Abstract
The Phenome-Wide Association Study (PheWAS) is increasingly used to broadly screen for potential treatment effects, e.g., IL6R variant as a proxy for IL6R antagonists. This approach offers an opportunity to address the limited power in clinical trials to study differential treatment effects across patient subgroups. However, limited methods exist to efficiently test for differences across subgroups in the thousands of multiple comparisons generated as part of a PheWAS. In this study, we developed an approach that maximizes the power to test for heterogeneous genotype–phenotype associations and applied this approach to an IL6R PheWAS among individuals of African (AFR) and European (EUR) ancestries. We identified 29 traits with differences in IL6R variant-phenotype associations, including a lower risk of type 2 diabetes in AFR (OR 0.96) vs EUR (OR 1.0, p-value for heterogeneity = 8.5 × 10–3), and higher white blood cell count (p-value for heterogeneity = 8.5 × 10–131). These data suggest a more salutary effect of IL6R blockade for T2D among individuals of AFR vs EUR ancestry and provide data to inform ongoing clinical trials targeting IL6 for an expanding number of conditions. Moreover, the method to test for heterogeneity of associations can be applied broadly to other large-scale genotype–phenotype screens in diverse populations.
Similar content being viewed by others
Introduction
Large-scale biobanks linked to electronic health records (EHR) offer a promising approach to screen for potential treatment effects1,2. In some cases, genetic variants are linked with altered protein expression resulting in an effect similar to a treatment3. One example is a missense variant in IL6R Asp(258)Ala, known to reduce membrane-bound IL6R expression and thus dampen IL-6 signaling4. The targeted therapies, tocilizumab and sarilumab, block the IL6R pathway. In a phenotypic screen performed in a Phenome-Wide Association Study (PheWAS), subjects carrying the Asp(258)Ala variant were found to have a phenotypic profile similar to those on drugs that block IL-6R; subjects with the IL6R variant have higher hemoglobin and lower high sensitivity C-reactive protein (CRP) compared to those without the variant5,6. The PheWAS is a study design in which the association between single-nucleotide polymorphisms or other types of genomic variants are tested for association across a broad range of phenotypes7. Thus, population-based biobanks may also provide an opportunity to query potential effects of treatments using genetic variants across a more diverse population than clinical trials. While large amounts of data are now available, limited methods exist to efficiently test for potential heterogeneity across subpopulations in large scale screens such as the PheWAS.
To expand upon the use of biobanks in generating evidence on treatment effects, the PheWAS is a promising approach that can systematically test for associations between a functional genetic variant which mimics the effect of a pharmaceutical agent with a wide spectrum of phenotypes5,8,9. Analyses may be stratified by genetic population strata to study heterogenous genotype–phenotype associations which may inform differential treatment effects across populations. Since a large number of hypotheses are being tested at the same time, correction for multiple testing must be undertaken. Traditional methods of multiple testing require large sample sizes, especially when detecting heterogeneity, or group effects from relatively weak signals such as genetic associations. Prior studies have deployed a modified Benjamini–Hochberg procedure (BHq)10,11,12,13 on the high dimensional heterogeneity or group effect test statistics for selective inference with false discovery rate (FDR) control. In the presence of imbalanced sample sizes across the subgroups, the power of this strategy could be largely impacted by the small sample sizes of the minority groups.
Recent work in adaptive multiple testing enables researchers to construct auxiliary statistics14 to increase the power. In one approach, multiple testing of two-sample mean differences with a high dimensional sparse structure used the overall mean statistics as auxiliary information to boost the power15,16. In this study, we build upon a new method for false discovery rate (FDR) controlled heterogeneity testing (hetFDR) under a more complicated PheWAS setup with two imbalanced subgroups.
To demonstrate the utility of our proposed hetFDR approach in discovering heterogeneous signals that correspond to potential differential treatment effects, we performed hetFDR on results from a PheWAS with the interleukin-6 receptor (IL6R) single nucleotide polymorphism (SNP) (rs2228145, Asp(358)Ala). This variant was selected for several reasons. First, it has been previously studied as a model for IL6R blockade17,18. Second, the functional impact of this variant, reduced IL6R expression has also been described4,19 where subjects with this variant have lower CRP, higher hemoglobin, and higher levels of soluble IL6R, changes also observed in subjects who receive IL6R blockade17,20,21. As well, known therapies exist for control of inflammatory conditions such as rheumatoid arthritis and giant cell vasculitis. More recently, IL-6 blockade has been used for the treatment of hospitalized COVID-19 and with ongoing studies blocking the IL-6 pathway to reduce cardiovascular disease in the general population. Clinical trials remain the gold standard for studying treatment effects but have known limitations in generalizing results to a more diverse population.
The objective was to develop and apply an approach to systematically identify potential heterogeneous genotype–phenotype associations in African (AFR) compared to European (EUR) populations, the two largest ancestries in a diverse mega-biobank cohort, as part of an IL6R PheWAS. We hypothesize that this large-scale screen will identify differential effects of the IL6R variant across phenotypes with implications for current and future trials targeting the IL6 pathway. Findings were validated in two independent biobank cohorts.
Materials and methods
Study design
We performed an IL6R PheWAS in the Veterans Affairs Million Veteran Program (MVP) cohort with data up to 09/30/2020. The VA MVP is a longitudinal, multi-institutional cohort study that collects clinical electronic health record (EHR) data, namely inpatient and outpatient data combined with genomic data from participants in approximately 50 Veterans Affairs facilities across the United States. Subjects were included in the MVP if they were 18 years of age or older; had a valid mailing address (to ensure the possibility of follow-up); were able to provide informed consent at the time of recruitment. All participants were required to provide written informed consent upon recruitment. They were asked to (1) complete baseline and lifestyle questionnaires, providing information such as self-reported race/ethnicity, dietary habits, and smoking/drinking status, as well as (2) provide blood samples for genotyping and biomarker studies.
Statistical analysis
PheWAS analysis for each ancestry
The PheWAS analysis was performed using a standardized published approach22. Briefly, we fitted a logistic regression for PheWAS analysis to test for association with phenotypes as defined by PheCodes and linear regression for the laboratory analysis. Since many of the laboratory measurements were highly skewed, we tested for association of the IL6R variant with log-transformed laboratory values. All models were adjusted for patient age, sex, length of EHR follow up, and health care utilization as measured by the log-total number of PheCodes.
Genetic ancestry was ascertained using previously published methods. Briefly, we trained a logistic regression classification algorithm using self-reported race as silver standard labels and 127 ancestry informative SNPs23. The cut-off of predicted probabilities for classification is chosen to guarantee sensitivity is above 0.975. We excluded related MVP participants (halfway between second-degree and third-degree relatives or closer) as measured by the Kinship-Based Inference for GWAS software (https://www.kingrelatedness.com/)24. We stratified all association analyses of the IL6R variant, rs2228145 (minor allele C; Asp358Ala), with disease phenotypes and laboratory test results by the predicted ancestry group. We focused the analyses on the two largest ancestry groups in MVP, African (AFR) and European (EUR) ancestry.
Within each ancestry group, we performed PheWAS analyses including 1875 phenotypes as defined by PheCodes25 and 69 routine laboratory measurements curated in prior studies at the VA, which includes complete blood count and lipid profiles. For each phenotype, a participant was defined as having the condition if they had at least 2 PheCodes, which is often recommended to attain a higher positive predictive value26. We excluded PheCodes with a prevalence of 0.5% or less from the analysis and excluded integer level (parent) PheCodes for which corresponding descendant PheCodes already existed, leaving a total of 660 remaining phenotypes. For example, we excluded the integer PheCode 250 (Diabetes mellitus) but included the descendant PheCodes such as 250.1 (Type 1 Diabetes) and 250.2 (Type 2 Diabetes). The screen was also performed on 69 adjudicated laboratory measurements available at VA. Values were defined by the median of all available measurements for each patient. A detailed list of the laboratory tests is in the Supplementary Materials Table S6. We first compared associations between IL6R and PheCodes and separately for the curated laboratory values in AFR vs EUR. Significant PheCodes/labs within each ancestry were determined with a false discovery rate (FDR) < 0.1 using the Benjamini–Hochberg procedure (BHq)13.
Heterogeneity testing with FDR control
Heterogeneity testing was conducted to identify phenotypes and laboratory values to detect a differential association between IL6R and phenotype among AFR vs EUR ancestries. To adjust for multiple testing, we developed a novel false discovery rate (FDR) controlled heterogeneity testing (hetFDR) procedure which leverages information from both the mean effect and the magnitude of heterogeneity under a prior assumption that heterogeneous effects are more likely to be present for phenotypes with non-zero mean effects across a large number of candidate phenotypes. The hetFDR procedure is a three-step procedure. In Step (I), for each phenotype, we construct (i) an overall mean effect test statistic as an inverse-variance weighted average effect estimate combining the regression coefficients (against the genetic variant of interest) from the two ancestry groups along with its associated p value; as well as (ii) a chi-square test statistic ascertaining the heterogeneity between the effects as observed from the regression coefficients of the two groups. The mean effect statistic and the heterogeneity statistic are designed to be asymptotically independent so that the validity of tests is ensured when incorporating the mean effect statistics to assist the heterogeneity testing. In Step (II), we use the mean effect statistics to weight the heterogeneity p values, assigning higher prior probabilities of null hypothesis rejection to those phenotypes with more significant mean effects, which corresponds to our prior assumption that phenotypes with non-zero mean effects are more likely to show heterogeneity across the considered ancestry groups. The weighting function is decided adaptively from the data through a regression-based approach. In the final Step (III), we adopt the multiple testing procedure of27 on the weighted heterogeneity p-values for detection with FDR control.
Simulation results were conducted, and showed that under different settings of the sample sizes, the heterogeneous effect magnitude, and the number of heterogeneous effects, our proposed hetFDR method controls FDR below 0.1 and shows substantial and consistent higher average power than the existing BHq13 and Storey’s procedures28. For example, when the sample size of the minority group is 25% of the majority group and the number of phenotypes with heterogeneous effects is 10 out of the totally 50 active ones, our method attains 0.4 higher power than the BHq and Storey’s procedures. Such power gain is also achieved in other settings of different numbers or magnitudes of heterogeneous signals. This is because our method leverages the mean effect statistics as additional information and assigns a higher chance of rejection to the phenotypes with a non-zero mean effect. In addition, the power improvement of our method is more significant in the setting with imbalanced sample sizes between the two ancestry groups, compared to one with equal sample sizes. This is a consequence of having more informative mean effect statistics when one group is larger than the other. A detailed description for the statistical method of heterogeneity multiple testing and the simulation studies are provided in the Supplementary Materials: Statistical Methodology.
Replication of laboratory results using UK Biobank and MGB Biobank Data
Findings were replicated in UK Biobank (UKB) and the Mass General Brigham (MGB) Biobank2,29,30. The UKB is a longitudinal cohort study that prospectively recruits patients to determine the effects of lifestyle, environmental, and genomic factors on disease outcomes over time. The study population includes approximately 500,000 volunteers recruited from the United Kingdom’s general population from 2006 to 2010. Measurements of 61 laboratory biomarkers and blood cell counts were ascertained for all UKB participants as part of a standardized baseline assessment. The MGB Biobank contains linked EHR, and genetic data anchored by two large tertiary care hospitals: Brigham and Women’s Hospital and Massachusetts General Hospital in Boston. The MGB Biobank data consist of 59,052 participants with both EHR data and genomic data available. Laboratory test results were extracted for these patients.
To validate heterogeneous IL6R-phenotype associations in AFR vs EUR observed in MVP, we performed analyses in UKB and MGB Biobank data. Due to the relatively smaller size of AFR in these cohorts, the analyses focused on traits with continuous values, i.e., laboratory results.
This study obtained institutional review board approval through the Veterans Affairs MVP under Central IRB #16-06 with title: Cardiovascular Disease Risk Factors, Prevalent Cardiovascular Disease, and Genetics in the Million Veteran Program, and the Mass General Brigham Institutional Review Board. All experiments were performed in accordance with relevant guidelines and regulations. All analyses were performed using R software. The code for analyzing the data is available on GitHub, https://github.com/wx202/HeterTestIL6R.git.
Results
In the MVP cohort, a total of 545,147 Veterans were included in the analysis, of which 91.3% were male, with a mean (SD) age of 62.1 (13.9) years and a mean (SD) follow-up time of 12.5 (5.7) years. Among these participants, 105,838 were classified as AFR and 439,309 were classified as EUR. In this study, we controlled for an FDR of 10%, which ensures that among the associations considered significant, at most 10% of the associations were false positives13. The frequency of the rs2228145 allele in MVP was 14% in AFR and 40% in EUR, in the UKB cohort was 16% in AFR and 41% in EUR, and in the MGB cohort was 17% in AFR and 40% in EUR.
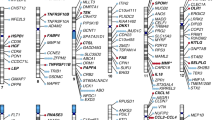
Overall, among phenotypes defined by PheCodes, we observed 10 with significant associations with IL6R among Veterans of AFR ancestry compared to 34 among Veterans of EUR ancestry, none of which were significant in both populations (Fig. 1). For laboratory measurements, we observed 30 measurements with significant associations with IL6R among Veterans of AFR ancestry compared to 28 among Veterans of EUR ancestry (Fig. 2). IL6R was significantly associated with 18 labs across both ancestries. As a positive control, based on prior knowledge of both the variant and the biologic function of blocking IL-6, we observed the expected association between the variant with lower C-reactive protein (CRP) and higher hemoglobin levels4,31,32 in both the EUR and AFR populations (Table S1).
The strongest associations within AFR subjects were related to white blood cell count (WBC), specifically, elevated WBC odds ratio (OR) 1.2, 95% confidence interval (CI), 1.1–1.3 (Fig. 1) by PheCode. The majority of IL6R-phenotype associations within EUR subjects pertained to vascular and cardiac disease. The phenotypes with the strongest association with IL6R were aortic aneurysm (AA) (OR 0.92; 95% CI, 0.90–0.94) as well as a specific type of aortic aneurysm, abdominal aortic aneurysm (AAA) (OR, 0.89; 95% CI, 0.87–0.90), coronary atherosclerosis and ischemic heart disease (CHD) (OR, 0.96; 95% CI, 0.95–0.97) (Fig. 1). The corresponding associations in AFR were similar but not significant [(AA) OR = 0.95 (0.87–1.03); (AAA) OR = 0.89 (0.80–1.00); (CHD) OR = 0.99 (0.95–1.02)].
After applying the test for heterogeneity, we observed 11 PheCodes translating to 7 conditions with differential association in AFR vs EUR: glaucoma, keratoconjunctivitis, periodontitis, type 2 diabetes, seborrheic dermatitis, walking difficulties, white blood cell count elevation (Fig. 3 and Table S2). IL6R was associated with reduced odds for glaucoma, keratoconjunctivitis, periodontitis, and type 2 diabetes among AFR with either no association or increased odds in EUR. The IL6R variant was associated with higher odds of an elevated white blood cell count in AFR (OR 1.21, 95% CI 1.12–1.30), and in line with this, a lower odds ratio for neutropenia in AFR (OR 0.80, 95% CI 0.72–0.89); these associations were not observed among EUR. IL6R was associated with seborrheic dermatitis and difficulty walking with increased odds in AFR and reduced odds in EUR.
Odds ratios for phenotypes with significant differential associations in AFR vs EUR ancestries (BH adjusted p value ≤ 0.1), see also Supplementary Table S2.
A comparison of laboratory values identified differences across 18 laboratory measurements (Fig. 4 and Table S3). In line with the significant difference in ICD codes related to WBC, the largest difference was observed in WBC whereby among individuals of AFR ancestry, each copy of the IL6R variant was associated with a higher WBC compared to those who did not carry the variant; no association was observed between IL6R and WBC among EUR. The higher value was observed across neutrophils, monocytes, eosinophils, and basophils, with the difference was most pronounced in absolute neutrophil count; the IL6R variant was associated with higher absolute values of neutrophils in AFR vs EUR. IL6R was also associated with higher triglyceride levels in AFR compared to EUR. The variant was associated with lower hemoglobin a1c (hba1c) in AFR with no significant association observed in EUR, in line with a lower odds ratio of T2D observed in AFR.
Comparison of standardized coefficients for associations between IL6R with laboratory values in AFR vs EUR (BH adjusted p value ≤ 0.1), see also Supplementary Table S3.
Due to the limited cohort size of individuals of AFR ancestry in the UKB and MGB, validation was focused on replicating laboratory values. The association and differences in WBC in AFR vs EUR remained the most significant finding. IL6R was associated with higher WBC among individuals of AFR vs EUR in both cohorts (Tables S4 and S5). IL6R was also associated with higher triglycerides in AFR vs EUR across the replication cohorts.
To understand the potential implications of the differential associations between IL6R with white blood cell phenotypes, we further tested the association between the variant and serious infection stratified by ancestry33. Overall, we observed an association between IL6R and a modest but significantly increased odds of serious infection in AFR but not EUR [AFR OR 1.03, 95% CI 1.01–1.04 vs EUR with OR 1.01, 95% CI 1.00–1.01]. Due to the small population size in UKB and MGB we did not have sufficient power to validate in these populations.
Discussion
This study provides a new roadmap for leveraging large biobanks to screen for differential associations between genetic variants and phenotypes across a diverse population. These data in turn can be used to inform potential differential effects of targeted therapies using an application designed to test for heterogeneity in large-scale genotype–phenotype screens to complement or inform clinical trials where populations are smaller and more homogeneous. We focused on a specific variant in IL6R with the known downstream effect of reducing IL-6 signaling with effects similar to existing therapies targeting IL-6R.
In this study using the most recent data from MVP, a biobank with the largest population of individuals of AFR ancestry to date, we observed 29 traits with heterogeneous associations, including WBC and T2D. The most significant heterogeneous signal observed was a lower odds ratio of neutropenia or higher WBC among Veterans of AFR descent compared to EUR; in EUR no association was observed between IL6R and WBC. The clinical significance of the association between IL6R and higher WBC, particularly neutrophil counts in AFR and EUR ancestry is unclear. To provide context, in a large population-based epidemiologic study, WBC was lower in Black compared White individuals34. As WBC are involved in host defense, in the present study, we tested the association between IL6R and serious infection and observed a modest but significant increased odds for serious infection among individuals of AFR descent where no association was observed in EUR. We were underpowered to validate these findings in UKB or MGB. In a review of the literature, we were unable to identify clinical trials of therapies targeting IL6 stratifying outcomes or adverse events by self-reported race (as genetic ancestry data are typically not available in trials). The majority of large observational studies for infection risk and IL6R blockade stems from studies of tocilizumab, the first IL6R antagonist approved for use in the US for RA. In these studies, risk of infection on tocilizumab is compared with another targeted therapy and overall, no difference has been observed33,35, however there were no data stratifying by self-reported race or ethnicity. Proposed follow-up analyses of a published study stratifying by self-reported race were underpowered since only a subgroup of their data had available information on race and ethnicity33,36. Based on findings from the present study, we anticipate that in studies with adequately sized populations, we would anticipate higher WBC among individuals of AFR ancestry on IL6R blockade, as well as a potential small increased odds for serious infection compared to EUR. Future trials and studies on the IL6 pathway can consider collecting data on WBC and neutrophil count, as well as stratifying infectious adverse events by self-reported race.
The heterogeneity test also identified an association between the IL6R variant with a reduced odds of T2D among Veterans of AFR descent, while no association was observed in EUR. Likewise, hba1c which reflects an average level of glucose over 2–3 months, was lower among individuals of AFR carrying the IL6R variant, while no association was observed among EUR in MVP. A lower hba1c was also observed among AFR carrying the IL6R variant compared to EUR in UKB. To our knowledge, glucose and hba1c levels were not reported in the randomized controlled trials in rheumatoid arthritis or giant cell arteritis37,38,39. However, the general association between the IL6R variant and lower odds of T2D was observed in meta-analysis examining the potential role of this pathway in the etiology of T2D40,41. Additionally, higher serum IL6 levels are associated with higher levels of hba1c, and increased risk of developing T2D in a large cohort study of women41,42. In an observational cohort study of RA patients with hba1c measurements before and after initiation of tocilizumab compared to a tumor necrosis factor inhibitor, a larger reduction in hba1c was observed in the tocilizumab group43. Thus, our study corroborates these findings and further anticipates that individuals of AFR descent either with T2D or at risk of T2D may derive more benefit from IL6R compared to individuals of EUR descent.
Notably, the strong associations observed between the IL6R variant and cardiovascular phenotypes, e.g. coronary heart disease, aortic aneurysms, peripheral arterial disease observed in prior studies was confirmed in EUR but not AFR6,17,44. This difference in association between IL6R and cardiovascular phenotypes in AFR vs EUR did not reach statistical difference with regards to heterogeneity. The hetFDR approach leverages information from both the mean effect and the magnitude of heterogeneity to determine the significance of the differences based on data from the entire population. Thus, in comparison to other phenotypes studied, the differential association with CV phenotypes were not considered heterogeneous and we would not anticipate a significant difference in the salutary effect of IL6R blockade for CV phenotypes in AFR vs EUR.
The hetFDR procedure applied in this study for multiple testing of heterogeneity fills an unmet need for methods that allow us to screen high-throughput data efficiently, such as PheWAS for differences across diverse patient populations. Compared with existing commonly used FDR control approaches such as BHq13 and Storey’s procedure28, our method is more powerful in detecting the phenotypes with heterogeneous effects. HetFDR takes advantage of the fact that among all phenotypes, only a small fraction has non-zero effects and nearly all those phenotypes with heterogeneous effect tend to have non-zero mean effects on the whole population, which can be characterized more effectively compared to the heterogeneity due to the larger sample size. This property was confirmed with our simulation results given in the Supplementary Materials. Specifically, we demonstrated in a simulation study using a similar scale of data and variable types as our current biobank datasets, the hetFDR achieved a satisfactory FDR control and a uniformly higher power compared to other existing methods. Lastly, in our study we use IL6R as an example, however, multiple other genetic variant-drug pairs exist that can benefit from further subgroup analysis. For example, studies on the proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitors identified an increased risk of type 2 diabetes, diastolic blood pressure, type 1 diabetes, peptic ulcer disease, and depression45.
Finally, we note that the IL6R Asp358Ala allele is of particular interest because the biochemical profile of subjects with this variant is similar to subjects receiving IL6R antagonist therapy. However, the precise mechanism of action differs. The IL6R variant leads to reduced expression on membrane-bound IL6R while, the IL6R antagonists tocilizumab and sarilumab block both soluble and membrane-bound IL6R. This highlights that these methods and the use of PheWAS to investigate potential drug effects are meant to generate hypotheses. Follow-up studies are needed to determine whether the potential heterogeneity is present among subjects actually on treatment.
Limitations
The population sizes for individuals of AFR ancestry were significantly lower in the UKB and MGB biobanks compared to MVP (UKB, AFR: n = 7,538; EUR: n = 459,315; MGB, AFR: n = 2922; EUR: n = 49,883; MVP, AFR: n = 105,838; EUR: n = 439,309). The smaller population resulted in limited power to replicate binary phenotypes, e.g., phecodes. Another potential limitation or difference between UKB and MVP is that UKB primarily contains inpatient codes and data from general medicine practices with less capture from other outpatient specialty practices in comparison to MVP and MGB. Importantly, this study did not include individuals of other ancestries.
This study focused on rs2228145, a relatively well-characterized loci and examined in prior studies as a potential proxy for IL6 blockade4,17,18. However, the majority of studies on genetic risk were performed in individuals of EUR ancestry, thus raising concerns regarding whether differences could be due to issues such as LD patterns. Given the limited existing data available regarding this locus in the AFR population, we believe this locus remains the best candidate to test for heterogeneity for the following reasons. In an eQTL mapping study in EUR and AFR populations, the top hit identified for IL6R was rs484652546. We identified that this SNP had a D′ of 1.0 with rs2228145 in AFR and EUR populations. Additionally, the anticipated biologic associations, lower CRP and higher hemoglobin was observed in AFR, thus confirming the known and expected downstream functional effects in both the AFR and EUR populations.
Conclusion
In summary, we leveraged 3 large population-based biobanks and applied a novel approach to test for heterogeneity identifying differential associations of the IL6R variant in AFR compared to EUR ancestry. Since the effect of the IL6R variant on phenotypic traits is known to parallel the effects of existing therapies targeting IL6R, findings from this study can inform ongoing and future trials targeting this pathway in the general population, particularly CVD. Our results suggest that targeting IL6R may be associated with higher WBC count and a potential modest signal for higher infection risk among individuals of AFR vs EUR descent. IL6R blockade may have a more beneficial effect for T2D with lower hba1c levels in AFR vs EUR, as well as potential beneficial effects for glaucoma, keratoconjunctivitis, and periodontitis. Notably, we observed a paucity of clinical trial data that were either sufficiently powered or reported data enabling post-hoc analyses of potential differences in effect across race and ethnicity. The increasing data available from more diverse populations such as MVP, along with the advancements in methods to analyze these data, can provide either complementary data or guidance on data elements to collect for pre-planned clinical trial subgroup analyses. Ultimately, these data together with approaches such as hetFDR can help us to design efficient trials that are powered to study the effectiveness of not just the primary outcome, but also potential beneficial and detrimental effects of a given therapy across a diverse population.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562(7726), 203–209 (2018).
Gaziano, J. M. et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 70, 214–223 (2016).
Finan, C. et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 9(383), eaag1166 (2017).
Ferreira, R. C. et al. Functional IL6R 358Ala allele impairs classical IL-6 receptor signaling and influences risk of diverse inflammatory diseases. PLoS Genet. 9(4), e1003444 (2013).
Cai, T. et al. Association of interleukin 6 receptor variant with cardiovascular disease effects of interleukin 6 receptor blocking therapy: A phenome-wide association study. JAMA Cardiol. 3(9), 849–857 (2018).
IL6R Genetics Consortium Emerging Risk Factors Collaboration. Interleukin-6 receptor pathways in coronary heart disease: A collaborative meta-analysis of 82 studies. Lancet 379(9822), 1205–1213 (2012).
Denny, J. C. et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 31(12), 1102–1111 (2013).
Diogo, D. et al. Phenome-wide association studies across large population cohorts support drug target validation. Nat. Commun. 9(1), 1–13 (2018).
Li, X. et al. MR-PheWAS: Exploring the causal effect of SUA level on multiple disease outcomes by using genetic instruments in UK Biobank. Ann. Rheum. Dis. 77(7), 1039–1047 (2018).
Tony Cai, T., Liu, W. & Xia, Y. Two-sample test of high dimensional means under dependence. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 76(2), 349–372 (2014).
Xia, Y., Cai, T. T. & Li, H. Joint testing and false discovery rate control in high-dimensional multivariate regression. Biometrika 105(2), 249–269 (2018).
Liu, M., Xia, Y., Cho, K. & Cai, T. Integrative high dimensional multiple testing with heterogeneity under data sharing constraints. J. Mach. Learn. Res. 22, 126–131 (2021).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 57(1), 289–300 (1995).
Zhang, M. J., Xia, F. & Zou, J. Fast and covariate-adaptive method amplifies detection power in large-scale multiple hypothesis testing. Nat. Commun. 10(1), 1–11 (2019).
Xia, Y., Cai, T. T. & Sun, W. Gap: A general framework for information pooling in two-sample sparse inference. J. Am. Stat. Assoc. 115, 1236–1250 (2019).
Tony Cai, T., Sun, W. & Wang, W. Covariate-assisted ranking and screening for large-scale two-sample inference. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 81(2), 187–234 (2019).
Interleukin-6 Receptor Mendelian Randomisation Analysis (IL6R MR) Consortium. The interleukin-6 receptor as a target for prevention of coronary heart disease: A mendelian randomisation analysis. Lancet 379(9822), 1214–1224 (2012).
Sarwar, N. et al. IL6R genetics consortium emerging risk factors collaboration. Interleukin-6 receptor pathways in coronary heart disease: A collaborative meta-analysis of 82 studies. Lancet 379(9822), 1205–1213 (2012).
Garbers, C. et al. The interleukin-6 receptor Asp358Ala single nucleotide polymorphism rs2228145 confers increased proteolytic conversion rates by ADAM proteases. Biochim. Biophys. Acta Mol. Basis Dis. 1842(9), 1485–1494 (2014).
Nishimoto, N. et al. Mechanisms and pathologic significances in increase in serum interleukin-6 (IL-6) and soluble IL-6 receptor after administration of an anti-IL-6 receptor antibody, tocilizumab, in patients with rheumatoid arthritis and Castleman disease. Blood J. Am. Soc. Hematol. 112(10), 3959–3964 (2008).
Maini, R. N. et al. Double-blind randomized controlled clinical trial of the interleukin-6 receptor antagonist, tocilizumab, in European patients with rheumatoid arthritis who had an incomplete response to methotrexate. Arthritis Rheum 54(9), 2817–2829 (2006).
Denny, J. C. et al. PheWAS: Demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics 26(9), 1205–1210 (2010).
Kosoy, R. et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum. Mutat. 30(1), 69–78 (2009).
Hunter-Zinck, H. et al. Genotyping array design and data quality control in the Million Veteran Program. Am. J. Hum. Genet. 106(4), 535–548 (2020).
Bastarache, L. Using phecodes for research with the electronic health record: From PheWAS to PheRS. Annu. Rev. Biomed. Data Sci. 4, 1–19 (2021).
Li, A. & Barber, R. F. Multiple testing with the structure-adaptive Benjamini-Hochberg algorithm. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 81(1), 45–74 (2019).
Storey, J. D. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 64(3), 479–498 (2002).
Sudlow, C. et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12(3), e1001779 (2015).
Karlson, E. W., Boutin, N. T., Hoffnagle, A. G. & Allen, N. L. Building the partners healthcare biobank at partners personalized medicine: Informed consent, return of research results, recruitment lessons and operational considerations. J. Pers. Med. 6(1), 2 (2016).
Atkins, M. B., Kappler, K., Mier, J. W., Isaacs, R. E. & Berkman, E. M. Interleukin-6-assoeiated anemia: Determination of the underlying mechanism. Blood 86(4), 1288–1291 (1995).
Nieken, J. et al. Recombinant human interleukin-6 induces a rapid and reversible anemia in cancer patients. Blood. 86, 900–905 (1995).
Pawar, A. et al. Risk of serious infections in tocilizumab versus other biologic drugs in patients with rheumatoid arthritis: A multidatabase cohort study. Ann. Rheum. Dis. 78(4), 456–464 (2019).
Beutler, E. & West, C. Hematologic differences between African-Americans and Whites: The roles of iron deficiency and α-thalassemia on hemoglobin levels and mean corpuscular volume. Blood 106(2), 740–745 (2005).
Grøn, K. L. et al. Overall infection risk in rheumatoid arthritis during treatment with abatacept, rituximab and tocilizumab; an observational cohort study. Rheumatology 59(8), 1949–1956 (2020).
https://www.google.com/books/edition/Personal_Communication_in_Human_Relation/X2QQAQAAIAAJ?hl=en.
Yazici, Y. et al. Efficacy of tocilizumab in patients with moderate to severe active rheumatoid arthritis and a previous inadequate response to disease-modifying antirheumatic drugs: The ROSE study. Ann. Rheum. Dis. 71(2), 198–205 (2012).
Nishimoto, N. et al. Study of active controlled monotherapy used for rheumatoid arthritis, an IL-6 inhibitor (SAMURAI): Evidence of clinical and radiographic benefit from an x ray reader-blinded randomised controlled trial of tocilizumab. Ann. Rheum. Dis. 66(9), 1162–1167 (2007).
Stone, J. H. et al. Trial of tocilizumab in giant-cell arteritis. N. Engl. J. Med. 377(4), 317–328 (2017).
Pradhan, A. D., Manson, J. E., Rifai, N., Buring, J. E. & Ridker, P. M. C-reactive protein, interleukin 6, and risk of developing type 2 diabetes mellitus. JAMA 286(3), 327–334 (2001).
Bowker, N. et al. Meta-analysis investigating the role of interleukin-6 mediated inflammation in type 2 diabetes. EBioMedicine 61, 103062 (2020).
Kado, S., Nagase, T. & Nagata, N. Circulating levels of interleukin-6, its soluble receptor and interleukin-6/interleukin-6 receptor complexes in patients with type 2 diabetes mellitus. Acta Diabetol. 36(1), 67–72 (1999).
Otsuka, Y. et al. Effects of tumor necrosis factor inhibitors and tocilizumab on the glycosylated hemoglobin levels in patients with rheumatoid arthritis; an observational study. PLoS ONE 13(4), e0196368 (2018).
Levin, M. G. et al. A missense variant in the IL-6 receptor and protection from peripheral artery disease. Circ. Res. 129(10), 968–970 (2021).
Nelson, C. P. et al. Genetic assessment of potential long-term on-target side effects of PCSK9 (Proprotein Convertase Subtilisin/Kexin Type 9) inhibitors. Circulation Genomic Precis. Med. 12(1), e002196 (2019).
Shang, L. et al. Genetic architecture of gene expression in European and African Americans: An eQTL mapping study in GENOA. Am. J. Hum. Genet. 106(4), 496–512 (2020).
Acknowledgements
This work was funded by the US Veterans’ Health Administration Million Veterans Program (MVP) and the NIH P30 AR072577. The Authors declare no Competing Financial or Non-Financial Interests.
Author information
Authors and Affiliations
Consortia
Contributions
T.C., and K.P.L. contributed to design and conceptualization of the study. X.W., I.E.N., M.L., T.C., X.X., C.L.B., H.Z., C.H., J.C., T.C., and K.P.L. contributed to methodology, data analysis or interpretation. K.D., L.C., Y.L.H. and K.C. contributed to data collection. All authors contributed to drafting the work or revising it critically for important intellectual content and approved the final version. All authors are responsible for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, X., Liu, M., Nogues, IE. et al. Heterogeneous associations between interleukin-6 receptor variants and phenotypes across ancestries and implications for therapy. Sci Rep 14, 8021 (2024). https://doi.org/10.1038/s41598-024-54063-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-54063-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.