Abstract
This study aimed to explore the potential of metal oxides such as Titanate Scrolled Nanosheets (TNs) in improving the radiosensitivity of sarcoma cell lines. Enhancing the response of cancer cells to radiation therapy is crucial, and one promising approach involves utilizing metal oxide nanoparticles. We focused on the impact of exposing two human sarcoma cell lines to both TNs and ionizing radiation (IR). Our research was prompted by previous in vitro toxicity assessments, revealing a correlation between TNs' toxicity and alterations in intracellular calcium homeostasis. A hydrothermal process using titanium dioxide powder in an alkaline solution produced the TNs. Our study quantified the intracellular content of TNs and analyzed their impact on radiation-induced responses. This assessment encompassed PIXE analysis, cell proliferation, and transcriptomic analysis. We observed that sarcoma cells internalized TNs, causing alterations in intracellular calcium homeostasis. We also found that irradiation influence intracellular calcium levels. Transcriptomic analysis revealed marked disparities in the gene expression patterns between the two sarcoma cell lines, suggesting a potential cell-line-dependent nano-sensitization to IR. These results significantly advance our comprehension of the interplay between TNs, IR, and cancer cells, promising potential enhancement of radiation therapy efficiency.
Similar content being viewed by others
Introduction
Radiation therapy (RT) is a pivotal cancer treatment used alongside surgery or chemotherapy, administered either before (neo-adjuvant) or after (adjuvant) treatment, to enhance its positive effects1,2. RT employs high-energy ionizing radiation (IR) to inhibit tumor growth. IR interacts with matter in two steps: (i) a physicochemical step involving atom excitation, ionization and water radiolysis, and (ii) a biological step, resulting in macromolecular alterations and cellular lesions that activate signaling pathways. External beam RT has the inherent potential to adversely affect healthy cells by inducing damage to their DNA or triggering cell death. The affected cells can undergo different outcomes such as recovery and return to normal function, cell death, or impaired repair, leading to DNA mutations and potential cancer development. For example, studies have shown that patients undergoing an RT treatment can face radiation-related risks, including cardiovascular complications, radiation pneumonitis, development of secondary cancers, and lymphedema3,4,5,6,7,8. The efficacy of RT hinges on delivering an effective dose to the tumor, with higher doses leading to better control, known as “dose escalation”. If the delivered RT dose fails to eliminate all cancer cells, there is a risk of future regrowth9,10. However, increasing the dose in the tumor tissue also affects the surrounding healthy tissue, leading to side effects that hinder treatment continuation. Therefore, ongoing developments in RT aim to refine the differential impact on tumors and healthy tissues. They encompass the exploration of innovative irradiation techniques based on ballistics and precise beam delivery alongside the exploration of the use of charged particles11,12,13,14,15.
Furthermore, research efforts concern new radio-sensitizing, radio-enhancing, or radioprotective agents against IR16,17,18,19. Among these, nanoparticles (NPs) have attracted significant attention in medicine due to their potential applications in drug delivery, contrast-enhanced imaging, and radio-sensitizing or radio-enhancement. The primary mechanism behind the ability of NPs to act as radiosensitizers relies on the increased absorption of X-rays, leading to the emission of secondary electrons and fluorescence photons20. This process involves photoelectric effect, Compton scattering, and pair production. At low energies (below 100 keV), the photoelectric effect governs the photon attenuation, while the Compton effect becomes the dominant process for energies between 100 keV and 10 MeV, and pair production dominates above 10 MeV21,22. Since clinical practice typically uses energies between 0.3 and 20 MeV, the Compton effect is the main contributor to energy deposition with a Compton cross-section proportional to Z. Thus, high atomic number elements received priority for radio-enhancement, with gold (Z = 79) widely studied for its biocompatibility and ability to enhance radiation therapy23,24,25,26. Other elements such as bismuth (Z = 83), platinum (Z = 78), and gadolinium (Z = 64) have also attracted interest as potential radiosensitizers27.
Gold NPs combined with X-rays improved survival rates in mice with mammary carcinomas25. Bismuth oxide NPs and bismuth selenide nanoplates exhibited dose enhancements both in vitro and in vivo28,29,30. Platinum NPs amplified gamma ray effects by over 40%, even in the highly radioresistant organism D. radiodurans31. Iron oxide NPs increased the radiotherapy effectiveness and the concentration of reactive oxygen species (ROS) in prostate carcinoma cells32,33. Gadolinium-based NPs, such as AGuIX, an ultrasmall formulation of polysiloxane and gadolinium chelates, proved in vitro efficacy on various cell lines, including a radioresistant head and neck squamous cell carcinoma34,35. The in vivo enhancement radiation sensitivity has also been demonstrated in glioblastoma, brain metastases, melanoma, pancreatic cancer, liver cancer, chondrosarcoma, head and neck cancer, and lung cancer34. Crystalline hafnium oxide NPs (Z = 72), referred to as NBTXR3, displayed benefits in survival, tumor growth delay, and local control when combined with radiation therapy (RT) in both mesenchymal and epithelial human tumor xenografts36. NBTXR3 gave promising results on sarcoma treatment in phase I and phase II–III of clinical trials37. Despite an uncertain mechanism of action, the phase II–III trial revealed a higher rate of pathological complete response in the NBTXR3 group compared to radiotherapy alone. The approval of NBTXR3 as a pioneering radiation enhancer for sarcoma treatment provided the potential to facilitate surgical resection. It can now serve as a model for enhancing RT in other solid tumors38.
In addition to the physical interactions mentioned above, NPs have the potential to induce a variety of biological effects. These responses include the generation of ROS, DNA damage, and changes in cellular metabolism39. Studies indicate that the NPs size influences ROS production and DNA damage, with high surface area to volume ratios leading to increased effects40,41. With IR, NPs can cause cell cycle arrest at the G2/M transition, leading to apoptosis42,43,44. Nanodiamonds also showed cell cycle arrest in G1/S followed by senescence45. Ghita M. et al., using soft X-ray microbeams (carbon K-shell, 278 eV) to achieve selective cytoplasmic and nuclear irradiation, demonstrated that gold NPs could induce DNA damage and mitochondrial depolarization46,47. Studies discovered potent radiosensitization effects in vitro with minimal NP quantities, suggesting the influence of factors beyond NP dosage40,48,49. Oxidative stress has emerged as a pivotal player, with gold NPs inhibiting redox regulators such as thioredoxin reductase and glutathione reductase in both cancer and normal cells48,50. Some authors suggested the catalytic nature of the NPs, as radiolytic reactants present on their surface might impact their radio-enhancing abilities51.
Titanium dioxide NPs (TiO2, Z = 22) are versatile, finding use in medical applications like disinfection, dopamine detection52,53, bone growth enhancement54, and dental composites55. Their cytotoxicity depends on factors like morphology, chemical composition, and functionalization56,57,58. The NP shape also affects their biodistribution since we and others showed that elongated organic NPs are more efficiently taken up by cells than spherical ones56,59,60. TiO2 NPs can induce ROS generation61,62,63; they are here explored to optimize RT through in vitro testing. In particular, titanate nanotubes containing water, oxygen, and hydroxides can escalate free radical production when exposed to IR. This interaction increases the production of free radicals such as OH⋅, H⋅, and HO2⋅, known as radiosensitizers44,64.
In the present study, our objective is to comprehensively assess how exposure to the combination of TNs and IR impacts cellular homeostasis, cell proliferation, and gene expression.
Results
Synthesis and physicochemical properties of titanate scrolled nanosheets (TNs)
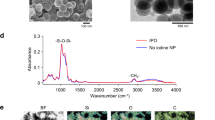
We synthesized titanate scrolled nanosheets (TNs) from AEROXIDE P25 NPs, which consist of 75% anatase and 25% rutile65,66,67. The alkaline-based hydrothermal synthesis developed by Kasuga et al. rapidly turned out to be the main non-templated method for the production of TNs68 or layered titanate nanomaterials65,66,67. P25 TiO2 is mixed into a 10 M NaOH aqueous solution, which is then submitted to hydrothermal treatment at 130 °C for 20 h, in a Teflon-lined autoclave. The product is washed with a 0.1N HNO3 solution, to reach a pH close to 7 at which point the slurry is filtered. TNs are either dried in air for analysis or kept in water for further use. The Transmission Electron Microscopy (TEM) image and XRD pattern of such synthesized TNs are shown in Fig. 1 and further detailed characteristics can be found in Simon et al.56 and Bai et al.69.
Titanate scrolled nanosheets (TNs). (a) Transmission electron microscopy images of synthesized Titanate scrolled nanosheets (Inset: high-resolution image showing the TN wall thickness. (b) XRD pattern of the TNs in (a). Histogram of the size distribution of the TNs measured on at least 300 NTs: (c) length in nm (d) diameter in nm.
The elongated nanoobjects are crystalline and consist of a thin curved structure of several layers of a single nanosheet wrapped on itself, along the c axis, leading to a nanotube with an average external diameter of about 9 ± 1.6 nm with a multiwall thickness of about 2 nm and a length of several hundred nanometers68. The transparency of the objects in their central part indicates a lower density and confirms their tubular morphology (Fig. 1a). The interlayer spacing of these nanotubes is 0.7 nm, close to reported values in the literature6,70,71,72,73. The average tube size is 172 ± 94 nm, indicating a high degree of dispersity. However, their diameter varies only slightly. The XR diffractogram of these nanosheets corresponds to a hydrogen titanate phase and the peaks have been indexed according to the H2Ti2O5⋅H2O phase (JCPDS No 47-0124), which is very close to the structure of nanosheets found in the literature74,75 (Fig. 1b). The crystal structure of these TNs is still a matter of debate. Several structures have been proposed for these objects, such as the monoclinic lattice H2Ti3O7, the lepidocrocite H0.7Ti1.825◊0.175O4, and finally the orthorhombic phase H2Ti2O567,75,76,77,78,79,80,81. However, the community as a whole is unanimous on obtaining a protonated form of titanium oxide or hydrogen titanate that can be written as H2mTinO2n+m82. Throughout the synthesis process, we kept the TNs in solution to prevent aggregation caused by drying. Despite their negative surface charge at physiological pH 7.4, TNs flocculated and quickly agglomerated after sonication even if easily re-dispersible. A stock solution with a concentration of 1 mg.ml-1 is used to achieve a cell exposure dose of 2 µg.cm−256.
Cell proliferation assay reveals differences in sarcoma cell lines based on exposure conditions: impact of TNs and ionizing radiation (IR)
Choice of sarcoma cells
Sarcomas represent a heterogeneous group of rare tumors, accounting for approximately 1% of adult cancers with more than 50 histological subtypes83. They are derived from mesenchymal tissue including bones, muscles, cartilage, and other connective tissues. Sarcoma etiology is unknown, but external RT is a well-established risk factor for soft tissue sarcoma. Indeed, sarcomas belong to the type of cancer that can be radiation-induced, and they present a higher resistance to conventional RT. We used two sarcoma cell lines83. The first, named IB115, was obtained from a dedifferentiated liposarcoma of a para-testicular tumor after surgery. IB115 has a simple genetic profile based on many known limited amplifications of the MDM2/CDK4 genes on chromosome 12q15 (Supplementary Information Fig. S1). The second, named IB106, is an unclassified sarcoma from the paravertebral mass with heterogenic pleomorphic cells with a complex genetic. Genomic stability was assessed over 50 passages via Comparative Genomic Hybridization (CGH). The CGH profiles remained unchanged, as evidenced by the overlapping CGH profiles in passages 30 and 50. This highlights their remarkable genomic stability and reinforces the interest in using these cell lines for in vitro studies83 (Supplementary Information, Fig. S2).
Sarcoma cell exposure to NTs alone
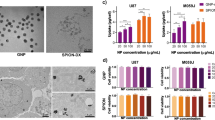
The toxicity of TNs has been assessed by cell proliferation assay. We based our choice on previous studies, which encompassed various cell lines such as primary keratinocytes, endothelial cells, and commercially available cancerous cell lines such as HeLa and HTB96 U2OS56,84. Figure 2 shows the results of a cell proliferation assay performed on IB106 and IB115 under different exposure conditions. One day after exposure to TNs, IB115 and IB106 displayed a decrease in cell proliferation of approximately 30% for IB106 and about 10% for IB115 at 0 Gy (Fig. 2b, d, white boxes). IB106 consistently exhibited higher sensitivity to TNs, as evidenced by the significant decrease in proliferation throughout the experiment. In contrast, IB115 showed a lower susceptibility to TNs exposure, with a weaker impact on the proliferation 24 h after the exposure and subsequent recovery of proliferative activity over several days (Fig. 2b, white boxes).
Cell proliferation assay on sarcoma cells under different exposure conditions. The relative number of cells per well per day was measured following an initial exposure to TNs for 20h at a dose of 2 µg.cm-2, followed by a second exposure by irradiation at different doses expressed in Gy (mean value ± standard deviation, n = 2). The irradiation doses ranged from 1 to 4 Gy, depicted in grayscale from white (control) to dark gray (4 Gy). Cell proliferation was evaluated over an 8-day post-irradiation. (a) IB115: irradiation without TNs. (b) IB115: irradiation in the presence of internalized TNs. (c) IB106: irradiation without TNs. (d) IB106: irradiation in the presence of internalized TNs.
Sarcoma cell exposure to IR alone
The effects of IR on cell proliferation are illustrated in Fig. 2a, c, which show the relative cell number per well per day during 8 days for irradiation doses ranging from 0 to 4 Gy. Notable differences in plating efficiency were observed between IB115 and IB106. One day after irradiation, IB115 exhibited a higher cell number in the irradiated 1 and 2 Gy conditions, suggesting a potential promotion of cell plating by irradiation (Fig. 2a).
Conversely, IB106 displayed a marked decrease in the cell number. These results suggest that IB106 is more sensitive to radiation than IB115. The proliferation rate of the two cell lines decreased upon irradiation, IB106 showing a rapid and significant decline with increasing dose (Fig. 2c), whereas IB115 demonstrated a progressive and delayed decrease, mainly at 2 and 4 Gy (Fig. 2a). A tendency to go back to a normal proliferation rate appeared after 6 days at a dose of 1 Gy, and both cell lines had a maximum decrease in proliferation at 6 days after irradiation (Fig. 2a, c).
Sarcoma cell exposure to IR in the presence of TNs once internalized in cells
IB115 and IB106 were first exposed to TNs for 20 h and to the different IR doses. Both cell lines exhibited alterations in their proliferation compared to the control (Fig. 2b, d, white column). After TNs internalization due to exposure to TNs at a dose of 2 µg.cm−2 for 20 h, the cells were subsequently irradiated at doses ranging from 1 to 4 Gy (6 MeV photon beams). The combined effect of internalized TNs and IR on cell proliferation induces a significant decrease in the cell number for both cell lines (Fig. 2b, d). This decrease reaches a maximum on day 6 for all the doses. Significantly, there is a clear contrast in sensitivity between the two cell lines. IB106 displays a slightly enhanced response to both TNs and IR, particularly at the 1 Gy dose. On days 6 and 8 in the absence of TNs, cells exposed to a radiation dose of 1 Gy displayed an increase in cell number. This indicated that their proliferation was not definitively impaired. However, in the presence of TNs, this increase in cell number was not observed, suggesting a potential interaction between TNs and the cellular response to radiation.
Effect of TNs and IR on sarcoma intracellular calcium content
We investigated both the intracellular content of titanium (Ti) and its influence on intracellular calcium concentration using micro-PIXE (Particle-Induced X-ray Emission) (Fig. 3).
TNs, irradiation, and calcium homeostasis. Quantitative analysis of the total intracellular titanium and calcium contents by micro-PIXE in sarcoma cell lines in the different tested experimental conditions, i.e., in the presence (or not) of 2 µg.cm−2 of TNs and combination (or not) of irradiation (0, 1, 2 and 4 Gy). Quantitative analysis was performed in the absence of TNs (red) or in the presence of TNs (blue). Sarcoma cell lines are depicted in rows (IB115-top; IB106-down). (a,c) Titanium content is expressed in ng.cm−2 for IB115 and IB106, respectively. (b,d) Calcium content is expressed in ng.cm−2 for IB115 and IB106, respectively.
The results depicted in Fig. 3 focus on the quantification at the single-cell level of titanium uptake (Fig. 3a, c) and calcium uptake (Fig. 3b, d), expressed as normalized values in ng.cm-2. First, the analysis revealed the presence of Titanium (Ti) with no notable difference in quantity and distribution between IB115 and IB106. No differences in the composition and distribution of chemical elements such as Na, Mg, Cl, P, K, and S were noticed across the various conditions (data not shown). Nevertheless, intracellular titanium content in both cell populations exhibited a marked heterogeneity, ranging from 50 to 250 ng.cm−2, as documented in previous studies84.
The intracellular calcium levels were also modified, considering not only the impact of both IR (Fig. 3b, d, red boxes, 1–4 Gy) and TNs (Fig. 3b, d, green box, 0 Gy ) acting separately but also the combined effect of these 2 exposures (Fig. 3b, d, green boxes, 1–4 Gy). Our results revealed a substantial presence of intracellular calcium in the two cell lines under baseline conditions, without TNs nor irradiation (ranging from 22 ± 5 to 26 ± 7 ng cm−2 in controls), with no significant difference between the cell lines. Exposure to TNs without irradiation significantly raised intracellular calcium content in IB115 (32 ± 14 ng.cm−2), whereas IB106 experienced no significant change (calcium content increase, 31 ± 9 ng.cm−2). When considering IB115 under irradiation alone, no difference in calcium content was observed between controls and those irradiated at doses of 1 and 2 Gy. A difference was only observed at a dose of 4 Gy. Moreover, the presence of both TNs and irradiation induced a marked increase in the calcium content within the same range as that observed for 4 Gy irradiation alone. These observations suggest that (i) irradiation has no impact on the calcium homeostasis in IB115, unlike TNs; (ii) IB115 cells exhibit resistance to irradiation at doses below 4 Gy; and (iii) the presence of TNs alone is sufficient to induce a similar effect as a 4-Gy irradiation.
In IB106, except for the condition at 4 Gy with a notable increase in calcium content observed, Fig. 3 illustrates that both irradiation and TNs resulted in a moderate increase in intracellular calcium content. The facts that (i) the presence of TNs consistently correlated with an increase in intracellular calcium content, and (ii) the irradiation alone did not have a significant effect could indicate that TNs induce a maximum increase in intracellular calcium content, and irradiation does not further enhance this effect under any condition. Unlike IB115, the differences in calcium content increase in IB106 were relatively low (31 ± 9 ng.cm−2). However, it should be noted that at a dose of 4 Gy, cells exposed to both TNs and irradiation could not be analyzed due to extensive cell death. This observation further supports the argument that IB106 cells were indeed sensitive to both IR and TNs and that a minimum irradiation dose is mandatory.
The changes in the intracellular calcium concentration induced by the presence of both TNs and irradiation have a minimal effect on the cell proliferation capacity of IB115 while in contrast, they significantly impact the response capacity of IB106 to TNs.
Transcriptomic analysis showed significant differential gene expression, supporting the hypothesis of TNs-induced sensitization to IR
We investigated the involvement of some specific cellular mechanisms in responses to TNs and radiation exposures. We examined the influence of these conditions on all the transcriptomic responses in both cell lines. Transcription analysis provided insight into the physiological status of the cell lines under different stress conditions. We performed principal component analysis (PCA) on expressed genes to identify which gene expression could discriminate between exposure conditions (Supplementary Information Figs. S3a and S4a). PCA is a mathematical method used to reduce the dimensionality of complex data while retaining important information. During the process, new variables, Principal Components (PC), are built from genes with the most variation among all samples. These genes are ranked into multiple PCs (PC1, PC2, PC3, etc.), each describing a decreasing proportion of the variability in the data. While PCA does not provide precise information on transcriptomic expression profiles, it does offer an overview of the global distribution of each cell line according to the experimental conditions. It is, therefore, performed on the non-normalized expression matrix to capture the major expression trends that differentiate the experimental conditions (Fig. 4).
Cluster map of Euclidean distances extracted from the PCA plot and upset plots. (a–c) Cluster map of Euclidean distances extracted from the PCA plot (a) IB115, (b) IB106. Samples with low Euclidean distances (black) are more similar in expression than samples with high Euclidean distances (white). (b–d) Upset plot showing intersections of differentially expressed genes between experimental conditions. (c) IB115, (d) IB106. Horizontal line: experimental condition. Vertical line: intersection size between conditions marked with a black dot.
We observed significant differences between the cell lines. The IB106 cluster map clearly shows three groups of samples with similar expressions: controls, irradiated, and TNs only. The absence of a discernible structure for IB115 indicates that no particular group stands out, making it impossible to discriminate between samples from a specific experimental condition. Lower eigenvalues of PC1 (19%) and PC2 (16%) for IB115 than for IB106 (29% and 26%) indicate a reduced percentage of variance within the set of samples explained by these principal components. This suggests that in IB115, it is more challenging for the algorithm to identify distinct expression trends. For the PC1 and PC2 of each cell line, we extracted the 100 genes with the highest weight. PC1s from both cell lines had only 17 genes in common, while PC2s shared 73 genes. This tends to confirm that the overall expression trends identified by the PCA in the two cell lines do not involve the same genes overall; with less than half of the genes in the PC1 and PC2 components being in common between the two cell lines.
We then performed a differential expression analysis on all conditions and compared them to controls to compute differentially expressed (DE) genes (Fig. 4c, d). The observed difference in the number of DE genes aligns with the findings from PCA, indicating a clear transcriptomic response in IB106 (239 to 469 DE genes) and the absence of such a response in IB115 (1 to 18 DE genes). In IB106, TNs alone exhibited 469 DE genes, the highest amount found across all conditions (Fig. 4d and Supplementary Information Fig. S4b). Although we cannot quantify the exact level of response based solely on the number of DE genes, it is noteworthy that the combined action of TNs and IR results in a decrease in the number of DE genes (1 Gy-TNs: 327 genes, 2 Gy-TNs: 330 genes) compared to the exposure to TNs alone, although higher than irradiation alone (1Gy: 239 genes, 2 Gy: 314 genes). This observation suggests that the cellular response to irradiation has a more pronounced influence than the response to TNs in this particular cell line (Fig. 4d and Supplementary Information, Fig. S4b).
In the case of IB115, only 1 DE gene is identified under the TNs condition. In the irradiation conditions (1 Gy: 8 genes, 1Gy-TNs: 14 genes, 2 Gy: 18 genes, 2 Gy-TNs: 5 genes), with 3 genes being common to irradiation and irradiation + TNs conditions (Fig. 4c and Supplementary Information, Fig. S3b). As the cellular response to the different experimental conditions in IB115 was weak, we focused on the DE genes identified in IB106. We were particularly interested in the difference in response between the irradiation conditions and the TNs-only condition. The PCA already showed that not only did the cellular response to irradiation differ significantly from the response to TNs but also that the response to irradiation seemed to eclipse that to TNs, with irradiation + TNs conditions grouped with irradiated conditions without TNs. This observation is confirmed in the DE genes identified in the different conditions of the IB106 line (Fig. 5).
Differential gene expression in IB106. (a) Venn diagram of common and condition-specific DE genes in TNs and irradiation samples from the IB106 cell line. (b) Number of DE genes identified per irradiation condition from the IB106 cell line. The hatched area represents the amount of DE genes common to all irradiation conditions.
Three main groups of DE genes can thus be identified: (i) DE genes common to all experimental conditions (99 genes), which should therefore correspond to general stress pathways, (ii) DE genes specific to TNs-only conditions (281 genes), which should correspond to the specific response to TNs, and (iii) DE genes common to all irradiation conditions (108 genes), with or without TNs, which should correspond to the specific response to irradiation. Not all DE genes were found in these three groups. For example, irradiation-specific DE genes accounted for less than half of all DE genes found in each irradiation condition (Fig. 5b). Despite their proportion, these genes effectively represented the two distinct expression profiles identifiable in these data. The high number of DE genes specific to the TNs-only condition, and therefore absent in the irradiation + TNs conditions, confirms the PCA observation that the cellular response to irradiation supersedes the response to TNs.
In IB106, we observe a positive correlation between the number of DE genes and the number of impacted Gene Ontologies (GOs) (Supplementary Information, Fig. S4c). In contrast, the low number of DE genes in IB115 limits our ability to classify GOs as significantly impacted, with only a few found in the 1Gy-TNs and 2Gy conditions (Supplementary Information, Fig. S3c).
Our analysis revealed four main relevant cellular pathways that impacted IB106: (i) protein metabolism, (ii) cell cycle, (iii) cellular respiration, and (iv) cell stress/death (Supplementary Information, Table S1–S4). As illustrated in Fig. 6, the majority of impacted ontologies undergo significant modification in response to the exposure to TNs. These suggest that TNs induced a global alteration of general metabolic pathways such as protein metabolism, cellular respiration, and cell cycle regulation. These observations have been partially described previously with the induction of the Endoplasmic Reticulum stress pathways (ER-Stress) and mitochondrial stress56. Nevertheless, these changes sign an unusual cellular status that could alter the radiation-induced response and thus promote the deleterious effects of IR.
Heatmap of target genes ontologies (biological processes) significantly impacted in the IB106 line. Genes ontologies (biological processes) significantly impacted (adjusted p-value < 0.05) related to the four identified metabolic pathways (GO:Protein metabolism, GO:Cellular respiration, GO:Stress/apoptosis, GO:Cell cycle) and to the different experimental conditions. Each line corresponds to a Gene Ontology (column for protein metabolism). Color corresponds to the negative log of the pvalue from the gprofiler significance test (red: lower p-values, yellow: higher p-values). White cases mean the Gene Ontology is not impacted in the corresponding experimental condition.
Discussion
Here, we focused on (i) synthesizing and characterizing titanate scrolled nanosheets (TNs) and (ii) assessing their effects in combination with ionizing radiation (IR) on two sarcoma cell lines. Micro-PIXE analysis confirmed that sarcoma cells internalize TNs as shown with TiO2 NPs by various cell types5,8,60. Quantification of intracellular titanium content revealed variation in cellular uptake, irrespective of their size, shape, and surface reactivity. Furthermore, Mirjolet et al. have provided evidence of TN internalization in glioblastoma cell lines44. Additionally, they showed that TNs were mainly located in the cytoplasm rather than in the nucleus. Since the primary effect of IR is DNA damage, it would be interesting to consider whether the localization of TNs facilitates or not the emergence of non-targeted effects of IR.
We then confirmed that the intracellular internalization of titanium (via TNs) did induce an alteration in the intracellular calcium content, as previously reported56. Interestingly, we did not observe any significant difference in the intracellular uptake of titanium (TNs) between the two cell lines. However, the presence of this intracellular titanium significantly influenced the intracellular calcium content, and this impact varied depending on the nature of the cell lines. Hence, we conclude that discrepancies in performed biological assessments, such as cell proliferation and transcriptomic analysis, could not be attributed solely to the variations in the intracellular TNs content within the studied cell populations. Instead, it seems that the alteration of intracellular calcium induced by the presence of intracellular titanium holds a central position in shaping calcium-dependent biological responses.
Our results revealed variations in cell proliferation, indicating the influence of TNs on both cell lines, with varying degrees of sensitivity; IB106 exhibiting the most pronounced response to TNs. Also, note that the intracellular content of titanium (TNs) is likely to decrease gradually within the cells as they proliferate and divide. Then, once TNs content has reached a minimal level, the cells regain their normal proliferation rate56. The fact that TNs induce a decrease in cell proliferation illustrates a potential fragilization of the cancer cells and supports the radiosensitization hypothesis, where TNs could enhance the effects of IR. The link between the intracellular content of TNs and the alteration in intracellular calcium homeostasis is a significant finding since it can influence various cellular and signaling mechanisms, including reactive oxygen species (ROS) production, mitochondrial metabolism, and inflammation. Of particular interest is the fact that the two cell lines, when exposed to IR, have a similar alteration in calcium homeostasis. TNs may reinforce this effect for the sarcoma cell lines studied. Since TNs and IR used separately had a detrimental effect on cancer cells, their combination could lead to an amplified disruption of calcium regulation within the cells, resulting in increased oxidative stress, inflammation, and potential damage to cellular structures. Our results highlight the complex interplay between TNs, IR, intracellular calcium homeostasis, and associated cellular responses.
The transcriptomic analysis, which provides valuable insights into the changes in gene expression that occur in response to the combination of TNs and IR, corroborates this hypothesis. We identified specific gene ontologies modulated by the interaction between TNs and IR. These include cellular respiration, apoptosis, cell cycle, and protein metabolism. All these pathways are known to be related to calcium homeostasis. The latter could be the expression of the ER-stress response as previously described in the context of TiO2 NPs toxicity evaluation56. Additionally, the distinct transcriptomic profiles observed in the two cell lines indicate that they respond differently to TNs and to the combination of TNs and IR. This helps us to understand the underlying mechanisms by which TNs enhance the effects of radiation and contribute to the observed cell sensitivity.
Indeed, in the case of IB115, the lack of significant induction in cell signaling indicates a reduced ability of these cells to respond to or sense the presence of TNs and the impacts of IR. As a result, the cellular response pathways associated with intracellular calcium homeostasis and related stress signaling in these cells are likely altered. This could result in a decreased sensitivity to the synergistic effects of TNs and IR, potentially contributing to cellular stress resistance. Understanding the factors influencing the resistance observed in IB115 not only provides insights into potential intervention targets but also sets the stage for developing strategies to enhance the sensitivity of cancer cells in the context of future therapy.
Conclusion
Our results, taken together, highlight the importance of considering the impact of TNs on intracellular calcium homeostasis. They provide a basis for further research to understand the underlying mechanisms responsible for these effects in different cell types, including sarcoma cells. This detrimental alteration in intracellular calcium levels in the cytoplasm opens new possibilities to optimize radiotherapy using sensitization with metal oxide nanoparticles (TNs in particular). Furthermore, intracellular calcium content may serve as a bioindicator of nanotoxicity and may help researchers and clinicians evaluate the potential safety and effectiveness of TNs or metal oxide nanoparticles as radiosensitizers in combination with radiation therapy. Further research is needed to validate these findings, establish the optimal treatment conditions, and better understand the biological mechanisms involved, ultimately paving the way for potential clinical applications in the future. This approach includes evaluating the dose-dependent sensitivity of various cancer cell types to the combination of TNs and different types of ionizing radiation (Energy, Linear Energy Transfer, irradiation modality). Finally, the development and optimization of TNs delivery and the specific targeting of cancer cells are essential steps to master their potential use as radiosensitizers and to promote their clinical application in combination with radiotherapy. To further progress in this direction, researchers need to develop and validate three-dimensional (3D) multicellular biological models. They should have such a design to fit with experiments under conditions of combined exposure to titanium oxide nanomaterials and ionizing radiation. 3D multicellular models would, indeed, provide a more realistic representation of the tumor microenvironment when compared to traditional 2D cell cultures. Such a model would constitute a valuable platform for studying the efficacy, safety, and underlying mechanisms of TNs as radiosensitizers.
Material and methods
Synthesis and characterization of titanate-scrolled nanosheets
P25 nanoparticles (P25, AEROXIDE) were kindly provided by Degussa/Evonik and used as a precursor for the synthesis of titanate-scrolled nanosheets (TNs). TNs were produced via the hydrothermal process described by Kasuga et al.67. Briefly, 2 g of P25 were introduced in a 50 mL Teflon-lined autoclave with 28 mL of 10 M sodium hydroxide solution, sealed and heated at 130 °C for 20 h. The white precipitate was washed with nitric acid (0.1 M) and water for neutralization and identified as TNs. The obtained white powder was finally washed with deionized water. Synthesized TNs were kept in an aqueous solution avoiding aggregation issues. Mass concentrations were measured by drying a known volume of solution and weighing the extracted powder. Suspensions with a concentration of 1 mg.mL−1 were finally produced, sonicated, and kept in the dark. The physicochemical properties of TNs were characterized by standard techniques. Specifically, the primary size and morphological features of TNs were observed by TEM and high-resolution transmission electron microscopy (HRTEM) using a Hitachi H-7650 transmission electron microscope (TEM, 120 kV, Hitachi High-Tech Company, Japan), and a JEOL 2200 FS equipped with a field emission gun, operating at 200 kV and with a point resolution of 0.23 nm. HRTEM micrographs were acquired with a GatanUltrascan CCD 2k–2k and digital diffractograms were calculated using the Gatan Digital Micrograph program. Moreover, to be representative and statistically meaningful, many images from several regions of various samples were recorded and the most characteristic results are presented here, and at least 300 NPs were treated. Powder X-ray diffraction was used to determine the phase composition and crystallite size of TNs and was performed with a Philips PW1820 diffractometer.
Sarcoma cell lines and culture conditions
Sarcoma cell lines, named IB106 and IB115, were characterized and obtained from Institut Bergonié (Bordeaux, France)83. The two cell lines were maintained in RPMI 1640 GlutaMAX (Thermo Fischer Scientific, Illkirch, France, Cat. No. 61870036) supplemented with Fetal Bovine Serum (10% v/v, FBS, Thermo Fischer Scientific, Illkirch, France, Cat. No. 16170-078) and streptomycin/penicillin (100 µg mL−1, Thermo Fischer Scientific, Illkirch, France, Cat. No. 15140-122). Cells were kept in a humidified atmosphere at 37 °C and 5% (v/v) CO2. For both electron and proton irradiation experiments, 20,000 cells were seeded in a drop in specific cell dishes 20 h before irradiation. As a control, a mock sample was used, which was treated in the same way except for irradiation.
Cell culture, titanate-scrolled nanosheets, and irradiation exposures
Sarcoma cell lines were grown in the defined medium at 37 °C in a 5% (v/v) CO2, humidified atmosphere, and passages were realized at 80% confluency. The suspensions of TNs were prepared in ultrapure water at a concentration of 1 mg mL−1. TNs were dispersed by intense sonication pulses of 1 min at RT (750 W, 20 kHz, with 30% amplitude) using a dedicated 3MM conical microprobe with Vibra-Cell™ (750 W, 28% amplitude, Sonics & Materials, Inc., Newton, CT, USA). Suspensions were hereby known as “stock suspensions”. Stock suspensions were diluted at the appropriate concentration in a defined culture medium to obtain an exposure suspension at 2 µg.cm−2 (final concentration). Briefly, 20,000 cells were seeded in a single drop in the middle of a 6-well plate for 24 h in an appropriate culture medium, and then exposed to TNs for 16 to 24 h. Cell irradiation was carried out at the Department of Radiotherapy, Institut Bergonié (Bordeaux, France) using a Clinical Linear Accelerator (CLINAC 21EX, Varian Medical Systems) used in the routine treatment of patients. Sarcoma cell lines were irradiated using 6 MV photon beams with 1, 2, and 4 Gy delivered at a dose rate of 2 Gy.min−1. These doses are selected as irradiation doses because 2 Gy is the fractionated dose used during patient treatments; 1 and 4 Gy are lower and higher doses compared to this standard value. Source Fantom Distance (SFD) of 100 cm was applied at the surface of a certain amount of equivalent water 30 × 30 cm2 slabs (RW3, PTW) providing sufficient backscatter conditions (> 15 cm) for a 6 MV photon beam (TRS 398, IAEA). The cell monolayer was covered by a total of 15 mm of water equivalent medium to achieve electronic equilibrium (10 mm of water in the wells and 5 mm of plastic slabs above). Interstices were also filled with water to prevent the creation of heterogeneities that could disturb the secondary particle fluences and hence alter the dose deposition precision. The cell monolayer was covered by 10 mm of medium to achieve electronic equilibrium. To maintain this depth without removing the TNs, 9.6 ml of the growth medium was added one hour before irradiation. The cell monolayer was covered by a total of 15 mm of water equivalent medium to achieve electronic equilibrium (10 mm of water in the wells and 5 mm of plastic slabs above). Interstices were also filled with water to prevent the formation of heterogeneities that could disturb the secondary particle fluences, and hence alter the dose deposition precision. The photon beam was collimated in a 15 × 15 cm2 square field at isocenter and irradiations were carried out with a single beam oriented at 0° (single vertical beam). The dose verification was performed on the day of the experiment as it is performed every day before clinical treatment, and the criterion of acceptability is that the variation in dose is less than 1% compared with the local reference. As a control, a mock sample was used, which was treated in the same way except for TNs exposure and/or irradiation.
Cellular proliferation assay
Sarcoma cells were harvested with trypsin–EDTA (0.05%, v/v, Thermo Fischer Scientific, Illkirch, France, Cat. No. 25300062) right away irradiation, and 2000 cells were seeded in a 12-well cell culture plate (Cellstar™, Grenier Bio-one, Dutscher Sas, Bernolsheim, France, Cat. No. 665180). Every 2-day the culture medium was removed, cells were paraformaldehyde fixed (4% w/v, Sigma-Aldrich, Saint-Quentin Fallavier, France, Cat. No. P6148-500G) in phosphate-buffered saline (PBS 1X, pH 7.4, Thermo Fischer Scientific, Illkirch, France, Cat. No. 10010023) for 15 min at room temperature and conserved at − 20 °C in ethanol 70% (v/v). Cell nuclei were stained for 10 min with Hoechst33342 (1 µM, Thermo Fischer Scientific, Illkirch, France, Cat. No. H3570). Data acquisitions were performed with a Zeiss AxioObserver Z1 microscope (Carl Zeiss Micro-Imaging S.A.S, Rueil-Malmaison, France, AxioObserver Z1). Two independent experiments were performed in duplicates.
Cell preparation for ion beam micro-analysis (IBA)
Sarcoma cells were cultured directly onto ion beam microprobe sample holders as adapted from previous studies85,86. Briefly, cells were directly grown on 2 µm-thick polycarbonate foil for 24 h in an appropriate culture medium and then exposed (or not) to TNs for 16 to 24 h before irradiation. Control cells were prepared similarly with no addition of TNs and no irradiation exposure. 24h after the irradiation sequence, cells were rinsed once in the culture medium and very briefly rinsed twice in ultrapure water to remove excess extracellular salts from the culture medium. Finally, cells were plunge-freeze at -150°C into liquid nitrogen chilled 2-methyl butane (ReagentPlus, ≥ 99%, Sigma-Aldrich) and freeze-dried using a freeze-dryer (Christ alpha, Thermo Fischer Scientific, Illkirch, France) in two phases: A phase of primary desiccation of 12 to 24 h (T°C = −99 °C, P = 0.001 mbar) followed by a phase of secondary desiccation of 24 h (T°C = + 40 °C, P = 0.001 mbar). With no contamination and no ionic diffusion, freeze-drying was found to be the most adapted process. Single-cell quantitative analysis of the chemical element (quantitative analysis and element distribution) was carried out with a high-resolution microprobe beamline (AIFIRA, LP2iB) using complementary ion beam analytical techniques (μ-STIM, μ-PIXE). Protocols are described in previous works84,86. Micro-PIXE involves the collision of incident protons with target atoms, resulting in the emission of X-rays. This technique provides valuable information about the chemical composition and two-dimensional elemental maps of the sample. It offers numerous advantages, including simple sample preparation, a large observable area with zooming capabilities, easy quantification of chemical elements expressed in ng.cm−2, and the ability to extract single-cell content within a cell population. It enables the simultaneous collection of trace elements from sodium to heavy metals, offering sub-cellular spatial resolution and high sensitivity.
Conversion of calcium content expressed in ng cm−2 into µM content
Indirect quantification of calcium content expressed in mole per liter is possible provided the hydrated cellular mass. Then, considering a water content in cultured cells of 97% and a dry cellular mass of 100 µg per square centimeter, the basal content of calcium in control cells, measured around 20 ng.cm−2 corresponds to a total intracellular calcium concentration of 5.4 µM.
Library preparation and sequencing (Oxford Nanopore, PCR-cDNA barcoding).
Two key considerations emerged regarding the experimental conditions. First, we encountered a multiplicity of conditions involving two cell lines, TN exposure at doses of 0 and 2µg cm−2, and exposure to IR at doses of 0, 1, and 2 Gy. Second, there is a limitation in the number of samples available for each condition. To address these challenges, we chose to use the Oxford Nanopore Technologies protocol, which allows for long-read and multiplexed sequencing of cDNA collections. The extracted transcriptomes were then prepared for sequencing using the PCR cDNA barcoding kit to sequence multiple conditions on a single chip. The biological triplicates were performed on three different Flow cells.
Total RNA was extracted from cells at different times (from 1 to 24 h) after exposure to NPs and/or irradiation using the RNeasy mini kit (Qiagen SAS, Courtaboeuf, France cat. no 74104) according to the manufacturer’s instructions. Whole-transcriptome cDNA libraries were first constructed from extracted mRNA using a PCR-cDNA barcoding kit (Oxford Nanopore Technologies, Oxford, UK, cat. no SQK-PCB109). Briefly, 50 ng total RNA (~ 1 ng (polyA)+ mRNA) from each condition was taken in RNase-free PCR tubes containing Maxima H Minus reverse transcriptase enzyme (Thermo Fischer Scientific, Illkirch, France, Cat. No. EP0751), VN primer (2 µM, variant of oligo (dT) with complementary nucleotides for annealing of barcode primers), 10 mM dNTPs (New England Biolabs, Evry, France Cat. No. N0447S), RNaseOUT (40 U/µl, Thermo Fischer Scientific, Illkirch, France, Cat. No. 10777019) and strand-switching primer (10 µM, Oxford Nanopore Technologies Oxford, UK). Products were sequentially added and prepared as recommended by the manufacturers. Samples were incubated at + 42 °C for 90 min followed by enzyme inactivation at + 85 °C for 5 min (one cycle) in a thermocycler for the generation of full-length cDNA from poly(A)+ messenger RNA.
cDNA libraries from each sample then underwent full-length amplification and sample barcoding using 14 cycles of PCR. Each PCR reaction mix consisted of 25 µl 2 × LongAmp Taq master mix (New England Biolabs, Evry, France Cat. No. M0287S), 1.5 µl barcoded primers (named BP01 to BP12), 18.5 µl nuclease-free water, and 5 µl (~ 0.25 ng) cDNA. The following PCR cycling conditions were used: initial denaturation at 95 °C for 30 s (1 cycle), denaturation at 95 °C for 15 s (14 cycles), annealing at 62 °C for 15 s (14 cycles), extension at 65°C for 15 s (14 cycles), and final extension at 65 °C for 6 min (1 cycle) (the long extension time of 6 min was to selectively amplify cDNAs up to ~ 5 kb in length). 1 µl of Exonuclease 1 (New England Biolabs, Evry, France, Cat. No. M0293S) was finally added in each PCR tube for incubation at + 37 °C for 15 min and followed by enzyme inactivation at + 80 °C for 5 min (one cycle). To clean up the cDNA libraries, PCR reactions with the same barcode (from BP01 to BP12) were pooled in two 1.5 ml DNA tubes and primer dimers, removed using 0.8 × volume equivalent Agencourt® AMPure® XP beads (Beckman Coulter, Villepinte, France, Cat. No. A63880). Briefly, beads (80 µl) were added to each pooled sample, incubated on a hula mixer for 5 min at room temperature, and spun and pelleted on a magnet. Supernatants were pipetted off and the resulting beads were washed with 70% (v/v) ethanol (200 µl freshly prepared using nuclease-free water) without disturbing the pellet. The ethanol was removed using a pipette and the beads were washed again with ethanol, and the pelleted beads spun down and placed back on the magnet. Residual ethanol was pipetted off and the beads were briefly allowed to dry. While the beads still appeared glossy (with no cracking) they were resuspended in 12 µl elution buffer (provided with the PCR-cDNA barcoding kit; Oxford Nanopore Technologies, Oxford, UK) to recover the cDNA libraries. Before loading the barcoded cDNA libraries onto the flow cell for long-read sequencing, 100 ng of each cleaned-up cDNA library were ligated with adapters. The barcoded libraries (100 fmol final) were pooled in a 1.5 ml DNA, combined with Rapid adapters (Oxford Nanopore Technologies, Oxford, UK, Cat. No. SQK-PCB109), and incubated at room temperature for 5 min. The adapter-ligated pooled cDNA library was transferred to a fresh tube and combined with loading beads and sequencing buffer (SQK-PCB109 kit) to form the sequencing mix. Three flow cells R9.4.1 (Oxford Nanopore Technologies, Oxford, UK, Cat. No. FLO-MIN106D) were primed with pre-mixed flush buffer and flush tether (Oxford Nanopore Technologies, Oxford, UK, cat. no. Flow Cell Priming kit EXP-FLP002) then loaded with the sequencing mixes and run for 24 h on a MinION Mk1C sequencing device (Oxford Nanopore Technologies, Oxford, UK). The sequencing libraries from one of the IB115 triplicates were of lower quality than the other libraries, resulting in fewer reads of poorer quality. It was therefore decided to exclude this IB115 triplicate from the rest of the analysis to avoid introducing a bias linked to sample preparation.
Transcriptome analysis
Upon completion, guppy v6.1.4 was used to perform high-accuracy base-calling and demultiplexing of the data (Raw fast5 files). Reads were mapped with minimap2 option '-ax map-ont' to the Homo sapiens GRCh38 transcriptome and pooled into an expression matrix via a dedicated Python script. Differential expression analysis was performed on R with edgeR and limma packages. Genes with expression intensity lower than 1 count per million (cpm) are considered non-expressed and discarded from the analysis.
PCA
Principal Component Analysis was performed with the plotMDS (limma) on the unnormalized expression matrix of all genes > 1cpm. The Euclidean distances were obtained from the coordinates position of the 2D representation of PC1 and PC2 values in all experimental conditions and presented via a cluster map (seaborn Python package). The top 100 genes with the most weight on PC1 and PC2 respectively were extracted for later comparison with differentially expressed genes.
Differential expression
Data are normalized (limma-voom) and transformed into log2, a linear model is then adjusted for each gene. The contrasts are then extracted and compared to the linear model according to the empirical Bayesian method, the obtained p values are adjusted by the Bonferroni method87 to obtain the final differential expression results.
Enrichment analysis
Enrichment analysis is performed with the gprofiler288 R package, which queries public APIs from several databases and determines statistically significant results based on the number of impacted genes and the total number of genes attached for each ontology. It is worth noting that we did not differentiate the DE genes based on whether they were under-expressed or over-expressed, as our primary focus was on identifying the types of impacted pathways in the cellular response to the exposure conditions.
Data availability
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation (contact H. Seznec, herve.seznec@cnrs.fr). The datasets generated and analysed during the current study are available in the Gene Expression Omnibus (GEO) repository [(GSE248011) (NCBI tracking system #24377339)].
References
Tang, L. et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J. Exp. Clin. Cancer Res. 37, 87 (2018).
Atun, R. et al. Expanding global access to radiotherapy. Lancet Oncol. 16, 1153–1186 (2015).
Sardar, P. et al. Long-term cardiovascular mortality after radiotherapy for breast cancer: A systematic review and meta-analysis. Clin. Cardiol. 40, 73–81 (2017).
Mazonakis, M. & Damilakis, J. Cancer risk after radiotherapy for benign diseases. Phys. Med. 42, 285–291 (2017).
Toma-Dasu, I., Wojcik, A. & Kjellsson Lindblom, E. Risk of second cancer following radiotherapy. Phys. Med. 42, 211–212 (2017).
Cheng, Y.-J. et al. Long-term cardiovascular risk after radiotherapy in women with breast cancer. J. Am. Heart Assoc. 6, e005633 (2017).
Taylor, C. W. & Kirby, A. M. Cardiac side-effects from breast cancer radiotherapy. Clin. Oncol. (R Coll. Radiol.) 27, 621–629 (2015).
Bledsoe, T. J., Nath, S. K. & Decker, R. H. Radiation pneumonitis. Clin. Chest Med. 38, 201–208 (2017).
Huang, R.-X. & Zhou, P.-K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target Ther. 5, 60 (2020).
Olivares-Urbano, M. A., Griñán-Lisón, C., Marchal, J. A. & Núñez, M. I. CSC radioresistance: A therapeutic challenge to improve radiotherapy effectiveness in cancer. Cells 9, 1651 (2020).
Paganetti, H. et al. Relative biological effectiveness (RBE) values for proton beam therapy. Int. J. Radiat. Oncol. Biol. Phys. 53, 407–421 (2002).
Choudhary, D., Srivastava, M., Sarma, A. & Kale, R. K. Effect of high linear energy transfer radiation on biological membranes. Radiat. Environ. Biophys. 37, 177–185 (1998).
Yock, T. et al. Proton radiotherapy for orbital rhabdomyosarcoma: Clinical outcome and a dosimetric comparison with photons. Int. J. Radiat. Oncol. Biol. Phys. 63, 1161–1168 (2005).
Wilson, R. R. Radiological use of fast protons. Radiology 47, 487–491 (1946).
Miller, J. Recent measurements for hadrontherapy and space radiation: Nuclear physics. Phys. Med. 17(Suppl 1), 45–49 (2001).
Ngwa, W., Kumar, R., Moreau, M., Dabney, R. & Herman, A. Nanoparticle drones to target lung cancer with radiosensitizers and cannabinoids. Front. Oncol. 7, 208 (2017).
Boateng, F. & Ngwa, W. Delivery of nanoparticle-based radiosensitizers for radiotherapy applications. Int. J. Mol. Sci. 21, 273 (2019).
Schaue, D. & McBride, W. H. Opportunities and challenges of radiotherapy for treating cancer. Nat. Rev. Clin. Oncol. 12, 527–540 (2015).
Ngwa, W. et al. Smart radiotherapy biomaterials. Int. J. Radiat. Oncol. Biol. Phys. 97, 624–637 (2017).
Retif, P. et al. Nanoparticles for radiation therapy enhancement: The key parameters. Theranostics 5, 1030–1044 (2015).
Currell, F. & Villagomez-Bernabe, B. Physical and chemical processes for gold nanoparticles and ionising radiation in medical contexts. In Gold Nanoparticles for Physics, Chemistry and Biology. 509–536 https://doi.org/10.1142/9781786341259_0015 (World Scientific (Europe), 2016).
Bilynsky, C., Millot, N. & Papa, A. Radiation nanosensitizers in cancer therapy—From preclinical discoveries to the outcomes of early clinical trials. Bioeng. Transl. Med. 7, e10256 (2021).
Chattopadhyay, N. et al. Molecularly targeted gold nanoparticles enhance the radiation response of breast cancer cells and tumor xenografts to X-radiation. Breast Cancer Res. Treat. 137, 81–91 (2013).
Jain, S., Hirst, D. G. & O’Sullivan, J. M. Gold nanoparticles as novel agents for cancer therapy. Br. J. Radiol. 85, 101–113 (2012).
Hainfeld, J. F., Slatkin, D. N. & Smilowitz, H. M. The use of gold nanoparticles to enhance radiotherapy in mice. Phys. Med. Biol. 49, N309-315 (2004).
Chithrani, D. B. et al. Gold nanoparticles as radiation sensitizers in cancer therapy. Radiat. Res. 173, 719–728 (2010).
Schuemann, J. et al. Roadmap for metal nanoparticles in radiation therapy: Current status, translational challenges, and future directions. Phys. Med. Biol. 65, 21RM2 (2020).
Chang, M.-Y. et al. Increased apoptotic potential and dose-enhancing effect of gold nanoparticles in combination with single-dose clinical electron beams on tumor-bearing mice. Cancer Sci. 99, 1479–1484 (2008).
Zhang, X.-D. et al. Metabolizable Bi2Se3 nanoplates: Biodistribution, toxicity, and uses for cancer radiation therapy and imaging. Adv. Funct. Mater. 24, 1718–1729 (2014).
Alqathami, M. et al. Enhancement of radiation effects by bismuth oxide nanoparticles for kilovoltage x-ray beams: A dosimetric study using a novel multi-compartment 3D radiochromic dosimeter. J. Phys. Conf. Ser. 444, 012025 (2013).
Li, S. et al. Platinum nanoparticles: An exquisite tool to overcome radioresistance. Cancer Nanotechnol. 8, 4 (2017).
Khoei, S., Mahdavi, S. R., Fakhimikabir, H., Shakeri-Zadeh, A. & Hashemian, A. The role of iron oxide nanoparticles in the radiosensitization of human prostate carcinoma cell line DU145 at megavoltage radiation energies. Int. J. Radiat. Biol. 90, 351–356 (2014).
Klein, S. et al. Superparamagnetic iron oxide nanoparticles as novel X-ray enhancer for low-dose radiation therapy. J. Phys. Chem. B 118, 6159–6166 (2014).
Lux, F. et al. AGuIX® from bench to bedside—Transfer of an ultrasmall theranostic gadolinium-based nanoparticle to clinical medicine. Br. J. Radiol. 92, 20180365 (2019).
Yong, Y. et al. Gadolinium polytungstate nanoclusters: A new theranostic with ultrasmall size and versatile properties for dual-modal MR/CT imaging and photothermal therapy/radiotherapy of cancer. NPG Asia Mater. 8, e273–e273 (2016).
Maggiorella, L. et al. Nanoscale radiotherapy with hafnium oxide nanoparticles. Future Oncol. 8, 1167–1181 (2012).
Marill, J. et al. Hafnium oxide nanoparticles: Toward an in vitropredictive biological effect?. Radiat. Oncol. 9, 150 (2014).
Bagley, A. F. et al. NBTXR3, a first-in-class radioenhancer for pancreatic ductal adenocarcinoma: Report of first patient experience. Clin. Transl. Radiat. Oncol. 33, 66–69 (2022).
Cameron, S. J., Sheng, J., Hosseinian, F. & Willmore, W. G. Nanoparticle effects on stress response pathways and nanoparticle–protein interactions. Int. J. Mol. Sci. 23, 7962 (2022).
Penninckx, S., Heuskin, A.-C., Michiels, C. & Lucas, S. Gold Nanoparticles as a potent radiosensitizer: A transdisciplinary approach from physics to patient. Cancers (Basel) 12, 2021 (2020).
Sun, H., Wang, X. & Zhai, S. The rational design and biological mechanisms of nanoradiosensitizers. Nanomaterials (Basel) 10, 504 (2020).
Ruan, J. et al. Graphene quantum dots for radiotherapy. ACS Appl. Mater. Interfaces 10, 14342–14355 (2018).
Reda, M. et al. PLK1 and EGFR targeted nanoparticle as a radiation sensitizer for non-small cell lung cancer. Cancer Lett. 467, 9–18 (2019).
Mirjolet, C. et al. The radiosensitization effect of titanate nanotubes as a new tool in radiation therapy for glioblastoma: A proof-of-concept. Radiother. Oncol. 108, 136–142 (2013).
Grall, R. et al. Impairing the radioresistance of cancer cells by hydrogenated nanodiamonds. Biomaterials 61, 290–298 (2015).
Li, N. et al. A mitochondria-targeted nanoradiosensitizer activating reactive oxygen species burst for enhanced radiation therapy. Chem. Sci. 9, 3159–3164 (2018).
Ghita, M. et al. A mechanistic study of gold nanoparticle radiosensitisation using targeted microbeam irradiation. Sci. Rep. 7, 44752 (2017).
Penninckx, S., Heuskin, A.-C., Michiels, C. & Lucas, S. Thioredoxin reductase activity predicts gold nanoparticle radiosensitization effect. Nanomaterials (Basel) 9, 295 (2019).
Liu, X. et al. The synergistic radiosensitizing effect of tirapazamine-conjugated gold nanoparticles on human hepatoma HepG2 cells under X-ray irradiation. Int. J. Nanomed. 11, 3517–3531 (2016).
Daems, N. et al. Gold nanoparticles affect the antioxidant status in selected normal human cells. Int. J. Nanomed. 14, 4991–5015 (2019).
Guerreiro, A., Chatterton, N., Crabb, E. M. & Golding, J. P. A comparison of the radiosensitisation ability of 22 different element metal oxide nanoparticles using clinical megavoltage X-rays. Cancer Nanotechnol. 10, 10 (2019).
Liu, A., Wei, M. D., Honma, I. & Zhou, H. Biosensing properties of titanate nanotube films: Selective detection of dopamine in the presence of ascorbate and uric acid. Adv. Funct. Mater. 16, 371–376 (2006).
Niu, L. et al. Titanate nanotubes: Preparation, characterization, and application in the detection of dopamine. J. Mater. Sci. 43, 1510–1514 (2008).
Brammer, K. S. et al. Improved bone-forming functionality on diameter-controlled TiO2 nanotube surface. Acta Biomater. 5, 3215–3223 (2009).
Khaled, S. M. Z., Miron, R. J., Hamilton, D. W., Charpentier, P. A. & Rizkalla, A. S. Reinforcement of resin based cement with titania nanotubes. Dent. Mater. 26, 169–178 (2010).
Simon, M. et al. In situ quantification of diverse titanium dioxide nanoparticles unveils selective endoplasmic reticulum stress-dependent toxicity. Nanotoxicology 11, 134–145 (2017).
Magrez, A. et al. Cellular toxicity of TiO2-based nanofilaments. ACS Nano 3, 2274–2280 (2009).
L’azou, B. et al. In vitro effects of nanoparticles on renal cells. Part Fibre Toxicol 5, 22 (2008).
Gratton, S. E. A. et al. The effect of particle design on cellular internalization pathways. Proc. Natl. Acad. Sci. USA 105, 11613–11618 (2008).
Decuzzi, P. et al. Size and shape effects in the biodistribution of intravascularly injected particles. J. Control Release 141, 320–327 (2010).
Aueviriyavit, S., Phummiratch, D., Kulthong, K. & Maniratanachote, R. Titanium dioxide nanoparticles-mediated in vitro cytotoxicity does not induce Hsp70 and Grp78 expression in human bronchial epithelial A549 cells. Biol. Trace Elem. Res. 149, 123–132 (2012).
Park, E.-J. et al. Oxidative stress and apoptosis induced by titanium dioxide nanoparticles in cultured BEAS-2B cells. Toxicol. Lett. 180, 222–229 (2008).
Saquib, Q. et al. Titanium dioxide nanoparticles induced cytotoxicity, oxidative stress and DNA damage in human amnion epithelial (WISH) cells. Toxicol. In Vitro 26, 351–361 (2012).
Papa, A.-L., Dumont, L., Vandroux, D. & Millot, N. Titanate nanotubes: towards a novel and safer nanovector for cardiomyocytes. Nanotoxicology 7, 1131–1142 (2013).
Kasuga, T., Hiramatsu, M., Hoson, A., Sekino, T. & Niihara, K. Formation of titanium oxide nanotube. Langmuir 14, 3160–3163 (1998).
Kasuga, T., Hiramatsu, M., Hoson, A., Sekino, T. & Niihara, K. Titania nanotubes prepared by chemical processing. Adv. Mater. 11, 1307–1311 (1999).
Kasuga, T. Formation of titanium oxide nanotubes using chemical treatments and their characteristic properties. Thin Solid Films 496, 141–145 (2006).
Nakahira, A., Kubo, T. & Numako, C. Formation mechanism of TiO2-derived titanate nanotubes prepared by the hydrothermal process. Inorg. Chem. 49, 5845–5852 (2010).
Bai, Q. et al. Hydrothermal transformation of titanate scrolled nanosheets to anatase over a wide pH range and contribution of triethanolamine and oleic acid to control the morphology. Inorg. Chem. 58, 2588–2598 (2019).
Yao, B. D. et al. Formation mechanism of TiO2 nanotubes. Appl. Phys. Lett. 82, 281–283 (2003).
Chen, Q., Du, G. H., Zhang, S. & Peng, L.-M. The structure of trititanate nanotubes. Acta Crystallogr. B 58, 587–593 (2002).
Du, G. H., Chen, Q., Che, R. C., Yuan, Z. Y. & Peng, L.-M. Preparation and structure analysis of titanium oxide nanotubes. Appl. Phys. Lett. 79, 3702–3704 (2001).
Bavykin, D. V., Friedrich, J. M., Lapkin, A. A. & Walsh, F. C. Stability of aqueous suspensions of titanate nanotubes. Chem. Mater. 18, 1124–1129 (2005).
Khan, S. et al. One-pot synthesis of anatase, rutile-decorated hydrogen titanate nanorods by yttrium doping for solar H2 production. ACS Omega 5, 23081–23089 (2020).
Nian, J.-N. & Teng, H. Hydrothermal synthesis of single-crystalline anatase TiO2 nanorods with nanotubes as the precursor. J. Phys. Chem. B 110, 4193–4198 (2006).
Gao, T., Wu, Q., Fjellvåg, H. & Norby, P. Topological properties of titanate nanotubes. J. Phys. Chem. C 112, 8548–8552 (2008).
Zhang, S. et al. Formation mechanism of H2Ti3O7 nanotubes. Phys. Rev. Lett. 91, 256103 (2003).
Tsai, C.-C. & Teng, H. Nanotube formation from a sodium titanate powder via low-temperature acid treatment. Langmuir 24, 3434–3438 (2008).
Wang, Y. Q., Hu, G. Q., Duan, X. F., Sun, H. L. & Xue, Q. K. Microstructure and formation mechanism of titanium dioxide nanotubes. Chem. Phys. Lett. 365, 427–431 (2002).
Weng, L.-Q., Song, S.-H., Hodgson, S., Baker, A. & Yu, J. Synthesis and characterisation of nanotubular titanates and titania. J. Eur. Ceram. Soc. 26, 1405–1409 (2006).
Yang, J. et al. Study on composition, structure and formation process of nanotube Na2Ti2O4(OH)2. Dalton Trans. https://doi.org/10.1039/B305585J (2003).
Protonated Titanates and TiO2 Nanostructured Materials: Synthesis, Properties, and Applications—Bavykin—2006—Advanced Materials—Wiley Online Library. https://doi.org/10.1002/adma.200502696 (2006).
Chibon, F. et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat. Med. 16, 781–787 (2010).
Simon, M., Barberet, P., Delville, M.-H., Moretto, P. & Seznec, H. Titanium dioxide nanoparticles induced intracellular calcium homeostasis modification in primary human keratinocytes. Towards an in vitro explanation of titanium dioxide nanoparticles toxicity. Nanotoxicology 5, 125–139 (2011).
Muggiolu, G. et al. In situ detection and single cell quantification of metal oxide nanoparticles using nuclear microprobe analysis. J. Vis. Exp. https://doi.org/10.3791/55041 (2018).
Le Trequesser, Q. et al. Single cell in situ detection and quantification of metal oxide nanoparticles using multimodal correlative microscopy. Anal. Chem. 86, 7311–7319 (2014).
Haynes, W. Bonferroni correction. In Encyclopedia of Systems Biology (eds. Dubitzky, W., Wolkenhauer, O., Cho, K.-H. & Yokota, H.). 154–154 https://doi.org/10.1007/978-1-4419-9863-7_1213 (Springer, 2013).
Raudvere, U. et al. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019).
Acknowledgements
The AIFIRA facility is financially supported by the CNRS, the University of Bordeaux, and the Région Nouvelle Aquitaine. We thank the technical staff members of the AIFIRA facility P. Alfaurt, J. Jouve, and S. Sorieul. The authors acknowledge the Evonik (Degussa) Company (Düsseldorf, Germany) for their generous gift (P25 TiO2 NPs). This work has been partly supported by the European Community as an Integrating Activity “Support of Public and Industrial Research Using Ion Beam Technology” (SPIRIT) under the EC contract no. 227012 and as an “Integrating Activity Supporting Postgraduate Research with Internships in Industry and Training Excellence” (SPRITE) under EC contract no. 317169. The CNRS, the French National Research Agency (ANR CES2010, no. CESA 009 01, TITANIUMS), and the Région Aquitaine (TOX-NANO—no. 20111201003/POPRA—no. 14006636-034) support the research program and the funding of QLT, GS, GM, MS.
Author information
Authors and Affiliations
Contributions
H.S. conceived and supervised the project. Q.L.T. and M.H.D. performed the synthesis and characterization of TNs. G.K., J.C., and M.A. prepared and realized irradiation experiments at the medical facility. M.S., G.M., F.V., L.P., and H.S. performed the cell culture, TNs exposure, radiation biology, and biological analyses; F.V., and H.S. performed RNA extraction, cDNA libraries preparation, and long read sequencing experiments (cDNA Barcoding). G.D., P.B., M.S., G.M., and H.S. performed the biological sample preparation for Micro-PIXE analysis. G.D., M.S., and Q.L.T. contributed to the micro-PIXE data analysis. P.B. performed the bioinformatic, statistical, and transcriptomic analysis. M.H.D., D.D. provided expert review and editing. The authors contributed to the data analysis and interpreted the results. Pi.B., G.D., P.B., M.H.D., and H.S. wrote the main manuscript and prepared the remaining figures. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Beaudier, P., Vilotte, F., Simon, M. et al. Sarcoma cell-specific radiation sensitization by titanate scrolled nanosheets: insights from physicochemical analysis and transcriptomic profiling. Sci Rep 14, 3295 (2024). https://doi.org/10.1038/s41598-024-53847-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-53847-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.