Abstract
This study aims to use spatial transcriptomics to characterize the cell-type-specific expression profile associated with the microscopic features observed in Wooden Breast myopathy. 1 cm3 muscle sample was dissected from the cranial part of the right pectoralis major muscle from three randomly sampled broiler chickens at 23 days post-hatch and processed with Visium Spatial Gene Expression kits (10X Genomics), followed by high-resolution imaging and sequencing on the Illumina Nextseq 2000 system. WB classification was based on histopathologic features identified. Sequence reads were aligned to the chicken reference genome (Galgal6) and mapped to histological images. Unsupervised K-means clustering and Seurat integrative analysis differentiated histologic features and their specific gene expression pattern, including lipid laden macrophages (LLM), unaffected myofibers, myositis and vasculature. In particular, LLM exhibited reprogramming of lipid metabolism with up-regulated lipid transporters and genes in peroxisome proliferator-activated receptors pathway, possibly through P. Moreover, overexpression of fatty acid binding protein 5 could enhance fatty acid uptake in adjacent veins. In myositis regions, increased expression of cathepsins may play a role in muscle homeostasis and repair by mediating lysosomal activity and apoptosis. A better knowledge of different cell-type interactions at early stages of WB is essential in developing a comprehensive understanding.
Similar content being viewed by others
Introduction
Wooden Breast myopathy (WB) has been a major cause for economic loss in the poultry industry due to deterioration to the muscle integrity and meat quality, rendering pale and stiff p. major muscle in the cranial to caudal regions1. High-throughput sequencing and omics techniques have previously revealed various factors and related expression patterns associated with WB such as energy metabolism, hypoxia, oxidative stress and cellular signaling2,3,4,5. Conducting upstream regulator analysis of WB using proteomic data, Bottje and colleagues predicted inhibition of regulators for mitochondrial biogenesis and cell death, and activation of those for angiogenesis6. From a perspective beyond skeletal muscle, a recent multi-omics study established the relationship between gut microbiota and WB through dynamic changes in plasma metabolic markers in commercial broilers7.
Nevertheless, skeletal muscle is a complex tissue where multiple cell populations reside and work in conjunction, from myofibers, vasculature, nerve fibers, and connective tissues including collagen producing fibrocytes, adipocytes, and immune cells. While single-cell RNA-seq surpasses whole transcriptome RNA-seq in yielding expression profiles at cell type level, the spatial relationships of interrelated cells within the tissue are lost upon processing. The advent of spatial transcriptomics has introduced a novel approach allowing mapping gene expression changes to individual architectural regions and structures and at times to individual cell populations histologically within tissue. In short, this state-of-the-art technique not only discloses different cell types present and their potential interactions, but also overcomes tissue dissociation bias resulted from over abundant cell types.
For instance, spatial transcriptomics techniques have been used to identify a plaque-induced gene network which constitutes a coordinated cellular response between astrocytes and microglia in the vicinity of amyloid plaques in Alzheimer’s Disease8. Furthermore, transcriptional signatures of functional anatomical domains were characterized in both murine muscle tissue after nerve crush injury9 and fish myotomal muscle10. Spatial analysis using a rabbit model of rotator cuff tear revealed heterogeneous myofibril lesions and identified markers for regenerating myofibers11. To the best of our knowledge, there are no published research performing spatial transcriptomics on chicken muscles, either with or without myopathic changes.
WB disease shows unique pathological features, starting with lymphoplasmacytic phlebitis progressing to complete venous occlusion with sparing of adjacent arteries12. Additionally, veins in myopathic regions frequently are lined or cuffed by clusters of lipid-laden macrophages. In degenerated myofiber regions, segmental disruption, fiber splitting and vacuolation are followed by fibrosis, or scarring, alongside multifocal myo-regeneration, characterized by myotube formation with nuclear rowing12. At present, it is uncertain which cells or tissue regions within p. major muscle are responsible for the omics changes identified. By elucidating molecular profile and changes localized to certain cells or tissue structures associated with histological features via spatial transcriptomics, we can further depict the pathophysiology and pathogenesis of WB. Furthermore, this investigation may provide novel and valuable insights for diagnosis, treatment or prevention. By overlaying transcriptome on top of histology, we have characterized expression pattern particular to resident cell types and their potential interactions within breast muscle in broilers. Consequently, this study aimed to improve our understanding on functional roles played by distinct cell types and tissue architectural elements during WB development.
Results
Sample classification and gene expression metrics
The stitched high-resolution images of H&E-stained samples are shown in Fig. S1. A more compact muscle tissue was observed in Sample 1 and 2 than in sample 3. Furthermore, sample 3 exhibited notable pathological lesions associated with WB, including myositis, evident as infiltration of immune cells in the interstitium (Fig. S1). Accordingly, sample 3 was classified as affected in contrast to sample 1 and 2 as unaffected. The presence of lipid laden macrophages (LLM) alone was not accompanied with nor an accurate indicator of myopathic changes as muscles both with and without myositis demonstrated small to moderate numbers of LLM accumulating around veins. Nevertheless, the presence of LLM in these samples could imply a certain degree of white striping and cellular changes, potentially culminating in the development of WB beyond the age at which samples were collected. In terms of histopathologic analysis, feature appearance ratio was calculated as number of tiles containing that feature divided by total number of tiles occupied by the sample. As a result, ratio of LLM increased from 8% for unaffected sample 1 and 2 to 27% and 42% for affected sample 3_1 and 3_2, respectively. Accumulation of LLM was observed adjacent to or circumferentially cuffing veins in both affected and unaffected samples (Fig. 1B), with noticeably increased number and severity in affected tissue. Similarly, area of connective tissue between myofiber bundles was seemingly larger with higher ratio in the WB affected samples (Fig. S1). Focal myositis was only observed in the affected muscle and manifested a sporadic pattern. The ratio of myositis increased from 20% in sample 3_1 to 60% in 3_2, corroborating that WB lesions happen sporadically, often in a multifocal polyphasic manner, and affect myofibers at different depth within the breast muscle.
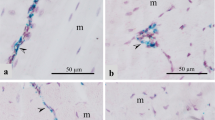
Representative spatial transcriptomics cell type clusters determined by K-means clustering. Clustering is identified by the presence of colored dots. Unmarked insets to the right of each region demonstrate the microscopic appearance of the cells/tissue structures in each region for visual histologic confirmation of cluster classification by cell type or feature. (A) Myofibers (orange and dark green dots) clustered separately from (B) Non-myofiber features (no dots). At higher magnification, clustering separated (C) vessels (blue dots) from perivascular lipid-laden macrophages (light green dots), (D) inflammatory infiltrates indicative of myositis (yellow dots), and (E) perimysial connective tissue (teal blue dots).
Sequencing and mapping performance measurements were satisfactory and comparable among samples (Fig. S2B and A). Despite a relatively lower sequencing depth, percentage of reads mapped to the genome and exons, as well as number of genes detected increased for sample 3 after changing permeabilization time to 6 min (Fig. S2B and C). Overall, these summary metrics suggested an adequate experiment for downstream feature classification.
Unsupervised K-means clustering
Unsupervised K-means clustering identified 5 different unique histologic features, cell types, or regions, including LLM, myositis, vessels (specifically veins), connective tissue, and myofibers (Fig. 1). Myofibers manifested heterogeneity in expression profiles, as identified by multiple clusters (marked by the orange and green dots in sample 3_2 in Fig. 1A) overlaying visually confirmed myofiber clusters. While K-means clustering of Visium spots identified LLM in all the samples, vascular regions were identified only in sample 1 and 3_2 by this analysis. Vessel walls were confirmed microscopically based upon observation of the endothelial-lined lumen, smooth muscle-lined vascular walls, and intraluminal red blood cells, when present. Connective tissue was also identified by K-means clustering in all the samples and confirmed by observation of discrete extracellular collagen fiber bundles between the myofibers or between vessels and muscle fascicles (Fig. 1E). In accordance with histopathologic analysis, myositis was unique to affected sample 3 (3_1 and 3_2) (Fig. 1D), in which distorted and smaller myofibers were infiltrated by immune cells such as histiocytes and heterophils.
Interestingly, some marker genes in one of the myofiber clusters of sample 1 and 2 were vasculature related, such as hemoglobin subunit alpha 1 (HBA1) and hemoglobin subunit epsilon 1 (HBBA). Therefore, these spots likely indicate the small capillaries abutting myofibers. Albeit virtually invisible histologically, capillary location was suggested by the linear pattern of the spatial transcriptomics spots (Fig. 2).
Marker genes for LLM, connective tissue, myositis and vasculature were reported in Table 1. These genes were selected as marker genes, because (i) they were up-regulated in the clusters identifying these features and (ii) they were overlapped across all 3 samples, except for “vasculature” with overlap in at least 2 samples and myositis in sample 3. Interestingly, markers for LLM were mostly involved in lipid metabolism, indicating their role in processing and storing lipids. In terms of myositis, marker genes encoding lysosomal proteins including cathepsins (CTSA, CTSK), IFI30 Lysosomal Thiol Reductase (IFI30) and CD74 Molecule (CD74) suggested active lysosome and immune cells activities in muscle turnover. As expected, marker genes related to extracellular matrix and smooth muscle were detected by the clusters identifying connective tissue and vascular regions, respectively.
Integration analysis
Integration analysis was performed to identify cell types and marker genes conserved in both WB affected and unaffected samples, as well as to compare between two conditions to find cell-type specific responses to the pathological state. In total, 8 clusters were identified, and UMAP revealed a good overlap of data points between the affected and unaffected conditions (Fig. 3A). Each cluster was then characterized based on their marker genes, resulting in LLM, connective tissue, vascular regions, and myofiber clusters. Identification of 4 distinct clusters characterized as myofibers (M1–M4) corroborated myofiber heterogeneity found by unsupervised K-means clustering. Heatmaps created using top 10 marker genes in each cluster showed a clear separation of the last 6 clusters (Fig. 3B), with M1 having only 3 markers. Considering the small number of markers in M1 and how scattered the spots in M1 were, this cluster probably contained the spots representative of background information and the ones overlapping across multiple cell types, for instance muscle and connective tissue (Fig. 3A). Moreover, there is a good overlap between these markers and the ones in Table 1, especially for LLM and connective tissue, having 8 and 7 overlapping genes, respectively, further supporting the M1 region as an overlap between myofibers and adjacent tissue.
Integration analysis using Seurat. (A) Scatter plot of Uniform Manifold Approximation and Projection results. (B) Heatmap of the top 10 marker genes of each cluster. In total, 8 clusters were identified by the integration analysis, and then characterized based on their marker genes as lipid leaden macrophages (LLM), connective tissue, vascular regions, myositis, and 4 distinct myofiber (M1–M4) clusters.
In the myopathic condition, connective tissue and vasculature exhibited divergence in expression of some marker genes (Fig. 4). In particular, type I and III collagens (COL1A1, COL3A1) showed comparable expression level in both WB affected and unaffected groups, whereas the fibril-associated collagen type XII alpha 1 chain (COL12A1) was highly expressed in the myopathic condition (Fig. 4A). Vasculature markers specific to the myopathic condition were EGF like domain multiple 7 (EGFL7) and monoamine oxidase A (MAOA) (Fig. 4B).
Violin plots of some marker genes in Wooden Breast affected and unaffected samples showed gene expression pattern specific to the myopathic condition. (A) Connective tissue and (B) Vasculature. These violin plots showed genes expressed specifically to the affected tissue, including collagen type XII alpha 1 chain (COL12A1) for connective tissue and EGF like domain multiple 7 (EGFL7) as well as monoamine oxidase A (MAOA) for vasculature.
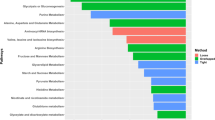
Pathway enrichment analysis identified three groups of pathways for LLM, including translation, lipid metabolism and immune response (Fig. S3A). Among pathways for lipid metabolism, the peroxisome proliferator activated receptor (PPAR) signaling pathway stood out by sharing 6 top marker genes: fatty acid binding protein (FABP3, FABP4), acyl-CoA synthetase long chain family member 1 (ACSL1), stearoyl-CoA desaturase (SCD), perilipin 1 (PLIN1) and lipoprotein lipase (LPL). Pathways for connective tissue (Fig. S3B) were mostly related to the extracellular matrix and collagen, suggestive of early tissue remodeling or initiation of fibrosis in WB tissue. As for myositis (Fig. S3C), enriched pathways included lysosome, heterophil degranulation, phagosome and apoptosis consistent with inflammation and clearance of damaged myofiber segments. Top-ranked pathways for myofiber cluster M2 were concentrated on energy metabolism, such as citric acid cycle, oxidative phosphorylation and glucose metabolism (Fig. S4A), whereas those for M4 were focused on cell cycle and apoptosis (Fig. S4B) suggesting possible clean-up/repair. Only three pathways were enriched for M3: (1) infectious disease, (2) rRNA processing and cooperation of Prefoldin and (3) TriC/CCT in actin and tubulin folding. Considering the metabolic pathways and the distance between M2 and M3–M4 clusters (Fig. 3A), M2 was presumably representing normal myofibers in contrast to damaged or reparative myofibers (M3 and M4) identified in the WB affected sample. As described previously, the M1 cluster genes overlapped with the connective tissue clustering and may demonstrate the myofiber/connective tissue interface.
Discussion
Lipid-laden macrophage accumulation
Lipid-laden macrophages (LLM), more colloquially known as “foam cells”, are monocyte/macrophage lineage cells with intracellular accumulation of lipid droplets13. This cell population was reported in a previous study to aggregate in perivascular connective tissue in more than 65% of chickens from a purebred commercial broiler line by one week of age12. Noticeably, the perivenous (i.e. external perivascular cuffing) location of these LLM in broiler chickens is rather different to that in atherosclerosis where LLM accumulate in subendothelial space and smooth muscle layers within the wall of arteries14.
PPARγ signaling activation in the cluster marking LLM, as indicated by the up-regulation of CD36 molecule (CD36), NPC intracellular cholesterol transporter 2 (NPC2), LPL, FABP4 and FABP3, suggest similarities between LLM accumulation in chicken breast muscle and metabolic activation of adipose tissue macrophages in human obesity15. It is possible that similar metabolic pathways are activated in broiler chickens due to intensive genetic selection for efficient and enhanced muscle growth in commercial meat production, even though the birds do not become obese via lipid deposition in adipocyte depots like their human counterparts. Induction of PPARγ activity in adipose tissue macrophages has been previously studied and linked to exposure to elevated levels of oxidized low-density lipoproteins (oxLDL) or palmitate15. Accumulation of palmitate and oxLDL were reported in WB2,4. Particularly, oxLDLs build-up was indicated by higher levels of its principal components 9-HODE and 13-HODE16. Additionally, using a murine alveolar macrophage cell line, Hou and colleagues17 reported induction of LLM formation along with elevated transcription of CD36 and PPARγ by oxLDL, with significant attenuation by either blocking CD36 or inhibiting the activity of PPARγ. Consequently, LLM formation in broiler chickens likely resulted from a similar process involving oxLDL-CD36 mediated signaling and PPARγ induced up-regulation of its transcriptional target genes16,17,18. A comparable LLM activation in these aforementioned varied tissue origins shared one similarity, a lipid rich extracellular environment.
Figure 5 depicts our proposed mechanism for LLM formation within the p. major muscle of broiler chickens. Basically, LPL preferentially hydrolyzes triacylglycerols (TAG) in extracellular space19. Upon oxLDL stimulation, scavenger receptors CD36 and NPC2 promote lipid uptake into the cells20. Specifically, oxLDL binds to its high-affinity receptor CD36 to enter the cells by endocytosis21. PPARγ could be activated by both oxLDL and fatty acids (FA), which in turn increase FA uptake by inducing expression of FABP4 and CD3621,22. Once inside, FABP4 then facilitates FA transport to intracellular vesicles and organelles to boost cholesterol ester and TAG accumulation23, participated by enzymes stearoyl-CoA desaturase (SCD)24 and diacylglycerol O-acyltransferase 2 (DGAT2)25 respectively. Expression of SCD and apolipoprotein 1 (APOA1), indicators of elevated cholesterol efflux and biosynthesis of mono unsaturated FA14, coincided with the significantly elevated lipid level in WB2,4,26. Unlike in adipose tissue of high feed efficiency broilers (low-fat phenotype), G0/G1 switch gene 2 (G0S2) was activated in the LLM, shifting towards a metabolism favoring lipid deposition27. Moreover, oxLDL-CD36 mediated signaling is associated with mitochondrial accumulation of long chain FA, possibly a consequence of enhanced FA oxidation and/or biosynthesis channeled by ACSL1, and suppression of LLM migration which could explain the perivascular accumulation and cuffing by these cells18,28. Finally, as a constitutive protein of lipid droplets, up-regulation of perilipins (PLIN1, PLIN4) was directly linked to increase in lipid droplets29. Interestingly, these lipid genes FABP4, G0S2 and PLIN1 showed a significantly higher expression in the cranial area than the caudal region30, suggesting a higher degree of lipid metabolism at play and consistent with the enhanced vulnerability to WB in the cranial region of the p. major muscle.
Proposed mechanism of lipid laden macrophage accumulation in the p. major muscle of broiler chickens. Cells are metabolically stimulated by oxidized low-density lipoproteins to activate PPARγ signaling pathway, which boosts lipid uptake and synthesis intracellularly to process excessive surrounding lipids. [oxLDL: oxidized low-density lipoproteins; FA: fatty acids; TAG: triacylglycerol; FC: free cholesterol; CE: cholesterol ester; CD36: CD36 molecule; NPC2: NPC intracellular cholesterol transporter 2; FABP4: Fatty Acid Binding Protein 4; PPARγ: peroxisome proliferator activated receptor gamma; PLIN1,4: Perilipin 1, 4; DGAT2: diacylglycerol O-acyltransferase 2; SCD: stearoyl-CoA desaturase; ACSL1: acyl-CoA synthetase long chain family member 1].
In general, these perivascular LLM in chicken breast muscle showed comparable gene expression pattern to atherosclerotic LLM, except for genes pertinent to pro-inflammatory cytokines nor M1 and M2 macrophages markers31,32. Previous research18,23,28 on peritoneal and myeloid monocyte-derived LLM in mice and in vitro cell culture studies discovered inflammatory activities producing interleukins, tumor necrosis factor (TNF-α), monocyte chemoattractant protein (MCP-1), etc. Nevertheless, according to a study by Kim et al.13, non-foamy macrophages are responsible for expressing interleukins and other inflammatory cytokines in the atherosclerotic intima. Since the identified marker genes were primarily related to a lipid metabolism, this LLM population in broiler chickens may primarily aid in processing excessive lipids entering the breast muscle mediated by LPL. Alternatively, given the elevated expression of LPL in LLM, it is possible that this cell population contributes to perivascular lipid infiltration, thereby establishing a positive feedback loop (i.e. further increasing perivascular lipid accumulation and perivascular LLM attraction) and playing a significant role in the development of the observed pathology. Up to 50% reduction of atherosclerotic lesions was reached in mice deficient for macrophage LPL33. Furthermore, considering its adjacent location to veins, continuous accumulation of LLM could compress the space occupied by veins or alter vascular flow, eventually leading to phlebitis as seen in older broiler chickens with WB12.
Connective tissue and veins
Myofibers are surrounded by and embedded in connective tissue, or extracellular matrix (ECM), composed of netted collagen fibers and other macromolecules34. As a signaling hub, the connective tissue modulates muscle growth, repair and contractile force transmission34, and thus is vital for muscle integrity. Naturally, the fibril-forming COL1A1, COL1A2, COL3A1 as well as microfibrillar collagen VI (COL6A2, COL6A3) were the conserved markers for connective tissue in this study. The unique expression of COL12A1 in the myopathic condition is consistent with prior bulk RNA-seq studies showing an up-regulation of this gene in WB affected chickens comparing to their unaffected counterparts starting as early as week 25,35. Since collagen XII binds the macromolecules in ECM to form bridges that organize adjacent collagen fibrils36, its deposition indicated a divergence in the structure and biomechanical properties of ECM between WB affected and unaffected tissues. An intense collagen XII deposition was previously reported in regenerative heart of Zebrafish, with involvement in tissue cohesion and regeneration37. In correspondence with the stiff characteristic of WB muscle, COL12A1 expression may be related to constant tensile strain and fibrosis36. In human and mice, mutations in COL12A1 led to joint hyperlaxity and muscle weakness due to a passive force-transducing elastic element in ECM38.
The enriched pathways of vascular marker genes such as leukocyte transendothelial migration suggested the presence of inflammation in the vasculature within the p. major muscle (Fig. S3D). Although examination of histological images for this study revealed no microscopically visible lesions of phlebitis, expression of some markers certainly imposed risks for venous inflammation in a later myopathic stage (Fig. 4B). One potentially important biomarker was von Willebrand factor (VWF), which mediates vascular inflammation via recruitment of immune cells including macrophages, platelets and leukocytes39. As a glycoprotein essential for blood clotting, VWF regulates platelet adhesion to endothelium, and thus its downregulation was associated with smaller atherosclerotic plaque and less macrophage accumulation in various animal models40. In particular, VWF release was enhanced by LDL presence in mice41, and macrophage released inflammatory cytokines42. Accordingly, VWF expressed in veins could be one of the major factors responsible for LLM cumulation in WB.
Furthermore, capillary endothelial FABP5 could play a role in boosting FA uptake through endothelial walls22, thus originating the enhanced lipid deposition in breast muscle of broilers. FABP5 may also stimulate cell proliferation by regulating PPARδ signaling43. A rather specific expression of EGFL7 in the myopathic condition (Fig. 4B) suggested hypoxia-induced angiogenic behavior or vessel regeneration44. In addition, adiponectin (ADIPOQ) expression in veins was found relevant to peroxidation substrates in the vascular wall45. Oxidative stress in the veins of WB affected muscle was inferred by marker genes involved in oxidative stress response pathway, MAOA, glutathione peroxidase (GPX1, GPX3) and cytochrome B-245 alpha chain (CYBA). While MAOA and CYBA are responsible for production of reactive oxygen species (ROS)46,47 and showed dominant expression in affected samples (Fig. 4B), GPX1 and GPX3 reduce ROS level to alleviate oxidative stress48. Moreover, expression of thymosin beta 4 (TMSB4X) is associated with phenotypic modulation of smooth muscle cells and angiogenesis49,50, whose down-regulation contributes to developing larger atherosclerotic plaques in mice49. Since TMSB4X was also up-regulated in WB chickens [2 (unpublished data)], its particular role in veins and muscle remains elusive and worth studying in the future. Overall, these results suggest a possible role of veins in generating ox-LDL through endothelial and/or smooth muscle cell-induced oxidation of LDL, in turn leading to perivascular lipid deposition and LLM accumulation. The findings further support the previous hypothesis that venous endothelial cells in WB exhibit an increased level of metabolic activity and potential dysfunction51.
Myositis
Lysosomes are the principal organelle in charge of protein turnover and tissue homeostasis, involving in various pathways such as metabolism, autophagy, and so on52. In alignment with functional analysis (Fig. S3C), overexpression of multiple cathepsins (CTSA, CTSB, CTSC, CTSD, CTSK, CTSS, CTSZ) and lysosomal membrane proteins (LAMP1, LAPTM5) in the myositis cluster strongly indicated active lysosomal function, suggesting a dynamic turnover of damaged or inflamed myofibers52,53. Specifically, CTSB hydrolyses various myofibrillar proteins, including myosin heavy chain, tropomyosin, troponin T and I52. The co-expression of CTSB and its inhibitor cystatin-A (CSTA) further implied localized inflammation, as observed in pancreatic tumor cells54. The existence of myofiber inflammation was also supported by expression of CD74 molecule (CD74), MHC class II beta chain 2 (BLB2) and ferritin heavy chain 1 (FTH1). As the receptor for macrophage migration inhibitory factor, CD74 regulates immune response as well as cell proliferation, and is implicated in other lipid-induced inflammatory diseases such as atherosclerosis55. Similarly, BLB2 is related to adaptive immune response in chickens through antigen processing and presentation56. And ferritin is a pro-inflammatory cytokine whose expression matches inflammatory activity and remains up-regulated during muscle regeneration57,58. Furthermore, ferritin was also up-regulated in response to muscle damage attributed to reactive oxygen species after denervation59, which was indicative of the oxidative stress related myofiber damage in WB. Also, the potential connection between FTH1 upregulation and ferroptosis, an iron-dependent programmed cell death, in WB is a worthwhile research direction that could contribute to our understanding of cellular responses to oxidative stress and programmed cell death.
Considering the high level of oxLDL and ubiquity of oxidative stress in WB1,2, lysosomal membrane permeabilization (LMP) could be induced and result in leakage of cathepsins and lysosome-mediated apoptosis60. Moreover, this muscle inflammation was supported by CTSS and CTSZ, whose expression compensates for the activity of CTSB and CTSC during LMP-regulated inflammation60. Such crosstalk between lysosome and cell death was also observed in bovine muscle postmortem, where ROS level was positively correlated with lysosomal membrane stability and activities of CTSB and CTSD61. Although little is known about the biological function of IFI30 lysosomal thiol reductase (IFI30) in muscle, IF130 was believed to dampen oxidative stress by recycling oxidized glutathione in hematopoietic stem cells62. That said, this spatial transcriptomic analysis displayed a possible linkage between lysosome-mediated muscle degeneration and inflammation probably under oxidative stress in WB, which warrants future study in a more advanced stage of this disease using molecular approaches.
Muscle heterogeneity
As stated by pathway analysis, myofiber cluster M2 manifested a normal expression pattern in accord with type II muscle fueled by glucose typical for avian pectoral muscle. Even though no pathologic lesions were observed in any of the histological images other than myositis, myofiber cluster M3 and M4 exhibited a profile consistent with WB denoted by markers identified in previous studies.
Majority of the top marker genes of the myofiber clusters M3 and M4 clusters were ubiquitously found up-regulated in the affected muscle samples and are involved in myogenesis, cell proliferation and regeneration3,63,64,65. For instance, expression level of myogenin (MYOG) and myosin heavy chain 15 (MYH15) increased linearly with WB severity to participate in skeletal muscle repair63. Cysteine and glycine rich protein 3 (CSRP3) and its grouped protein myozenin 2 (MYOZ2) are related to repair mechanism and fiber-type switching in WB affected muscle3,64. Musculoskeletal, embryonic nuclear protein 1 (MUSTN1), ankyrin repeat domain 1 (ANKRD1), HOP homeobox (HOPX) and PDZ and LIM domain 3 (PDLIM3) were up-regulated in WB birds at 7 weeks65. Particularly, MUSTN1 showed differential expression between slow growing chickens and those affected with WB at 6 weeks of age66. Their up-regulation in affected samples at both 6–7 weeks and 23 days post hatch implied a constant myogenesis and regeneration process in broiler breast muscle probably due to the fast muscle growth. Meanwhile, the resulted muscle hypertrophy should be accompanied with cytoskeleton assembly in order to maintain a structural organization and contractibility67. That being said, a gene in charge of cytoskeleton reorganization, muscle RAS oncogene homolog (MRAS) was also up-regulated in WB affected chickens of 7 weeks5. Overall, the results of this study reaffirm the importance of these WB-specific markers and identify their heightened expression specifically within myofibers.
Conclusions
Via spatial transcriptomics, distinct histologic features were successfully characterized based on their gene expression profiles, including LLM, myofibers, myositis, connective tissue and vasculature. In our prior bulk RNA-seq studies, we observed increased expression of lipid metabolism genes during the early stages of WB35,65,68. However, the specific cell type and regions responsible for this gene expression signature remained largely unknown. Our results from spatial transcriptomics revealed LLM as the key cell population responsible for altered lipid metabolism during early stages of WB. Building upon prior findings of elevated 9-HODE and 13-HODE in WB4, we propose that the formation of LLM in broiler chickens occurs as a cellular cascade stimulated by oxLDLs. Additionally, we report discrepancies in gene expression within connective tissue and vascular regions, leading to discoveries of potential WB specific biomarkers and generating insights in muscular disease pathology. We also confirmed existence of gene expression heterogeneity within chicken pectoralis major muscle, despite visually indistinguishable histological presentation across intact myofibers. In inflamed myofibers, active participation by lysosomes in muscle homeostasis was suggested by the up-regulation of numerous lysosomal genes. Overall, this study revealed potential gene expression biomarkers related not only to myopathic muscle but also to various cell types present in healthy pectoralis major muscle of broiler chickens.
Materials and methods
Chickens and tissue collection
As part of an ongoing genetic study of WB, 114 Cobb500 broiler chickens were raised in a chicken house at the University of Delaware in accordance with the Animal Care and Use Handbook 2019 of the College of Agriculture and Natural Resources, University of Delaware and allowed free access to feed and water. The current experiment utilized p. major muscle tissues from three randomly-sampled chickens at 23 days of age, harvested post-euthanasia, and examined for gross lesions during necropsy. Specifically, chickens were euthanized by cervical dislocation and examined for muscle firmness by manual palpation of the p. major muscle, and each bird was given a wooden breast score, as described in a previous work from our laboratory12. Approximately, 1 cm3 muscle samples were dissected from the cranial part of the right pectoralis major muscle and immediately embedded in Optimal Cutting Temperature (OCT) compound media, flash frozen in a bath of isopentane and liquid nitrogen before being stored at − 80 °C until further processing. The animal conditions and experimental procedures used in this scientific study were approved by the University of Delaware Institutional Animal Care and Use Committee (protocol number: 120R-2021-AP). The study was carried out in compliance with the ARRIVE guidelines.
Permeabilization experiments
The fresh frozen samples in OCT media were sectioned to 10 μm using a cryostat and mounted on 10X Visium slides. Tissue permeabilization (TP) experiments were conducted on both longitudinal- and cross-sectional muscle tissues as per manufacturer instructions using 10 x Genomics Visium Tissue Optimization Kit (PN-1000193), yielding optimal TP time of 18 and 6 min, respectively. In the subsequent steps (section below), since samples 1 and 2 were largely longitudinal; TP time for this samples was set at 18 min during spatial RNA sequencing library preparation (i.e., the following section). However, for sample 3, which was an oblique section, the entire spatial transcriptomic experiment was replicated with 6 and 18 min TP time, mainly because the orientation of individual myofibers (obliquity) varied considerably (from cross-sectional to longitudinal) across different areas within the same tissue section. Therefore, repeating the experiment under optimal TP time for obliquity affected sample allowed capture of as much useful information as possible.
Spatial RNA sequencing library preparation
The fresh frozen samples in OCT media were sectioned to 10 μm using a cryostat and mounted on 10X Visium slides and stained with hematoxylin and eosin (H&E). The H&E stained samples underwent high-resolution imaging on the confocal Dragonfly Spinning Disk and Super Resolution Microscope at the Bioimaging center of Delaware Biotechnology Institute (DBI) and were subsequently visualized using Fiji software69. After imaging, muscle samples were permeabilized and cDNA libraries were generated following the manufacturer’s instructions using Visium Spatial Gene Expression Slide & Reagent Kit (10x Genomics, PN-1000187; Pleasanton, CA). Quality analysis of the extracted total RNA and cDNA was conducted using the fragment analyzer to verify both quality and quantity, including Qubit quantification. The libraries were then sequenced in paired-end 150 bp mode on an Illumina Nextseq 2000 system (Illumina, San Diego,CA), with sequencing depth determined by the percentage capture areas covered by each muscle sample.
Histopathologic analysis and tile annotation
Microscopic analysis and annotation were conducted on a tile-by-tile basis for each image file captured per specimen by co-author P. Khondowe. The classification of samples into WB affected and unaffected categories was based on histopathologic features identified by co-author E. Brannick, an ACVP certified veterinary anatomic pathologist as described previously12. Specifically, tiles were annotated for evidence of perivascular lipid-laden macrophages (LLM), inflammatory infiltrates within the interstitium or myofibers (myositis) or myofiber degeneration typical of WB. Structurally, regions of connective tissue, blood vessels, and white adipose were also labeled. Once all tiles were annotated, the veterinary pathologist (co-author E. Brannick) reviewed all tiles/annotations for accuracy and compiled data regarding the microscopic lesions and architectural elements present in each tile for subsequent data analysis and interpretation with regional molecular expression profiles as described below.
Statistical analysis
The fiducial frames were manually aligned for each slide using Loop Browser v6.2.0 software (10x Genomics). Subsequently, sequencing data were aligned to the chicken reference genome Gallus_gallus-6a (Ensembl, database version 99) and overlaid onto the histological images using the SpaceRanger v2.0.0 (10x Genomics) to quantify gene expression and obtain the feature-by-barcode expression matrix. Unbiased k-means clustering was performed using Loup Browser v6.2.0 to determine feature clusters with k set as 10 to increase the power of cell type differentiation. Marker genes for each identified cluster were selected as up-regulated genes (adjusted P-value < 0.1).
Integration analysis was conducted using Seurat v4.3.070 in R v4.1.071, in a way that all datasets were anchored together to align the same cell types across conditions. Briefly, the 2000 most variant genes were identified as shared sources of variation across all the samples were identified by the canonical correlation analysis72. Before integration, anchors between datasets were extracted based on gene expression values followed by similarity score correction to remove incorrect anchor pairs72. This time cell type clusters were generated by the unsupervised shared nearest neighbor (SNN) graph-based clustering with resolution parameter set at 0.5 as it yielded clusters with meaningful biological interpretation without over-clustering the data72,73. Uniform Manifold Approximation and Projection (UMAP) was constructed to visualize integrated data and clusters using Seurat v4.3.070 in R v4.1.071. Marker genes were extracted for each cluster using default Wilcoxon Rank Sum test in Seurat v4.3.070 to select only up-regulated genes (adjusted P-value < 0.05) expressed in at least 25% of Visium spots in a cluster compared to all other spots. Pathway enrichment analysis was conducted using the ToppFun function of ToppGene Suite, with a false discovery rate threshold of 0.0574.
Data availability
Spatial transcriptomic RNA-Seq data are currently in the process of submission to NCBI- Sequence Read Archive (SRA) under submission ID SUB13828000.
References
Kuttappan, V. A., Hargis, B. M. & Owens, C. M. White striping and woody breast myopathies in the modern poultry industry: A review. Poult. Sci. 95, 2724–2733 (2016).
Wang, Z., Brannick, E. & Abasht, B. Integrative transcriptomic and metabolomic analysis reveals alterations in energy metabolism and mitochondrial functionality in broiler chickens with wooden breast. Sci. Rep. 13, 4747 (2023).
Mutryn, M. F., Brannick, E. M., Fu, W., Lee, W. R. & Abasht, B. Characterization of a novel chicken muscle disorder through differential gene expression and pathway analysis using RNA-sequencing. BMC Genom. 16, 399 (2015).
Abasht, B., Mutryn, M. F., Michalek, R. D. & Lee, W. R. Oxidative stress and metabolic perturbations in wooden breast disorder in chickens. PLoS One 11, e0153750 (2016).
Malila, Y. et al. Insights into transcriptome profiles associated with wooden breast myopathy in broilers slaughtered at the age of 6 or 7 weeks. Front. Physiol. 12, 691194 (2021).
Bottje, W. G. et al. Upstream regulator analysis of wooden breast myopathy proteomics in commercial broilers and comparison to feed efficiency proteomics in pedigree male broilers. Foods 10(1), 104 (2021).
Kang, K. et al. Multi-omics analysis of the microbiome and metabolome reveals the relationship between the gut Microbiota and wooden breast myopathy in broilers. Front. Vet. Sci. 9, 922516 (2022).
Chen, W.-T. et al. Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell 182, 976–991 (2020).
D’Ercole, C. et al. Spatially resolved transcriptomics reveals innervation-responsive functional clusters in skeletal muscle. Cell Rep. 41, 111861 (2022).
Liu, G. et al. Zebrafish danio rerio myotomal muscle structure and growth from a spatial transcriptomics perspective. Genomics 114, 110477 (2022).
Ruoss, S. et al. Spatial transcriptomics tools allow for regional exploration of heterogeneous muscle pathology in the pre-clinical rabbit model of rotator cuff tear. J. Orthop. Surg. Res. 17, 440 (2022).
Papah, M. B., Brannick, E. M., Schmidt, C. J. & Abasht, B. Evidence and role of phlebitis and lipid infiltration in the onset and pathogenesis of Wooden Breast Disease in modern broiler chickens. Avian Pathol. 46, 623–643 (2017).
Kim, K. et al. Transcriptome analysis reveals nonfoamy rather than foamy plaque macrophages are proinflammatory in atherosclerotic Murine models. Circ. Res. 123, 1127–1142 (2018).
Javadifar, A. et al. Foam cells as therapeutic targets in atherosclerosis with a focus on the regulatory roles of non-coding RNAs. Int. J. Mol. Sci. 22, 2529 (2021).
Kratz, M. et al. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 20, 614–625 (2014).
Nagy, L., Tontonoz, P., Alvarez, J. G., Chen, H., & Evans, R. M. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell. 93, 229–240 (1998).
Hou, X. et al. Lipid uptake by alveolar macrophages drives fibrotic responses to silica dust. Sci. Rep. 9, 399 (2019).
Vogel, A., Brunner, J. S., Hajto, A., Sharif, O. & Schabbauer, G. Lipid scavenging macrophages and inflammation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1867, 159066 (2022).
Rader, D. J., & Puré, E. Lipoproteins, macrophage function, and atherosclerosis: Beyond the foam cell? Cell Metab. 1, 223–230 (2005)
van Eijk, M. & Aerts, J. M. F. G. The Unique Phenotype of Lipid-Laden Macrophages. Int. J. Mol. Sci. 22 (2021).
Yu, X.-H., Fu, Y.-C., Zhang, D.-W., Yin, K. & Tang, C.-K. Foam cells in atherosclerosis. Clin. Chim. Acta. 424, 245–252 (2013).
Iso, T. et al. Capillary endothelial fatty acid binding proteins 4 and 5 play a critical role in fatty acid uptake in heart and skeletal muscle. Arterioscler. Thromb. Vasc. Biol. 33, 2549–2557 (2013).
Furuhashi, M., Saitoh, S., Shimamoto, K. & Miura, T. Fatty acid-binding protein 4 (FABP4): Pathophysiological insights and potent clinical biomarker of metabolic and cardiovascular diseases. Clin. Med. Insights Cardiol. 8, 23–33 (2014).
Miyazaki, M., Kim, Y. C., Gray-Keller, M. P., Attie, A. D., & Ntambi, J. M. The biosynthesis of hepatic cholesterol esters and triglycerides is impaired in mice with a disruption of the gene for stearoyl-coa desaturase 1. J. Biol. Chem. 275, 30132–30138 (2000).
Hung, Y.-H., Carreiro, A. L. & Buhman, K. K. Dgat1 and Dgat2 regulate enterocyte triacylglycerol distribution and alter proteins associated with cytoplasmic lipid droplets in response to dietary fat. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862, 600–614 (2017).
Liu, R. et al. Dominant changes in the breast muscle lipid profiles of broiler chickens with wooden breast syndrome revealed by lipidomics analyses. J. Anim. Sci. Biotechnol. 13, 93 (2022).
Wang, Z., Özçam, M. & Abasht, B. 3’UTR-Seq analysis of chicken abdominal adipose tissue reveals widespread intron retention in 3’UTR and provides insight into molecular basis of feed efficiency. PloS One 17, 0269534 (2022).
Chen, Y. et al. Mitochondrial metabolic reprogramming by CD36 signaling drives macrophage inflammatory responses. Circ. Res. 125, 1087–1102 (2019).
Brasaemle, D. L. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48, 2547–2559 (2007).
Brothers, B., Zhuo, Z., Papah, M. B. & Abasht, B. RNA-seq analysis reveals spatial and sex differences in pectoralis major muscle of broiler chickens contributing to difference in susceptibility to wooden Breast Disease. Front. Physiol. 10, 764 (2019).
Martinez, F. O., Gordon, S., Locati, M., & Mantovani, A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J.Immunol. 177, 7303–7311 (2006).
Guerrini, V., & Gennaro, M. L. Foam cells: one size doesn't fit all. Trends Immunol. 40, 1163–1179 (2019).
Van Eck, M., Zimmermann, R., Groot, P. H., Zechner, R. & Van Berkel, T. J. Role of macrophage-derived lipoprotein lipase in lipoprotein metabolism and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 20, E53–62 (2000).
Csapo R, Gumpenberger M, Wessner B. Skeletal muscle extracellular matrix - what do we know about its composition, regulation, and physiological roles? A narrative review. Front Physiol 11, 253. https://doi.org/10.3389/fphys.2020.00253 (2020).
Papah, M. B., Brannick, E. M., Schmidt, C. J., & Abasht, B. Gene expression profiling of the early pathogenesis of wooden breast disease in commercial broiler chickens using rna-sequencing. PLoS One. 13, 0207346 (2018).
Chiquet, M., Birk, D. E., Bönnemann, C. G. & Koch, M. Collagen XII: Protecting bone and muscle integrity by organizing collagen fibrils. Int. J. Biochem. Cell Biol. 53, 51–54 (2014).
Marro, J., Pfefferli, C., de Preux Charles, A.-S., Bise, T. & Jaźwińska, A. Collagen XII contributes to epicardial and connective tissues in the zebrafish heart during ontogenesis and regeneration. PLoS One 11, e0165497 (2016).
Zou, Y. et al. Recessive and dominant mutations in COL12A1 cause a novel EDS/myopathy overlap syndrome in humans and mice. Hum. Mol. Genet. 23, 2339–2352 (2014).
Gragnano, F. et al. The role of von Willebrand factor in vascular inflammation: From pathogenesis to targeted therapy. Mediators Inflamm. 2017, 1–13 (2017).
Manz, X. D., Bogaard, H. J. & Aman, J. Regulation of VWF (Von Willebrand factor) in inflammatory thrombosis. Arterioscler. Thromb. Vasc. Biol. 42, 1307–1320 (2022).
Ozawa, K. et al. Proteolysis of Von Willebrand factor influences inflammatory endothelial activation and vascular compliance in atherosclerosis. JACC Basic Transl. Sci. 5, 1017–1028 (2020).
Mussbacher, M. et al. Cell type-specific roles of NF-κB linking inflammation and thrombosis. Front. Immunol. 10, 85 (2019).
Yu, C.-W. et al. Dual role of fatty acid-binding protein 5 on endothelial cell fate: a potential link between lipid metabolism and angiogenic responses. Angiogenesis 19, 95–106 (2016).
Nichol, D., & Stuhlmann, H. Egfl7: a unique angiogenic signaling factor in vascular development and disease. Blood 119, 1345–52 (2012).
Margaritis, M. et al. Interactions between vascular wall and perivascular adipose tissue reveal novel roles for adiponectin in the regulation of endothelial nitric oxide synthase function in human vessels. Circulation 127, 2209–2221 (2013).
Manzella, N. et al. Monoamine oxidase-A is a novel driver of stress-induced premature senescence through inhibition of parkin-mediated mitophagy. Aging Cell 17, e12811 (2018).
Kumar, R. et al. CYBA (p22phox) variants associate with blood pressure and oxidative stress markers in hypertension: a replication study in populations of diverse altitudes. Hypertens. Res. 38, 498–506 (2015).
Pei, J., Pan, X., Wei, G., & Hua, Y. Research progress of glutathione peroxidase family (gpx) in redoxidation. Front Pharmacol. 14, 1147414 (2023).
Munshaw, S., Redpath, A. N., Pike, B. T., & Smart, N. Thymosin β4 preserves vascular smooth muscle phenotype in atherosclerosis via regulation of low density lipoprotein related protein 1 (lrp1). Int. Immunopharmacol. 115, 109172 (2023).
Kim, J.-H. et al. Thymosin β4-enhancing therapeutic efficacy of human adipose-derived stem cells in mouse ischemic hindlimb model. Int. J. Mol. Sci. 21, 2166 (2020).
Abasht, B., Papah, M. B., & Qiu, J. Evidence of vascular endothelial dysfunction in wooden breast disorder in chickens: insights through gene expression analysis, ultra-structural evaluation and supervised machine learning methods. PLoS One 16, 0243983 (2021).
Bechet, D., Tassa, A., Taillandier, D., Combaret, L. & Attaix, D. Lysosomal proteolysis in skeletal muscle. Int. J. Biochem. Cell Biol. 37, 2098–2114 (2005).
Gumpper, K., Sermersheim, M., Zhu, M. X. & Lin, P.-H. Skeletal muscle lysosomal function via cathepsin activity measurement. Methods Mol. Biol. 1854, 35–43 (2019).
Komura, T. et al. Clinical features of cystatin A expression in patients with pancreatic ductal adenocarcinoma. Cancer Sci. 108, 2122–2129 (2017).
Martín-Ventura, J. L. et al. Increased CD74 expression in human atherosclerotic plaques: contribution to inflammatory responses in vascular cells. Cardiovasc. Res. 83, 586–594 (2009).
Wang, X. et al. Salvia miltiorrhiza polysaccharides alleviate florfenicol-induced inflammation and oxidative stress in chick livers by regulating phagosome signaling pathway. Ecotoxicol. Environ. Saf. 249, 114428 (2023).
Corna, G. et al. The repair of skeletal muscle requires iron recycling through macrophage ferroportin. J. Immunol. 197, 1914–1925 (2016).
Kernan, K. F. & Carcillo, J. A. Hyperferritinemia and inflammation. Int. Immunol. 29, 401–409 (2017).
Kitahara, T., Kiryu, S., Takeda, N., Kubo, T. & Kiyama, H. Up-regulation of ferritin heavy chain mrna expression in the rat skeletal muscle after denervation: Detected by means of differential display. Neurosci. Res. 23, 353–360 (1995).
Yadati, T., Houben, T., Bitorina, A., & Shiri-Sverdlov, R. The ins and outs of cathepsins: physiological function and role in disease management. Cells 9, 1679–1679 (2020).
Zhang, J., Li, M., Yu, Q., Han, L. & Ma, Z. Effects of lysosomal-mitochondrial apoptotic pathway on tenderness in post-mortem bovine longissimus muscle. J. Agric. Food Chem. 67, 4578–4587 (2019).
Cacialli, P. et al. A connexin/ifi30 pathway bridges HSCs with their niche to dampen oxidative stress. Nat. Commun. 12, 4484 (2021).
Praud, C. et al. Molecular phenotyping of white striping and wooden breast myopathies in chicken. Front. Physiol. 11, 633 (2020).
Marciano, C. M. M. et al. Differential expression of myogenic and calcium signaling-related genes in broilers affected with white striping. Front. Physiol. 12, 712464 (2021).
Lake, J. A., Papah, M. B., & Abasht, B. Increased expression of lipid metabolism genes in early stages of wooden breast links myopathy of broilers to metabolic syndrome in humans. Genes, 10, 746 (2019).
Pampouille, E. et al. Differential expression and co-expression gene network analyses reveal molecular mechanisms and candidate biomarkers involved in breast muscle myopathies in chicken. Sci. Rep. 9, 14905–14905 (2019).
Stromer, M. H. The cytoskeleton in skeletal, cardiac and smooth muscle cells. Histol. Histopathol. 13, 283–291 (1998).
Papah, M. B., & Abasht, B. Dysregulation of lipid metabolism and appearance of slow myofiber-specific isoforms accompany the development of wooden breast myopathy in modern broiler chickens. Sci. Rep. 9, 17170 (2019).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. https://doi.org/10.1016/j.cell.2021.04.048 (2021).
Ripley, B. D. The R project in statistical computing. MSOR Connections 1, 23–25 (2001).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell, 177, 1888-1902.e21 (2019).
Xu, C., & Su, Z. Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics 31, 1974–1980 (2015).
Chen, J., Bardes, E. E., Aronow, B. J. & Jegga, A. G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–11 (2009).
Acknowledgements
This research is supported by U.S. Department of Agriculture, Agriculture, and Food Research Initiative competitive grant number 2021-67015- 34543. We would like to thank Jeffery Caplan, Jean Ross, and staff from the University of Delaware Bio-imaging center; along with Bruce Kingham and Mark Shaw from University of Delaware Sequencing and Genotyping Center at the Delaware Biotechnology Institute for their expertise and assistance with sample processing, imaging and RNA sequencing. We also thank Dr. Carl Schmidt for his insightful input during manuscript preparation.
Author information
Authors and Affiliations
Contributions
B.A. conceived the study, designed and planned the experiments, procured funding support, provided advisory guidance and molecular biology laboratory training, contributed to interpretation of the results, revised and edited the manuscript for intellectual content. E.B. conducted tile and histology analyses, contributed to the interpretation of histological data and revised the manuscript for content and clarity. P.K. contributed to the spatial transcriptomic experiments including sample preparation and data collection, conducted tile analysis. Z.W. contributed to the spatial transcriptomic experiments, including sample preparation and data collection. Z.W. performed data curation, and played a key role in interpreting the results. Z.W. took the lead in drafting and writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, Z., Khondowe, P., Brannick, E. et al. Spatial transcriptomics reveals alterations in perivascular macrophage lipid metabolism in the onset of Wooden Breast myopathy in broiler chickens. Sci Rep 14, 3450 (2024). https://doi.org/10.1038/s41598-024-53904-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-53904-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.