatal circuitry3, the involvement of other brain areas and peripheral tissues/cells plays a substantial role in the decline of quality of life, extending the repertoire of HD symptoms4,5,6,7,8. However, our current toolbox of biomarkers with potential application in clinics is still limited and cannot explain the large variability in the pleiotropic manifestation of symptoms9,10,11. This fact justifies the search for novel biomarkers with prognostic value in HD to enable the evaluation of therapeutic responses and to facilitate decision-making during clinical management.
The retina is a highly organized tissue of the central nervous system (CNS) characterized by a high cellular diversity arranged in discrete layers. This organization determines the functionality of the retina, in which photoreceptor neurons (rods and cones) transduce light stimuli into the complex network of interneurons (amacrine, bipolar and horizontal cells) to finally converge into ganglion cells, which axons form the optic nerve12; the retina also contains a specialized immune system with resident microglia and macroglia (astrocytes, Müller glia), which together with other nonneural type cells provide homeostatic and regulatory support to this network13. The retina can reproduce the disruption of key mechanisms that compromise cell function and viability of several neurodegenerative disorders, therefore the accessibility of this tissue to noninvasive techniques offers unique opportunities to infer the health status of inner regions of the CNS. In patients, the suitability of retinal imaging in diagnosis is being investigated in Alzheimer's disease, Parkinson's disease, Lewy body dementia, frontotemporal dementia and multiple sclerosis, with promising prospects14,15. HD is not an exception; there are selective abnormalities (e.g., thickness reduction of the temporal retinal nerve fibre layer (RNFL), colour vision impairment) that can be tentatively correlated with performance on the Unified Huntington’s Disease Rating Scale (UHDRS). These anomalies can be accompanied by additional alterations of the visual pathway, i.e., reduced amplitude of visual evoked potentials and impaired temporal contrast sensitivity (reviewed in Ref.16). In the case of HD mouse models, mutant retinas express mHTT, display deeply reduced photoexcitability responses and undergo a cellular remodelling17,18,19,20,21 that coincides with early stages of motor impairment: therefore, retinal dysfunction could be correlated with striatal impairment in HD.
However, the molecular mechanisms that compromise the retinal structure and visual function in HD mice and their corresponding correlates with inner brain events have not yet been investigated. This study reports for the first time a comprehensive analysis of the concomitant molecular alterations in the retina and striatum of HD mouse models, the transgenic R6/1 and the knock-in (KI) zQ175 strains, in which glial activation becomes a distinctive feature between both tissues during pathology progression.
Materials and methods
Animals
Transgenic R6/122 and KI zQ17523, together with control wild-type (wt) animals, were maintained on a pure C57BL/6 J background under a 12-h light/dark cycle with food and water provided ad libitum. In all experiments, we pooled females and males in similar proportions (not exceeding 60% towards any sex), except for the RNA-seq analysis in which only males were used to avoid cost increase of the experiment. All animals were euthanized by cervical dislocation by well-trained personnel, followed by immediate tissue dissection for subsequent procedures. Experimental protocols were approved by the Comité de Ética de Experimentación Animal—Órgano Habilitado de la Universidad de Cádiz and authorized by the Dirección General de la Producción Agrícola y Ganadera de la Junta de Andalucía according to European and regional laws. This study was in compliance with the ARRIVE guidelines.
Nucleic acid extraction and PCR assays
After sacrifice, the striatum and retina were immediately dissected and submerged in RNAlater (Thermo Fisher, Madrid, Spain) until processing. Total RNA and genomic DNA were sequentially extracted using TRIzol (Thermo Fisher, Madrid, Spain), and we next followed the procedures for RT-qPCR and CAG repeat analysis described in Ref.24 with minor modifications: qPCR was performed on the Rotor-Gene 6000 Detection System (Corbett, Hilden, Germany) and QuantStudio 12 K Flex (Thermo Fisher, Madrid, Spain), and each independent reaction was normalized to the level of Tbp, since its expression in the retinal samples was less variable across time points (coefficient of variation CV = 0.51) than other housekeeping genes (e.g., Gapdh, CV = 0.69; Actb, CV = 1.00); in addition, the latter genes were found to be significantly altered in the RNA-seq analysis (see Supplementary Material). Fold changes were estimated using the ΔΔCT method. The sequences of all primer pairs are provided in Supplementary Table S1.
RNA-seq analysis, external datasets and bioinformatics
After the TRIzol procedure, striatal and retinal RNA from the same R6/1 mice was pooled (3–4 samples per genotype) and further processed using the clean-up protocol of the RNeasy Mini Kit (Qiagen, Hilden, Germany), which also included on-column DNase I treatment. A total of three pools per genotype were analyzed. DNA libraries were produced for mRNA using the TruSeq Stranded mRNA kit (Illumina, San Diego, CA, USA) and subsequently sequenced using a NovaSeq apparatus (Illumina, San Diego, CA, USA) at STAB-VIDA facilities in a 150-bp paired-end configuration. The resulting reads (> 40 M/sample) were mapped onto the mouse genome GRCm38/mm10 using “Salmon” software25. Genes with < 10 counts in all samples were removed. The normalization of read counts and differential expression analysis was conducted using the “DEseq2” package26. Differentially expressed genes (DEGs) were filtered with an FDR threshold (adjusted p-value) of 0.05. See Supplementary Table S2 for complete results. To retrieve differentially spliced genes, we combined the results of three programs: vast-tools27 (dPSI > 0.95), rMATS28 (lncLevelDifference, FDR < 0.05) and SUPPA229 (dPSI, p-value < 0.05). See Supplementary Table S3 for complete results. The RNA-seq data can be downloaded from the Gene Expression Omnibus (GEO) database using the accession number GSE216520.
To determine the most affected cell subtypes by the expression of mHTT in the differential expression between R6/1 and wt littermates, we used the cell-specific signatures obtained from mouse retinas in physiological conditions30, i.e., the genes that define the different subpopulations (clusters) of cells identified by scRNA-seq (Supplementary Table S4 of the original publication30). Amacrine and bipolar cells were defined by several clusters that we pooled for our study. Because rod cells constitute ~ 70% of the total retinal cells, contamination from rod cytoplasmic mRNA was present in nearly all clusters31; therefore, we filtered out the rod signature of the cell-specific signatures prior to the analysis of marker expression.
To investigate the presence of glial activation markers in our RNA-seq datasets, we used the most significant DEGs (top250): (i) between control and activated Aldh1h+-astrocytes (isolated from 1 to 7 days after different in vivo injuring protocols32), as calculated by applying the “affy”33 and “limma”34 packages, and (ii) among CD11b+-CD45+ microglial cells isolated from the neurodegenerative model CK-p25 at different time points after induction of the transgene (from immediately to 6 weeks), as presented in Supplementary Table S4 of the original publication35. Astrocytic panmarkers and specific markers for neurotoxic (A1) and neuroprotective (A2) astrocytes were obtained from a previous report36. Related to autophagy, we also used two classifications of autophagic-related genes as compiled by a recent study37: one consisted on “mTOR and upstream pathways”, “autophagy core”, “autophagy regulators”, “mitophagy”, “docking and fusion”, “lysosome” and “lysosome-related”, and another one consisted on “autophagy induction” and “lysosomal biogenesis” genes. Other additional bioinformatic tools included Venny (http://bioinfogp.cnb.csic.es/tools/venny/) for the identification of overlapping genes between multiple lists of genes, DAVID 2021 Update38 for overrepresentation analysis of Gene Ontology (GO) terms related to biological processes, Pscan39 for overrepresentation analysis of transcription factor binding sites (TFBS) at the promoter regions (-950/ + 50) from the Jaspar 2020_NR database, and the native R environment for statistical analysis (ANOVA and Student’s t-test).
Western blotting assays
Whole retinas were homogenized in lysis buffer containing 125 mM Tris–HCl pH 6.9, 2% SDS, and 1 mM DTT supplemented with protease inhibitors (cOmplete EDTA-free, Sigma-Aldrich, Darmstadt, Germany). All debris was removed by centrifugation at 14,000 × g for 10 min at 4 °C and the protein concentration was quantified using the Bio-Rad protein assay with BSA as a standard. Equivalent amounts of protein were resolved using denaturing sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE), followed by transfer to PVDF membranes (Merck Millipore, Cork, Ireland). Membranes were blocked using 5% nonfat dried milk or 3% BSA in 10 mM Tris-HCl pH 7.5, 150 mM NaCl, and 0.1% Tween-20, and incubated overnight with several primary antibodies (1:1000) in fresh blocking solution. After incubation with secondary antibodies (1:5000), immunoreactive bands were visualized using enhanced chemiluminescence reagent (Bio-Rad, Hercules, CA, USA). The fold change relative to the basal condition is shown. Blots were quantified by scanning densitometry. Primary antibody against LC3 (#2775) were purchased from Cell Signaling Technology (Danvers, CA, USA); primary antibody against α-tubulin (T5168) and secondary antibodies (A0545, anti-rabbit; A9044, anti-mouse IgG-peroxidase) were purchased from Sigma-Aldrich (Darmstadt, Germany).
Immunohistochemistry assays
Eye balls and whole retinal explants were fixed in 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4, at 4 °C overnight and subsequently put in a solution of PBS with 25% sucrose for another 24 h and cryopreserved in Tissue-Tek (4583, Sakura Finetek, Barcelona, Spain) at − 80 °C until cryosectioning. For immunofluorescence analysis, cryosections were incubated with a permeabilization solution containing 0.1 M TBS, 2% Triton X-100, blocked for 2 h in TBS containing 3% BSA and 1% Triton X-100, and incubated overnight in a humid chamber at 4 °C with rabbit anti-glial fibrillary acidic protein (GFAP) antibody (1:500, Z0334, DAKO, Agilent, Glostrup, Denmark), rabbit anti-ionized calcium binding adaptor molecule 1 (Iba-1) (1:500, 019-19741, WAKO, FUJIFILM Cellular Dynamics, Madison, WI, USA) or mouse anti-huntingtin EM48 (1:300, Merck Millipore, Burlington, MA, USA) in blocking solution. Next, sections were washed and incubated for 2 h with secondary antibodies conjugated to Alexa-488 (1:2000; Molecular Probes, Thermo Fisher Scientific, Waltham, MA, USA). After washing, both the retinal explants and sections were mounted with medium (Fluoromount G, Southern Biotech, Thermo Fisher Scientific, Waltham, MA, USA) containing 4’-6-diamidino-2-phenylindole (DAPI). Staining was observed with an inverted laser confocal microscope Axio Observer LSM900 (Carl Zeiss Microscopy GmbH, Göttingen, Germany). Cell nuclei labelled with DAPI were counted in the outer nuclear layer (ONL) using ImageJ software on retinal section images at 40 × magnification. In each mouse, we measured a total of six retinal cross-sections made through the optic nerve head, and averaged for each animal. For the EM48 antibody we followed the Diaminobenzidene (DAB) staining produce described for the 3,3′-Diaminobenzidine (DAB) Liquid Substrate System tetrahydrochloride (Sigma-Aldrich, Darmstadt, Germany) following the manufacturer’s instructions.
Electroretinogram recordings
All electroretinogram (ERG) procedures have been described previously40. Dark-adapted mice were anaesthetized with ketamine (95 mg/kg) and xylazine (5 mg/kg) under dim red light and the eyes were instilled as in the SD-OCT procedure. All mice were placed in a Faraday cage. Light intensity was measured in the eye with a photometer (Mavo Monitor USB, Nuremberg, Germany). Flash-induced ERG responses were recorded from the right eye in response to 4 to 64 consecutive light stimuli produced with a Ganzfeld stimulator: from − 4 to 1.5 log Cd × s × m−2 for rod-mediated responses, from − 1.5 to 0.5 log Cd × s × m-2 for mixed rod- and cone-mediated responses, from − 0.5 to 2 log Cd × s × m−2 on a rod saturating background of 30 Cd × s × m−2 for cone-mediated responses, with intervals between flashes of 10 s (dim flashes, scotopic condition), up to 60 s (high intensity flashes, scotopic condition) and 1 s (photopic condition). For recording, we used corneal electrodes, a reference electrode (in the mouth) and a ground electrode (in the tail) (Burian-Allen electrode, Hansen Ophthalmic Development Laboratory, Coralville, IA, USA). The recorded signals were amplified and band-filtered between 0.3 and 10.000 Hz with a Grass amplifier (CP511 AC amplifier; Grass Instruments, Quincy, MA, USA), and digitized at 10 kHz with a Power-Lab data acquisition board (AD Instruments, Chalgrove, UK). Wave amplitudes were calculated blinded to the animal genotype.
Spectral-domain optical coherence tomography (SD-OCT)
R6/1 (25-week-old), zQ175 (12-month-old) and matched-age wt mice were anaesthetized and maintained on a heated pad at 37 °C. The eyes were instilled with a topical drop of 1% tropicamide (Colircusí Tropicamida, Alcon Cusí SA, Barcelona, Spain) for pupil dilation and 2% Methocel (Ciba Vision AG, Hetlingen, Switzerland). OCT images were obtained using a Micron IV rodent imaging system (Phoenix Research Labs, Pleasanton, CA, USA) as described elsewhere41. A B-scan including the maximal retinal thickness (in the centre of the retina) was segmented between the inner limiting membrane and the base of the retinal pigment epithelium using the Insight software package (Phoenix Research Labs). The thickness of both eyes (containing average raw data from 624 A-scans from each eye after removing the first and last 200 scans) was averaged per animal for statistical analysis.
Results
The retina and striatum of R6/1 mice showed extensive transcriptional dysregulation
Based on the retinal impairments reported in HD mouse models17,18,19,20,21, we examined the molecular alterations in the R6/1 retina and their relationships with the dysfunction of the most affected brain area in HD, the striatum. Basal ganglia are well documented to manifest the most dramatic transcriptional dysregulation in patients and HD mouse models compared to the rest of the brain42,43 but a direct comparison with retinal tissue is still lacking. Once demonstrated that R6/1 retina effectively expressed mHTT (Supplementary Fig. S1A) and showed an altered visual function (Supplementary Fig. S1B), we screened by RNA-seq the whole mRNA expression of the retinas and striata obtained from the same mutant and wt mice in an earlier pathological stage (13–15 weeks-old). The differential expression analysis (adjusted p-value < 0.05) revealed extensive dysregulation of the R6/1 retina transcriptome related to wt littermates (1078 downregulated and 575 upregulated genes), resulting in more DEGs compared to the striatum of the same animals (763 downregulated and 176 upregulated genes) although comparable in magnitude and significance (Fig. 1A,B). Using our significance cut-off, most of the DEGs in our RNA-seq analysis were specific to the retina and the striatum: 87.7% and 78.3%, respectively. In fact, downregulation generally affected genes that were highly expressed in the corresponding neural tissue: i.e., downregulated retinal genes were physiologically more highly expressed in the retina than in the striatum, and vice versa (Fig. 1C), in agreement with the strong tissue-enriched component of HD-associated transcriptional dysregulation (Refs.44,45 and references therein). This tissue specificity was also confirmed at the functional level (Fig. 1D), since we detected that downregulated genes in the R6/1 retina (subset a) were enriched in GO terms genes associated with visual perception, phototransduction and retinal development, whereas downregulated genes in the striatum (subset c) were enriched with genes linked to addiction, locomotion, and medium spiny neurons, among the expected pathways affected in HD (e.g., cAMP and G-protein-dependent). Notably, we retrieved circadian functions among the commonly altered genes in both tissues (subset b) that might be related to the circadian-like modulation of GABAergic interneurons and dopamine in diverse brain areas, including the retina and striatum46,47. This observation should be explored in future studies considering the circadian disruption in HD patients and mice5,48. All subsets of downregulated genes were enriched with neuronal functions (e.g., synaptic transmission, ion transport, neuronal development) (Fig. 1D). In contrast, upregulation was linked to distinctive phenomena in each tissue, as evidenced by the retrieval of nonoverlapping associated functions (i.e., immune response in retinal subset d and nonneural development in striatal subset f, Fig. 1D) and different putative regulatory mechanisms (Supplementary Fig. S2A,B). Since RNA splicing is also documented to be disturbed in HD49,50, we also investigated differential spliced genes between genotypes in our RNA-seq datasets. We observed a similar behaviour as in differential expression: high tissue specificity with a prominent retinal deregulation (Fig. 1E). The affected genes were enriched in mRNA processing functions (e.g., splicing in the retina and translation in the striatum) (Fig. 1F).
RNA-seq analysis in the retina and striatum of R6/1 mice. (A) Venn diagram showing the number of DEGs in pairwise comparisons between wt and R6/1 mice (adj. p-value < 0.05, n of pools = 3 per genotype). Downregulated genes were indicated as a (exclusive retinal), b (common to both tissues) and c (exclusive striatal), and upregulated genes as d (exclusive retinal), e (common) and f (exclusive striatal). (B) Absolute values of log2 fold change (left) and − log adj. p-value (right) in the R6/1 retina and striatum: outliers > 5 and > 30 were removed from each panel respectively to enable visualization of medians and quartiles of the values. (C) Normalized basal expression of DEGs in wt retinas and striata. **p-value < 0.005, Student’s t-test between tissues. (D) GO enrichment analysis of DEGs in the retina and the striatum of R6/1 mice (p-value < 0.001 (DAVID)); not significant results were retrieved for subset e. Letters indicate the subsets of genes defined in (A). Data are expressed as mean ± s.e.m. Vm membrane potential, cAMP cyclic AMP, MSN medium spiny neurons, IFN interferon, Ag antigen. (E) Venn diagram showing the genes affected by differential alternative splicing (diffAS) in R6/1 retina and striatum compared to wt littermates. Pie charts indicate the type of aberrant splicing compiled in vast-tools, rMATS and SUPPA2: A5, alternative 5’ splice-site; A3, alternative 3’ splice-site; RI, retained intron; Excl, excluded exon (mutually exclusive exons, alternative first and last exon); SE, skipped exon. (F) GO enrichment analysis of genes with diffAS in the retina and the striatum of R6/1 mice (p-value < 0.001 (DAVID)). Numbers besides bars indicate the number of genes contained in GO categories. (G) Retinal DEGs were ranked according to their significance and direction of change, and divided in bins of 100 genes. On the left, percentage of retinal cell-specific markers counted per bin. For “Other neuronal” we represent the average of counts from amacrine, bipolar and retinal ganglion cells, for “Macroglia” the averaged counts from Müller glia and astrocytes, and for “Other non-neuronal” the averaged counts from pericytes and perivascular fibroblasts. On the right, the same data were represented as grouped percentages in a bar graph, besides the genes affected by aberrant alternative splicing (diffAS).
To explore in further detail the retinal transcriptional profile of the R6/1 strain, we examined the cell types that were the most affected according to our differential gene expression analysis. To this end, we used the markers obtained from a scRNA-seq analysis performed in the mouse retina that were assigned to specific neuronal and nonneuronal cells30 (see “Materials and methods” for further details). The presence of these markers among the DEGs inferred the prominent alterations of photoreceptor cells because specific genes of cones (e.g., Opn1sw, Arr3) and rods (e.g., Rho, Gnat1) were the most downregulated genes in R6/1 retinas compared to wt littermates, followed by the upregulation of microglia (e.g., Trf, H2-K1, Ctss) and macroglia markers (e.g., Gfap) (Fig. 1G), in agreement with the GO enrichment of innate immune response in Fig. 1D. Aberrant splicing was less selective but still showed a trend towards photoreceptor cells (Fig. 1G).
Progressive molecular alterations in the R6/1 retina and striatum
To establish the time-frame of molecular impairments in the R6/1 retinas, we further analysed the transcript levels of photoreceptors and gliosis markers together with other down- and upregulated genes at different stages of the pathology: prodromal (7 weeks old), symptomatic (13–15 weeks old) and advanced symptomatic (25–28 weeks old). In general, downregulation preceded upregulation in the R6/1 retina; nonetheless, all examined genes were significantly altered in the intermediate age of sampling (Fig. 2A,B). A similar behaviour was observed in the R6/1 striatum, except for the lack of upregulation of the gliosis-related genes A2m and Gfap compared to wt striatum (Fig. 2C,D), confirming previous results from our laboratory24 at the timings of sampling.
Time-course profiling of molecular alterations in the R6/1 retina and striatum. (A,B) Time-course analysis of the downregulation (A) and upregulation (B) of selected genes in the R6/1 retina compared to wild-type littermates. 7 weeks, n = 6 (wt) and n = 8 (R6/1); 13–15 weeks, n = 5 (wt) and n = 7 (R6/1); 25–28 weeks, n = 3 per genotype. (C,D) The same analysis in the R6/1 and wild-type striata. 7 weeks, n = 7 (wt) and n = 8 (R6/1); 13–15 weeks, n = 7 (wt) and n = 9 (R6/1); 25–28 weeks, n = 8 per genotype. Data are expressed as mean ± s.e.m. *p < 0.05, **p < 0.005, genotype effect; §p < 0.05, §§p < 0.005, age effect; #p < 0.05, ##p < 0.005, interaction effect; ~p < 0.1 in any effect, ANOVA test. (E,F) Protein extracts from the retina (E) and striatum (F) of R6/1 and wt littermates were analyzed by western blotting assays with antibodies against LC3II/I and α-tubulin as loading control. Retina early, 7/9/15-week-old, n = 4 (wt) and n = 6 (R6/1); Retina late, 21/25-week-old, n = 3 (wt) and n = 4 (R6/1); striatum early, 5/13-week-old, n = 6 (wt) and n = 9 (R6/1); striatum late, 25-week-old, n = 4 (wt) and n = 5 (R6/1). Data are expressed as mean ± s.e.m. *p < 0.05 genotype effect; §p < 0.05 age effect; #p < 0.05 interaction; ~p < 0.1 in any effect, ANOVA test in R6/1 and wt littermates. More blots are shown in Supplementary Fig. S3.
On the basis that autophagy is known to be altered in HD51 and is involved in inflammation resolution due to its clearance role52,53, we compared the dysfunctions in autophagy processes between the retina and the striatum of R6/1 mice and their wt littermates. To this end, we analysed by western blotting assays the conversion of the cytosolic form of LC3 (LC3-I) into the LC3-phosphatidylethanolamine conjugate (LC3-II), which is recruited to autophagosomal membranes, as a marker of autophagy activation. Based on their similarity, we grouped the results for R6/1 at early (5–15 weeks old) and advance stages (21–25 weeks old). We detected a significant increase in the LC3-II / LC3-I ratio in the retinas from early R6/1 mice compared with age-matched wt mice in response to the mHTT insult, but the LC3-II / LC3-I ratio was later similar in advanced R6/1 mice (Fig. 2E and Supplementary Fig. S3A), indicating that autophagic influx was delayed54. The inability to activate the autophagic influx in the mutant striatum was already observed in early stages, leading to the accumulation of LC3-I in advanced stages (Fig. 2F and Supplementary Fig. S3B) in agreement with other reports55,56. After inspecting the RNA-seq results, we confirmed the significant deregulation of several components of the autophagic system in nearly all the categories summarized in Ref.37 in both the retina and the striatum of 13–15 weeks-old R6/1 mice, although the most notable difference was observed in genes involved in lysosomal biogenesis that were significantly upregulated in the R6/1 retina in comparison with the striatum of the same mice (Supplementary Fig. S4).
Overall, these results indicated that the progression of the neuronal alterations was similar between the retina and striatum of the R6/1 mice whereas neuroinflammation and autophagy was differentially impaired between both tissues.
R6/1 retinas showed glial activation
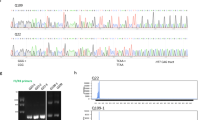
Our previous analyses suggested that local inflammatory processes took place in the retina, but not in the striatum, of symptomatic R6/1 mice, in agreement with a TFBS prediction in which DNA motifs for inflammatory transcription factors (i.e., IRFs and STATs)57 were highly enriched in the genes that were upregulated in the R6/1 retina (Supplementary Fig. S2C). This retina-accelerated neuroinflammation was not due to either a higher CAG instability of the transgene (Supplementary Fig. S5A) or higher expression of the R6/1 transgene in the mutant retina related to the striatum (Supplementary Fig. S5B), which might exacerbate the progression of the pathology in the former tissue.
Since retinal markers used in Fig. 1G were retrieved in the basal state, this analysis did not differentiate the activation states of resident glial cells. To determine whether the retinal neuroinflammation was associated with glial activation, we used external datasets to obtain the most significant genes (top250) in the pairwise comparison between isolated astrocytes (Aldh1l1+) from control and injured mice (combining LPS injection and middle cerebral artery occlusion32): whereas the upregulation signature was linked to astrocytic activation and gliosis, downregulated genes indicated overexpression in resting astrocytes; these markers might be shared between retinal astrocytes and Müller glia as they trigger similar molecular changes after injury58. In a similar manner, we also obtained the top250 genes from isolated microglia (CD11b+-CD45+) at different stages of neurodegeneration triggered by p25 induction; thus, we retrieved the signatures for homeostatic/basal, early and late activated microglia under ongoing neurodegenerative processes35. We identified examples of all sets of genes related to basal and activated glia to be significantly upregulated in the R6/1 retina compared to wt littermates, suggesting the coexistence of different stages of glial activation (Fig. 3A, top panels) in HD progression. In contrast, the R6/1 striatum did not show any increase in glial markers (Fig. 3A, bottom panels). To gain further insights regarding the type of glial activation, we examined the changes occurring in markers ascribed to two recently described subtypes of reactive astrocytes: neurotoxic A1 and neuroprotective A236. We found in the retina of R6/1 mice the activation of 25% of A1-astrocytic and 38.5% pan-astrocytic markers, indicating that the detected gliosis was associated with an inflammatory deleterious response (Fig. 3B).
A glial activation signature is only present in the R6/1 retina. (A) Plots showing the upregulated genes in 13–15 weeks-old R6/1 retina (top panels) and striatum (bottom panels) according to magnitude (log2 fold change) and significance (− log adj. p-value) of change in the RNA-seq analysis that are markers for astrocytes (left panels) and microglia (right panels) at different stages of activation (see text for further details). For striatal genes no significance cut-off was applied to permit the analysis of the same number of genes as in retina to facilitate the comparison between both tissues. (B) Heatmap plot of the fold changes of DEGs belonging to the A1, A2 and panastrocytic signatures (see text). ¶, adjusted p-value < 0.05 from the RNA-seq analysis. *p-value < 0.05, Student’s t-test between the fold changes means of both tissues.
Retinal neuroinflammation was apparently restricted to glial cells since additional inflammatory markers were not induced in our RNA-seq data. For example, most adaptive immune markers for T- and B cells were not detected in the retinal transcriptomes or were expressed at low levels, independently of the genotype (Supplementary Fig. S6A). In addition, other genes associated with inflammation were also lowly expressed (such as the classical proinflammatory cytokines Il1b, Il6 and Tnf) or did not show differences between genotypes (such as the inducible heme oxygenase by mitochondrial dysfunction Hmox1), as confirmed by RT-qPCR assays in independent samples (Supplementary Fig. S6B).
Because our gene expression analyses were performed using bulk homogenates, we investigated the expression of GFAP by immunofluorescence to obtain spatial sensitivity of Müller cells and astrocytes activation (Fig. 4A,B). Reactive GFAP was detected in retinal sections from R6/1 mice beginning at seven weeks of age with increased labelling in later time points, first in the feet of the ganglion cell layer (GCL), and later extended into the inner nuclear (INL) and inner plexiform layers (IPL) as glial reactivity increased in advance stages (Fig. 4A). We also analysed the microglial activation, as a potential contributor in retinal neuroinflammation detected in R6/1 mice, by determining the time-course staining pattern of the specific reactive microglial marker Iba-1. Immunopositive Iba-1 cells were detected mainly in both plexiform retinal layers of the HD mouse model (Fig. 4C). We did not observed a significant increase in the total number of microglial cells during pathology progression but, interestingly, there was a prominent shift towards an amoeboid-migrating phenotype detriment to ramified-resting morphology in the retinas of 25-week-old R6/1 mice, indicating ongoing progression in microglial activation (Fig. 4C) associated to neuroinflammatory responses.
Morphological alterations associated with neuroinflammation in the R6/1 retinas. (A) Representative immunofluorescence stainings across the indicated time points of the gliosis marker GFAP in R6/1 and wt littermates. (B) Representative whole mounts of R6/1 and wt retinas with GFAP staining. (C) Upper panels, representative immunofluorescence staining of the microglia marker Iba-1 for R6/1 and wt littermates. Lower panel, quantification of the Iba-1+ cells distinguishing their morphology in ramified and amoeboid in R6/1 retinas compared to wt littermates. N = 2–4 per genotype, age and model; n = 3–5 slices per animal, n = 3 fields per slice; field area = 84,100 μm2. Data are expressed as mean ± SD. *p-value < 0.05, Student’s t-test between Iba-1+ cell subtypes. Green, glial marker; blue, DAPI staining. Scale = 20 μm. ONL outer nuclear layer, OPL outer plexiform layer, INL inner nuclear layer, IPL inner plexiform layer, GCL ganglion cell layer, Ramif ramified, Amoeb amoeboid. (D) Representative SD-OCT images from the fundus of a R6/1 mouse and wt littermates. Red inset, magnified image indicating hyperreactive spots (*) and thicknesses of INL and ONL (yellow and green lines, respectively). Arrows denote the B-scanned transects for thickness calculations. Scale bars = 100 μm. (E) Quantifications of the retinal thickness in 25-week-old R6/1 (n = 5) and matched-age wt (n = 5). Data are shown as mean ± SD. *p < 0.05; **p < 0.005, Student’s t-test between genotypes. (F) Quantification of DAPI-stained cells in the retinal ONL of mutant mice compared to wt across different time points, N = 3–4 for each genotype, n = 2–5 slices per animal. Data are shown as mean ± SD. **p < 0.005 genotype effect; §§p < 0.005 age effect; ##p < 0.005 interaction effect from ANOVA test in R6/1 and wt littermates.
These results, together with those retrieved from the gene expression analysis (Fig. 2,3), determined that older R6/1 mice showed an increased retinal neuroinflammation. This is in agreement with the presence of aberrant fundus images due to the presence of bright white or hyperreflective spots, as already reported20, which were indicative of inflammatory processes59,60,61 (Fig. 4D). This finding was accompanied by a reduction in the total retinal thickness in R6/1 mice (Fig. 4E), and a decrease in the number of nuclei compared to matched-age controls (Fig. 4F). In addition, the localization of activated glial cells identified the OPL as the most damaged layer, potentially explaining the loss of synaptic connection between the ONL and the INL, as determined by ERG recordings (Supplementary Fig. S1B).
The zQ175 strain also shows retinal neuroinflammation
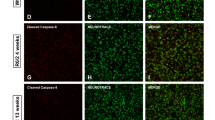
The R6/1 strain is a rapidly progressive model that may exhibit extreme pathological phenotypical traits. To demonstrate that retinal inflammation is not a particularity of this transgenic model, we extended our analysis to heterozygous zQ175 KI mice, which better resemble the pathology in humans since the disease progression is much slower and comprises the loss-of-function component23. We demonstrated a significant downregulation of neuronal genes in both tissues but a specific induction of gliosis-related markers in the zQ175 retina, in which changes in 12-month-old mice were comparable to those occurring in R6/1 mice 13–15 weeks old (Fig. 5A,B); this induction was corroborated in immunohistochemistry assays against GFAP (Fig. 5C). In addition, microglia also exhibited a prominent amoeboid phenotype at the same age (Fig. 5D), together with aberrant hyperreactive spots in the retinal fundus (Fig. 5E) and a remarkable thinning (Fig. 5F) and nuclei loss (Fig. 5G) of the ONL, where the cell bodies of photoreceptors reside, as observed in the R6/1 retinas. Overall, both symptomatic zQ175 and R6/1 mice developed a similar behaviour of inflammatory markers in their retinas and striata, suggesting that our observations can be generalized to other HD mouse models.
Retinal neuroinflammation is also detected in the zQ175 retinas. (A,B) The same analysis as in Fig. 2A and B for zQ175 and wt littermates. At the early time point (7 months) we included homozygous mice for the CAG expansion to check whether there was a possible exacerbation of the transcriptional dysregulation that was only observed for downregulated striatal genes. 7 months in each tissue, n = 5 (wt), n = 4 (zQ175+/−) and n = 4 (zQ175+/+); 12 months in striatum, n = 6 per genotype; in retina, n = 5 (wt) and n = 7 (zQ175+/−). Data are expressed as mean ± s.e.m. *p < 0.05, **p < 0.005, genotype effect; §p < 0.05, §§p < 0.005, age effect; ##p < 0.005, interaction effect; ~ , p < 0.1 in any effect, ANOVA test. (C) Representative whole mounts of zQ175+/− and wt retinas with GFAP staining. (D) The same analysis as in Fig. 4C for zQ175 and wt littermates. N = 2 per genotype, age and model; n = 3–5 slices per animal, n = 3 fields per slice; field area = 84,100 μm2. Data are expressed as mean ± SD.*, p-value < 0.05, Student’s t-test between Iba-1+ cell subtypes. Green, microglial marker; blue, DAPI staining. Scale = 20 μm. ONL outer nuclear layer, OPL outer plexiform layer, INL inner nuclear layer, IPL inner plexiform layer; GCL ganglion cell layer, Ramif ramified, Amoeb amoeboid. (E) Representative SD-OCT images from the fundus of a zQ175 mouse and wt littermate. Red inset, magnified image indicating hyperreactive spots (*) and thicknesses of INL and ONL (yellow and green lines, respectively). Arrows denote the B-scanned transects for thickness calculations. Scale bars = 100 μm. (F) Quantifications of the retinal thickness in 12-month-old zQ175 (n = 2) and matched-age wt (n = 4). Data are shown as mean ± SD. *p < 0.05; **p < 0.005, Student’s t-test between genotypes. (G) Quantification of DAPI-stained cells in the retinal ONL of 12-month-old mutant mice compared to wt. N = 3 for each genotype, n = 2–4 slices per animal. Data are shown as mean ± SD. **p < 0.005, Student’s t-test between wt and zQ175 mice.
Discussion
The present work defines for the first time the correlation between the molecular alterations in the retina and striatum in HD mouse models. Retinopathy in HD has been described as a late event in the pathology, based on the normal fundus and minimal ERG anomalies in young R6/1 animals17,20. However, degeneration of specific cell types and altered retinal responses to light can occur in the R6/2 model prior to the onset of motor impairment and body weight loss18,19. Another polyQ disorder caused by an aberrant expansion of CAG triplets in the ATXN7 gene, spinocerebellar ataxia 7 (SCA7), is characterized by retinal degeneration and dystrophy, leading to visual anomalies preceding motor symptoms62. HD and SCA7 animal models display common features of retinal anomalies at the level of morphology, light response and gene expression20,63,64, retinal dysfunction may share similar progression in both disorders and might manifest in early symptomatology. Notably, our gene expression analysis in the KI zQ175 strain suggested that photoreceptor genes can be affected prior to striatal-specific genes (Fig. 5).
Although gliosis has been previously reported in the R6/1 retina17, we provided for the first time evidence of microglial activation in the retina of two different HD models as a marker of neuroinflammation. Microglia comprise the resident phagocyte population in the CNS that can have both protective and deleterious effects, with anti-inflammatory and repair responses during the early phase of neurodegeneration but with harmful effects on neurons in prolonged activation in chronic inflammation65,66,67,68. In our RNA-seq analysis, we were able to detect different states of microglial activation, consistent with the hyperreactivity reported in microglia derived from pluripotent stem cells and blood-isolated myeloid cells of HD patients69,70. Reactive microglia induce the proinflammatory A1 astrocyte subtype, which is found in patients’ brains affected by different neurodegenerative disorders including HD36. In the R6/1 retina, we observed the induction of some of these astrocytic-related markers that mostly occurred in Müller cells, considering the GFAP staining pattern observed in the HD retinas. In any case, glial diversity falls beyond the dichotomous classification in A1 and A2, as reported in patients71.
Our striatal results were in agreement with the absence of overt neuroinflammation in HD models24,72. However, in HD patients activated glia and induction of inflammatory markers are well detected in cortical and basal ganglia regions43,73,74. This discrepancy is exemplified in a recent study that compared the gene expression profiles of murine R6/1 and zQ175 striata with those of human caudate nuclei from HD patients at a single nucleus resolution75. The induction of pan- and A1-astrocytic markers in human HD samples was reminiscent of our own results in the retina of R6/1 mice, suggesting that mHTT-expressing retinas might better reproduce the conditions of HD-associated neuroinflammatory processes, or at least might serve as experimental settings to study the HD-associated neuroinflammation thanks to the acceleration of these processes. Tightly connected to inflammatory processes, autophagy delivers aberrant organelles and macromolecules in double-membrane vesicles to lysosomes for degradation and recycling54. Deficits in this process can trigger neuroinflammation and cellular damage76 by modulating the functionality of immune and glial cells77. In HD autophagy becomes severely disrupted at several steps (e.g., cargo recognition, autophagosome formation, maturation and fusion to lysosomes), not only due to the toxic effects of the polyQ peptide but also because of the reduced regulatory activity of physiological HTT over autophagy, affecting the clearance of mHTT and contributing to its accumulation in cells51. We confirmed that autophagy was altered in the retina and striatum of R6/1 mice, contributing to the worsening of the disease, but apparently showed different rates of autophagosome accumulation, in which autophagic influx was stopped earlier in the R6/1 striatum than in the retina. The latter was transiently able to respond to mHTT. Whether the upregulation of lysosomal components (including the microglial cathepsin Ctss) is part of a compensatory mechanism for the HD-associated deficiency inautophagydeserves further exploration.
Based on our results, we propose that the retinas of HD mouse models can serve as a model to analyse HD-linked neuroinflammation, allowing for the use of ex vivo retina cultures from HD mouse models to elucidate mechanisms of neurodegeneration and to evaluate potential therapeutic approaches78, as research models that are evolutionarily closer than HD Drosophila ommatidia79,80,81,82.
In conclusion, the retinas of HD mouse models show profound morphological and functional abnormalities that are accompanied by a dramatic transcriptional dysregulation and glial activation, suggesting that retinal dysfunction may be of higher relevance in HD than envisaged. Studying this activation can provide novel insights regarding the role of macroglia and microglia during HD neuroinflammation.
Data availability
The RNA-seq data can be downloaded from the Gene Expression Omnibus (GEO) database using the accession number GSE216520.
Abbreviations
- A2m :
-
Alpha-2-macroglobulin
- Actb :
-
Actin beta
- Arr3 :
-
Arrestin 3
- cAMP:
-
Cyclic adenosine monophosphate
- CNS:
-
Central nervous system
- Ctss :
-
Cathepsin S
- DAPI:
-
4’-6-Diamidino-2-phenylindole
- DEG:
-
Differentially expressed gene
- ERG:
-
Electroretinogram
- FDR:
-
False discovery rate
- Gapdh :
-
Glyceraldehyde-3-phosphate dehydrogenase
- GCL:
-
Ganglion cell layer
- GFAP:
-
Glial fibrillary acidic protein
- Gnat1 :
-
G-Protein subunit alpha transducin 1
- H2-K1 :
-
H-2 class I histocompatibility antigen, K-B alpha chain
- HD:
-
Huntington’s disease
- Hmox1 :
-
Heme oxygenase 1
- HTT:
-
Huntingtin
- Iba-1:
-
Ionized calcium binding adaptor molecule 1
- Il1b :
-
Interleukin 1b
- Il6 :
-
Interleukin 6
- INL:
-
Inner nuclear layer
- IPL:
-
Inner plexiform layer
- IRF:
-
Interferon regulator factor
- KI:
-
Knock-in
- ONL:
-
Outer nuclear layer
- OPL:
-
Outer plexiform layer
- Opn1sw :
-
Opsin 1, short wave sensitive
- Rho :
-
Rhodopsin
- RNFL:
-
Retinal nerve fibre layer
- RT-qPCR:
-
Retrotranscription and quantitative PCR
- scRNA-seq:
-
Single cell RNA sequencing
- SD-OCT:
-
Spectral domain optical coherence tomography
- STAT:
-
Signal transducer and activator of transcription
- Tbp :
-
TATA-box binding protein
- TFBS:
-
Transcription factor binding site
- Tnf :
-
Tumor necrosis factor
- Trf :
-
Transferrin
- UHDRS:
-
Unified Huntington’s disease rating scale
- wt:
-
Wild-type
References
Rawlins, M. D. et al. The prevalence of Huntington’s disease. Neuroepidemiology 46, 144–153. https://doi.org/10.1159/000443738 (2016).
Bates, G., Harper, P. & Jones, L. in Oxford Monographs on Medical Genetics (2004).
Vonsattel, J. P., Keller, C. & Cortes Ramirez, E. P. Huntington’s disease—Neuropathology. Handb. Clin. Neurol. 100, 83–100. https://doi.org/10.1016/B978-0-444-52014-2.00004-5 (2011).
Glikmann-Johnston, Y., Carmichael, A. M., Mercieca, E. C. & Stout, J. C. “Real-life” hippocampal-dependent spatial memory impairments in Huntington’s disease. Cortex 119, 46–60. https://doi.org/10.1016/j.cortex.2019.04.006 (2019).
Gomez-Jaramillo, L. et al. A new perspective on Huntington’s disease: How a neurological disorder influences the peripheral tissues. Int. J. Mol. Sci. 23, 6089. https://doi.org/10.3390/ijms23116089 (2022).
Padron-Rivera, G. et al. Cerebellar degeneration signature in Huntington’s disease. Cerebellum 20, 942–945. https://doi.org/10.1007/s12311-021-01256-5 (2021).
van Wamelen, D. J. & Aziz, N. A. Hypothalamic pathology in Huntington disease. Handb. Clin. Neurol. 182, 245–255. https://doi.org/10.1016/B978-0-12-819973-2.00017-4 (2021).
van der Burg, J. M., Bjorkqvist, M. & Brundin, P. Beyond the brain: Widespread pathology in Huntington’s disease. Lancet Neurol. 8, 765–774. https://doi.org/10.1016/S1474-4422(09)70178-4 (2009).
Byrne, L. M. & Wild, E. J. Cerebrospinal fluid biomarkers for Huntington’s disease. J. Huntingtons Dis. 5, 1–13. https://doi.org/10.3233/JHD-160196 (2016).
Mitchell, C. T. et al. Longitudinal expression changes are weak correlates of disease progression in Huntington’s disease. Brain Commun. 2, fcaa172. https://doi.org/10.1093/braincomms/fcaa172 (2020).
Marti-Martinez, S. & Valor, L. M. A glimpse of molecular biomarkers in Huntington’s disease. Int. J. Mol. Sci. 23, 5411. https://doi.org/10.3390/ijms23105411 (2022).
Demb, J. B. & Singer, J. H. Functional circuitry of the retina. Annu. Rev. Vis. Sci. 1, 263–289. https://doi.org/10.1146/annurev-vision-082114-035334 (2015).
Benfey, N., Foubert, D. & Ruthazer, E. S. Glia regulate the development, function, and plasticity of the visual system from retina to cortex. Front. Neural Circuits 16, 826664. https://doi.org/10.3389/fncir.2022.826664 (2022).
Christinaki, E. et al. Retinal imaging biomarkers of neurodegenerative diseases. Clin. Exp. Optom. 105, 194–204. https://doi.org/10.1080/08164622.2021.1984179 (2022).
Doustar, J., Torbati, T., Black, K. L., Koronyo, Y. & Koronyo-Hamaoui, M. Optical coherence tomography in Alzheimer’s disease and other neurodegenerative diseases. Front. Neurol. 8, 701. https://doi.org/10.3389/fneur.2017.00701 (2017).
Dhalla, A., Pallikadavath, S. & Hutchinson, C. V. Visual dysfunction in Huntington’s disease: A systematic review. J. Huntingtons Dis. 8, 233–242. https://doi.org/10.3233/JHD-180340 (2019).
Batcha, A. H. et al. Retinal dysfunction, photoreceptor protein dysregulation and neuronal remodelling in the R6/1 mouse model of Huntington’s disease. Neurobiol. Dis. 45, 887–896. https://doi.org/10.1016/j.nbd.2011.12.004 (2012).
Lin, M. S. et al. Degeneration of ipRGCs in mouse models of Huntington’s disease disrupts non-image-forming behaviors before motor impairment. J. Neurosci. 39, 1505–1524. https://doi.org/10.1523/JNEUROSCI.0571-18.2018 (2019).
Ragauskas, S. et al. Early retinal function deficit without prominent morphological changes in the R6/2 mouse model of Huntington’s disease. PLoS One 9, e113317. https://doi.org/10.1371/journal.pone.0113317 (2014).
Helmlinger, D. et al. Progressive retinal degeneration and dysfunction in R6 Huntington’s disease mice. Hum. Mol. Genet. 11, 3351–3359. https://doi.org/10.1093/hmg/11.26.3351 (2002).
Li, M. et al. Intravitreal administration of HA-1077, a ROCK inhibitor, improves retinal function in a mouse model of huntington disease. PLoS One 8, e56026. https://doi.org/10.1371/journal.pone.0056026 (2013).
Mangiarini, L. et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493–506. https://doi.org/10.1016/s0092-8674(00)81369-0 (1996).
Menalled, L. B. et al. Comprehensive behavioral and molecular characterization of a new knock-in mouse model of Huntington’s disease: zQ175. PLoS One 7, e49838. https://doi.org/10.1371/journal.pone.0049838 (2012).
Gallardo-Orihuela, A. et al. Transcriptional correlates of the pathological phenotype in a Huntington’s disease mouse model. Sci. Rep. 9, 18696. https://doi.org/10.1038/s41598-019-55177-9 (2019).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. https://doi.org/10.1038/nmeth.4197 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Tapial, J. et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 27, 1759–1768. https://doi.org/10.1101/gr.220962.117 (2017).
Shen, S. et al. MATS: A Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. 40, e61. https://doi.org/10.1093/nar/gkr1291 (2012).
Trincado, J. L. et al. SUPPA2: Fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 19, 40. https://doi.org/10.1186/s13059-018-1417-1 (2018).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. https://doi.org/10.1016/j.cell.2015.05.002 (2015).
Fadl, B. R. et al. An optimized protocol for retina single-cell RNA sequencing. Mol. Vis. 26, 705–717 (2020).
Zamanian, J. L. et al. Genomic analysis of reactive astrogliosis. J. Neurosci. 32, 6391–6410. https://doi.org/10.1523/JNEUROSCI.6221-11.2012 (2012).
Gautier, L., Cope, L., Bolstad, B. M. & Irizarry, R. A. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315. https://doi.org/10.1093/bioinformatics/btg405 (2004).
Smyth, G. K., Michaud, J. & Scott, H. S. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21, 2067–2075. https://doi.org/10.1093/bioinformatics/bti270 (2005).
Mathys, H. et al. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 21, 366–380. https://doi.org/10.1016/j.celrep.2017.09.039 (2017).
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487. https://doi.org/10.1038/nature21029 (2017).
Bordi, M. et al. A gene toolbox for monitoring autophagy transcription. Cell Death Dis. 12, 1044. https://doi.org/10.1038/s41419-021-04121-9 (2021).
Sherman, B. T. et al. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221. https://doi.org/10.1093/nar/gkac194 (2022).
Zambelli, F., Pesole, G. & Pavesi, G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 37, W247-252. https://doi.org/10.1093/nar/gkp464 (2009).
Barhoum, R. et al. Functional and structural modifications during retinal degeneration in the rd10 mouse. Neuroscience 155, 698–713. https://doi.org/10.1016/j.neuroscience.2008.06.042 (2008).
Yamauchi, K. et al. The spectral-domain optical coherence tomography findings associated with the morphological and electrophysiological changes in a rat model of retinal degeneration, rhodopsin S334ter-4 rats. Biomed. Res. Int. 2018, 5174986. https://doi.org/10.1155/2018/5174986 (2018).
Langfelder, P. et al. Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat. Neurosci. 19, 623–633. https://doi.org/10.1038/nn.4256 (2016).
Hodges, A. et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet. 15, 965–977. https://doi.org/10.1093/hmg/ddl013 (2006).
Hervas-Corpion, I. et al. Early alteration of epigenetic-related transcription in Huntington’s disease mouse models. Sci. Rep. 8, 9925. https://doi.org/10.1038/s41598-018-28185-4 (2018).
Alcala-Vida, R., Awada, A., Boutillier, A. L. & Merienne, K. Epigenetic mechanisms underlying enhancer modulation of neuronal identity, neuronal activity and neurodegeneration. Neurobiol. Dis. 147, 105155. https://doi.org/10.1016/j.nbd.2020.105155 (2021).
Jasinska, M. & Pyza, E. Circadian plasticity of mammalian inhibitory interneurons. Neural Plast. 2017, 6373412. https://doi.org/10.1155/2017/6373412 (2017).
Korshunov, K. S., Blakemore, L. J. & Trombley, P. Q. Dopamine: A modulator of circadian rhythms in the central nervous system. Front. Cell Neurosci. 11, 91. https://doi.org/10.3389/fncel.2017.00091 (2017).
Colwell, C. S. Defining circadian disruption in neurodegenerative disorders. J. Clin. Investig. 131, e148288. https://doi.org/10.1172/JCI148288 (2021).
Krach, F. et al. An alternative splicing modulator decreases mutant HTT and improves the molecular fingerprint in Huntington’s disease patient neurons. Nat. Commun. 13, 6797. https://doi.org/10.1038/s41467-022-34419-x (2022).
Elorza, A. et al. Huntington’s disease-specific mis-splicing unveils key effector genes and altered splicing factors. Brain 144, 2009–2023. https://doi.org/10.1093/brain/awab087 (2021).
Martin, D. D., Ladha, S., Ehrnhoefer, D. E. & Hayden, M. R. Autophagy in Huntington disease and huntingtin in autophagy. Trends Neurosci. 38, 26–35. https://doi.org/10.1016/j.tins.2014.09.003 (2015).
Harris, J. Autophagy and cytokines. Cytokine 56, 140–144. https://doi.org/10.1016/j.cyto.2011.08.022 (2011).
Matsuzawa-Ishimoto, Y., Hwang, S. & Cadwell, K. Autophagy and Inflammation. Annu. Rev. Immunol. 36, 73–101. https://doi.org/10.1146/annurev-immunol-042617-053253 (2018).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. https://doi.org/10.1080/15548627.2015.1100356 (2016).
Ramirez-Jarquin, U. N., Sharma, M., Zhou, W., Shahani, N. & Subramaniam, S. Deletion of SUMO1 attenuates behavioral and anatomical deficits by regulating autophagic activities in Huntington disease. Proc. Natl. Acad. Sci. U. S. A. https://doi.org/10.1073/pnas.2107187119 (2022).
Bailus, B. J. et al. Modulating FKBP5/FKBP51 and autophagy lowers HTT (huntingtin) levels. Autophagy 17, 4119–4140. https://doi.org/10.1080/15548627.2021.1904489 (2021).
Platanitis, E. & Decker, T. Regulatory NETWORKS INVOLVING STATs, IRFs, and NFkappaB in inflammation. Front. Immunol. 9, 2542. https://doi.org/10.3389/fimmu.2018.02542 (2018).
Yoo, H. S., Shanmugalingam, U. & Smith, P. D. Harnessing astrocytes and muller glial cells in the retina for survival and regeneration of retinal ganglion cells. Cells 10, 1339. https://doi.org/10.3390/cells10061339 (2021).
Vujosevic, S. et al. Optical coherence tomography as retinal imaging biomarker of neuroinflammation/neurodegeneration in systemic disorders in adults and children. Eye (London) 37, 203–219. https://doi.org/10.1038/s41433-022-02056-9 (2022).
Vujosevic, S. et al. Subthreshold Micropulse Laser in Diabetic Macular Edema: 1-Year Improvement in OCT/OCT-Angiography Biomarkers. Transl Vis Sci Technol 9, 31. https://doi.org/10.1167/tvst.9.10.31 (2020).
Dannhausen, K. et al. Acid sphingomyelinase (aSMase) deficiency leads to abnormal microglia behavior and disturbed retinal function. Biochem. Biophys. Res. Commun. 464, 434–440. https://doi.org/10.1016/j.bbrc.2015.06.133 (2015).
La Spada, A. R. Spinocerebellar Ataxia Type 7. GeneReviews, NBK1256 [bookaccession] (1993).
Yvert, G. et al. Expanded polyglutamines induce neurodegeneration and trans-neuronal alterations in cerebellum and retina of SCA7 transgenic mice. Hum. Mol. Genet. 9, 2491–2506. https://doi.org/10.1093/hmg/9.17.2491 (2000).
Abou-Sleymane, G. et al. Polyglutamine expansion causes neurodegeneration by altering the neuronal differentiation program. Hum. Mol. Genet. 15, 691–703. https://doi.org/10.1093/hmg/ddi483 (2006).
Hanisch, U. K. Microglia as a source and target of cytokines. Glia 40, 140–155. https://doi.org/10.1002/glia.10161 (2002).
Streit, W. J. Microglia as neuroprotective, immunocompetent cells of the CNS. Glia 40, 133–139. https://doi.org/10.1002/glia.10154 (2002).
Graeber, M. B. & Streit, W. J. Microglia: Biology and pathology. Acta Neuropathol. 119, 89–105. https://doi.org/10.1007/s00401-009-0622-0 (2010).
Butovsky, O. & Weiner, H. L. Microglial signatures and their role in health and disease. Nat. Rev. Neurosci. 19, 622–635. https://doi.org/10.1038/s41583-018-0057-5 (2018).
O’Regan, G. C. et al. Human Huntington’s disease pluripotent stem cell-derived microglia develop normally but are abnormally hyper-reactive and release elevated levels of reactive oxygen species. J. Neuroinflamm. 18, 94. https://doi.org/10.1186/s12974-021-02147-6 (2021).
Miller, J. R. et al. RNA-Seq of Huntington’s disease patient myeloid cells reveals innate transcriptional dysregulation associated with proinflammatory pathway activation. Hum. Mol. Genet. 25, 2893–2904. https://doi.org/10.1093/hmg/ddw142 (2016).
Al-Dalahmah, O. et al. Single-nucleus RNA-seq identifies Huntington disease astrocyte states. Acta Neuropathol. Commun. 8, 19. https://doi.org/10.1186/s40478-020-0880-6 (2020).
Francelle, L., Lotz, C., Outeiro, T., Brouillet, E. & Merienne, K. Contribution of neuroepigenetics to Huntington’s disease. Front. Hum. Neurosci. 11, 17. https://doi.org/10.3389/fnhum.2017.00017 (2017).
Sapp, E. et al. Early and progressive accumulation of reactive microglia in the Huntington disease brain. J. Neuropathol. Exp. Neurol. 60, 161–172. https://doi.org/10.1093/jnen/60.2.161 (2001).
Labadorf, A. et al. RNA sequence analysis of human huntington disease brain reveals an extensive increase in inflammatory and developmental gene expression. PLoS One 10, e0143563. https://doi.org/10.1371/journal.pone.0143563 (2015).
Lee, H. et al. Cell type-specific transcriptomics reveals that mutant huntingtin leads to mitochondrial RNA release and neuronal innate immune activation. Neuron 107, 891-908 e898. https://doi.org/10.1016/j.neuron.2020.06.021 (2020).
Harris, H. & Rubinsztein, D. C. Control of autophagy as a therapy for neurodegenerative disease. Nat. Rev. Neurol. 8, 108–117. https://doi.org/10.1038/nrneurol.2011.200 (2011).
Cho, M. H. et al. Autophagy in microglia degrades extracellular beta-amyloid fibrils and regulates the NLRP3 inflammasome. Autophagy 10, 1761–1775. https://doi.org/10.4161/auto.29647 (2014).
Schnichels, S. et al. Retinal organ cultures as alternative research models. Altern. Lab. Anim. 47, 19–29. https://doi.org/10.1177/0261192919840092 (2019).
Steffan, J. S. et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413, 739–743. https://doi.org/10.1038/35099568 (2001).
Joshi, A. S., Singh, V., Gahane, A. & Thakur, A. K. Biodegradable nanoparticles containing mechanism based peptide inhibitors reduce polyglutamine aggregation in cell models and alleviate motor symptoms in a drosophila model of Huntington’s disease. ACS Chem. Neurosci. 10, 1603–1614. https://doi.org/10.1021/acschemneuro.8b00545 (2019).
Zhu, Y. et al. Nmnat restores neuronal integrity by neutralizing mutant Huntingtin aggregate-induced progressive toxicity. Proc. Natl. Acad. Sci. U. S. A. 116, 19165–19175. https://doi.org/10.1073/pnas.1904563116 (2019).
Karpuj, M. V. et al. Prolonged survival and decreased abnormal movements in transgenic model of Huntington disease, with administration of the transglutaminase inhibitor cystamine. Nat. Med. 8, 143–149. https://doi.org/10.1038/nm0202-143 (2002).
Acknowledgements
We thank Rocío Pérez-González for critical reading of the manuscript and José J. Lucas for providing mice to establish our zQ175 colony.
Funding
L.M.V. research is supported by Programa Estatal de Generación de Conocimiento y Fortalecimiento del Sistema Español de I + D + i and financed by Instituto de Salud Carlos III and Fondo Europeo de Desarrollo Regional 2014–2020 (grants PI16/00722 and PI19/00125), Ayuda a la Investigación 2021 financed by Fundación Navarro-Tripodi, Ayudas para el Apoyo y Fomento de la Investigación financed by ISABIAL (grants 2021-0406 and 2022-0338). L.M.V. is the recipient of a Miguel Servet II contract (CPII20/00025), financed by Instituto de Salud Carlos III and Fondo Social Europeo 2014–2020, Programa Estatal de Promoción del Talento y su empleabilidad en I + D + i. A.I.A. research is supported by Programa Estatal de Generación de Conocimiento y Fortalecimiento del Sistema Español de I + D + i and financed by Instituto de Salud Carlos III and Fondo Europeo de Desarrollo Regional 2014–2020 (grant PI18/01287), Consejería de Salud de la Junta de Andalucía (grant PI-0123-2018), Consejeria de Universidad, Investigación e Innovación de la Junta de Andalucía (grant PI-01331-2020) and Convocatoria de Subvenciones para la Financiación de la Investigación y la Innovación Biomédica y en Ciencias de la Salud en el Marco de la Iniciativa Territorial Integrada 2014–2020 para la Provincia de Cádiz, Fondos ITI-FEDER (PI-0012-2019). A.I.A is the recipient of a Nicolás Monardes contract financed by Consejería de Salud, Programa de Excelencia de la Junta de Andalucía. Funding sources had no involvement in study design, in the collection, analysis and interpretation of data, in the writing of the report, and in the decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
Conceptualization, Funding acquisition, Project administration, Resources and Supervision: L.M.V. and A.I.A. Data curation, Formal analysis and Software: L.M.V., S.O.-M. and F.C.-C. Investigation: F.C.-C., F.M.-L., A.G.-O., M.C.G.-M., I.H.-C., P.V., L.R.-M., J.N.-C., L.G.-J., A.I.A. and L.M.V. Methodology: A.I.A., L.M.V. and P.V. Software: F.C.-C., S.O.-M. and L.M.V. Validation: F.M.-L., M.C.G.-M. and L.R.-M. Visualization: L.M.V., F.M.-L. and A.I.A. Writing—original draft: L.M.V. Writing—review & editing: A.I.A., M.C.G.-M., F.C.-C., F.M.-L., I.H.-C., A.G.-O., L.G.-J. and L.M.V.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cano-Cano, F., Martín-Loro, F., Gallardo-Orihuela, A. et al. Retinal dysfunction in Huntington’s disease mouse models concurs with local gliosis and microglia activation. Sci Rep 14, 4176 (2024). https://doi.org/10.1038/s41598-024-54347-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-54347-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.