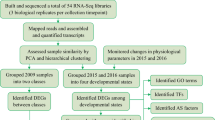
Abstract
Oilseed rape (Brassica napus) is an important global oil crop, with spring and winter varieties grown commercially. To understand the transcriptomic differences between these varieties, we collected transcriptomes from apex and leaf tissue from a spring variety, Westar, and a winter variety, Tapidor, before, during, and after vernalisation treatment, until the plants flowered. Large transcriptomic differences were noted in both varieties during the vernalisation treatment because of temperature and day length changes. Transcriptomic alignment revealed that the apex transcriptome reflects developmental state, whereas the leaf transcriptome is more closely aligned to the age of the plant. Similar numbers of copies of genes were expressed in both varieties during the time series, although key flowering time genes exhibited expression pattern differences. BnaFLC copies on A2 and A10 are the best candidates for the increased vernalisation requirement of Tapidor. Other BnaFLC copies show tissue-dependent reactivation of expression post-cold, with these dynamics suggesting some copies have retained or acquired a perennial nature. BnaSOC1 genes, also related to the vernalisation pathway, have expression profiles which suggest tissue subfunctionalisation. This understanding may help to breed varieties with more consistent or robust vernalisation responses, of special importance due to the milder winters resulting from climate change.
Similar content being viewed by others
Introduction
Plants change growth and development in response to their environment1. Different varieties of plants have evolved different life strategies, flowering and setting seed at particular times of the year to adapt to their surroundings2. This is of particular economic importance for crop species, allowing growers to choose varieties well suited to the climate and their cropping schedule; oilseed rape (OSR; Brassica napus), for instance, has spring and winter varieties that are both grown commercially3. Understanding how signals from the environment are integrated by the plant is important to breed new crop varieties with favourable characteristics, particularly given the projected effects of unpredictable temperature variation on OSR yield4.
Flowering time is a key adaptive trait that is affected by climate5. The timing of the floral transition, where the apex transitions from vegetative to floral growth, is one component which affects the final flowering time of a plant. Control of the floral transition depends on multiple environmental cues, such as photoperiod and temperature, which are integrated with internal signals to determine when the plant should flower6. The flowering of some plants is accelerated by experiencing an extended period of cold temperature, which is called vernalisation7. In the field, such plants can be planted before winter and the vernalisation requirement ensures that the plants do not flower until spring. In the model species Arabidopsis thaliana (Arabidopsis), the delay of flowering in the absence of vernalisation is due to expression of FLOWERING LOCUS C (FLC)8,9. FLC delays flowering by repressing expression of the flowering activator FT10,11. FT is downstream of both the vernalisation pathway, via FLC, and the photoperiod pathway, which senses day length12,13,14,15. As external signals are sensed in different plant organs, communication between them is important for the signals to be properly integrated into a coherent gene regulatory response. FT is expressed in the phloem companion cells, with the FT protein transported in the plant vasculature from leaves to the apex to promote flowering16,17,18,19. A gene which acts antagonistically to FT in the floral transition is TERMINAL FLOWER 1 (TFL1), which represses the floral state of the shoot apical meristem20,21,22. TFL1 and FT are closely related proteins23 and both bind DNA in complex with the bZIP transcription factor FD24,25. As a result, both influence the expression of similar sets of genes in the gene regulatory network controlling flowering26. SUPPRESSOR OF CONSTANS 1 (SOC1) is also a floral integrator downstream of the photoperiod and vernalisation pathways, directly downstream of FLC10,27,28,29. In addition, roles have been found for SOC1 in several other flowering pathways, such as the gibberellic acid pathway30, intermittent cold-sensing pathway31, and the age-dependent pathway32.
Although both members of the Brassicaceae family, the direct application of Arabidopsis genetic understanding to OSR is not straightforward. Brassica napus is an amphidiploid derived from ancestral allopolyploidisation of B. oleracea and B. rapa, related mesohexaploids33,34,35. This has resulted in multiple copies of many genes in OSR that are represented as single copies in Arabidopsis36, with flowering time genes overrepresented among those genes retained37. Gene multiplication provides opportunities for gene neo- and subfunctionalisation, for example, in tissue-specificity, in response to stimuli, and in location in the gene regulatory network38. Despite these challenges, knowledge of the floral pathways in Arabidopsis has been translated to Brassica crops36,39. In the same way as Arabidopsis, there is variation in the vernalisation requirement between OSR varieties. Indeed, whether a variety is a spring variety (sown in the spring and harvested in the same year) or a winter variety (sown in the summer or autumn and harvested the following year) is a commercially relevant trait, with winter varieties being preferred in Europe and Asia and spring varieties grown in Australia, Canada, and northern Europe3. This variation in vernalisation requirement has been found to be associated with copies of BnaFLC40,41,42, with BnaFLC copies on chromosome A10 and A2 exhibiting the strongest association42,43,44, although variation across multiple BnaFLC copies have been found to drive diversity within Brassica crop types45. BnaFT genes in OSR have also been found to exhibit different expression patterns, with variation in the effect on flowering time present between copies46,47. In particular, the BnaFT.A2 gene is strongly associated with flowering time differences between two winter OSR varieties44. Within other flowering time integrators, a specific promoter variant of BnaSOC1.A5 is predominantly found in winter OSR varieties48. These findings point towards variation in known flowering time genes from Arabidopsis being responsible for flowering time diversity between OSR, particularly in terms of vernalisation requirement.
Perhaps because of the known roles of OSR homologues of Arabidopsis flowering time genes in determining flowering time, many previous transcriptomic studies in OSR have tended to focus on individual gene copies49,50,51. However, understanding the similarity of transcriptomes on a global level allows for more general expression dynamics to be elucidated52. To determine if gene regulation is coordinated between leaves and the growing apex during the floral transition in OSR, we collected a transcriptomic time series from the first true leaf and apex across development. To understand how vernalisation affects gene expression in both tissues, we sampled tissue from both a winter OSR variety, Tapidor, and a spring OSR variety, Westar. Comparisons between the transcriptomes between tissues suggests that transcriptomic changes in the apex are aligned to the developmental state of the plant, whereas in the leaf the transcriptomic changes are aligned based on the age of the plant. This is reflected by the apex transcriptome exhibiting large changes during flower development, whereas the leaf transcriptome changes coincide with leaf senescence. Comparing between Westar and Tapidor, we find two copies of BnaFLC (BnaFLC.A2 and BnaFLC.A10) as being the best candidates for mediating the stronger vernalisation requirement of Tapidor relative to Westar. Copies of BnaSOC1, a key floral integrator, exhibit expression patterns which are dependent on tissue, suggesting that copies have undergone sub-functionalisation.
Results
Developmental differences between varieties are reflected in the apex transcriptomes, but not the leaf transcriptomes
To understand the transcriptomic differences underlying the different flowering strategies of Tapidor, a winter OSR variety, and Westar, a spring OSR variety, the apex and the first true leaf were sampled across development, from 22 days post sowing to when floral buds were visible, for both varieties (Fig. 1). Plants from both varieties were subjected to a cold treatment, which began 23 days post sowing and ended after day 64 of the time series. Although Westar, the spring variety, does not have a vernalisation requirement, it does exhibit a vernalisation response53. Therefore, subjecting both varieties to the same treatment allows for clearer comparisons to be made between them. Transcriptomic time series were generated for both varieties and both tissues37,54. To ensure that comparable periods of development were captured between varieties, the BBCH scale was used to assess developmental stage55. At the first time point sampled (22 days post sowing), plants of both varieties were at BBCH stage 13 (3 leaves unfolded), but subsequent development deviated in time: Westar plants reached BBCH stage 51 (Flower buds visible from above) at day 72 of the time series, whereas Tapidor plants reached the same developmental stage at day 83.
Cartoon of the developmental stages of Tapidor and Westar at each time point. Plant tissue was sampled on the days indicated by red dotted lines and numbers. The plant silhouettes represent the approximate number of full leaves at the indicated points in development, allowing the developmental stage of the plants to be estimated. On day 69, only Westar apex samples were sequenced.
To evaluate whether these differences in development were reflected in global gene expression changes, we compared the transcriptomes over time using the Euclidean distance (Materials and Methods). The maximum similarity between transcriptomes can be visualised by the lines overlayed on the distance heatmaps, which indicate the minimum distance relative to each time point (Fig. 2). Using all OSR gene models (155,240) to calculate the distances, the first time point for both tissues is very similar between varieties, as are the transcriptomes sampled during the cold treatment on days 43 and 64 (Fig. 2a,b). After cold, however, the minima diverge in the two tissues. In the apex, the Westar time points post-cold are most similar to the day 22 time point in Tapidor, except for the final time point which is most similar to the final time point in Tapidor (blue, horizontal lines; Fig. 2a). For Tapidor, while the first time point post-cold is most similar to the first Westar time point (day 22), the day 67 and 72 time points in Tapidor are most similar to the day 67 time point in Westar, with the final time points once again being most similar, despite being sampled on different days (red, vertical lines; Fig. 2a). This is consistent with the developmental differences observed between plants (Fig. 1). The most similar time points between varieties in the leaf are those sampled at the same time, with the day 83 time point in Tapidor being most similar to the day 72 time point in Westar (Fig. 2b). These results show that the best agreement between the leaf transcriptomes of these two varieties is aligned with time (age), whereas the apex samples show a more complex relationship dictated by developmental stage.
Flowering genes show a different time alignment compared to all OSR genes. Euclidean distances were calculated between the time points from different varieties for both the apex (a, c) and leaf (b, d). The distances were calculated using all OSR genes (a, b) and only those OSR genes showing homology to Arabidopsis genes which are in the FLOR-ID database56 (c, d). The minimum distances are shown by coloured points and lines, with the vertical, red lines showing, for each Tapidor time point, the Westar time point with the smallest distance to it, and the horizontal, blue lines showing, for each Westar time point, the Tapidor time point with the smallest distance to it. The two time points collected during the vernalisation treatment (day 43 and 64) are indicated with a blue background.
To determine whether these results were affected by the different flowering regimes of the two varieties, we calculated the distances over time between a subset of 1380 OSR genes that showed homology to Arabidopsis flowering genes, defined as those genes included in the FLOR-ID database56. The FLOR-ID database is a list of 346 Arabidopsis genes which have been well characterised in the literature as being involved with flowering or influencing flowering time. The age alignment observed between the global leaf transcriptomes is not seen in the set of flowering genes (Fig. 2d). This suggests that the differences observed between tissues at the global scale are not defined by floral genes. For both tissues, the Westar samples all show highest similarity (lowest Euclidean distance) with Tapidor samples taken after the cold treatment (blue, horizontal lines; Fig. 2c,d). The early Tapidor samples (day 22 to 65) show highest similarly to the day 64 and 65 time points in Westar, which correspond to the final day of cold treatment and first day post-treatment respectively. Given that the cold treatment will have blocked flowering in the Westar plants, this is consistent with flowering being repressed in the Tapidor plants prior to being vernalised. There is also a marked block of similarity in both tissues, beginning on day 67 of the Tapidor time series and day 65 of the Westar time series. This similarity starts immediately after the vernalisation treatment in Westar, and after a slight delay after the vernalisation treatment in Tapidor. These findings suggest that the Westar plants were competent to flower at the very beginning of the time series, a state the Tapidor plants only reached after cold treatment, consistent with the vernalisation requirements of the two varieties.
Within the leaf samples, there are more genes expressed specifically in Tapidor
We next assessed gene level expression differences between Tapidor and Westar. To do this, all 155,240 OSR genes were classified as expressed if the maximal expression level of the gene in at least one time point across the time series was equal to or exceeded 2.0 FPKM. We find that the vast majority of expressed genes (37,752 in the leaf and 40,076 in the apex, representing 84% and 85% of the total number of expressed genes considered respectively) are expressed in both varieties (Fig. 3a,b). This overall pattern is maintained even when only the 1380 OSR flowering genes (Fig. 3c,d) and 79,014 OSR genes with Arabidopsis homologues (Supplementary Fig. 1) are considered. The number of genes that are specifically expressed in each variety is similar in the apex for all subsets of genes considered (Fig. 3a,c), but in the leaf there are consistently more genes specifically expressed in Tapidor relative to Westar (4147 versus 2909 for all OSR genes and 64 versus 34 for OSR flowering genes; Figs. 3b,d).
There is high overlap between varieties in the sets of expressed genes. OSR genes were regarded as expressed if their maximal expression level across the transcriptomic time series was greater than, or equal to, 2.0 FPKM. The gene subsets used to calculate the overlaps in each case are: (a, b) All OSR genes; (c, d) OSR genes that show sequence similarity to Arabidopsis genes in the FLOR-ID database of floral genes56. Percentages have been rounded to the closest integer.
To functionally characterise the variety-specific genes, gene ontology (GO) term enrichment was conducted (Supplementary Tables 1 and 2). In the apex, the Westar-specific genes were enriched for defence related GO terms (GO:0009751 response to salicylic acid; GO:0031348 negative regulation of defence response), whereas the Tapidor-specific genes were enriched for terms related to nitrogen and ion transport (GO:1901698 response to nitrogen compound; GO:0006820 anion transport). In the leaf, the Westar-specific genes did not show high enrichment (p-value < 10–5) for any term, while the Tapidor-specific genes were enriched for DNA replication terms (GO:0006275 regulation of DNA replication; GO:0006260 DNA replication).
The higher number of OSR flowering genes expressed only in Tapidor, relative to genes expressed only in Westar, may be due to certain sets of homologous genes. To assess this, OSR flowering genes were first grouped into homologue families based on their sequence similarity to genes in Arabidopsis. Then, the number of expressed copies within each OSR homologue family were compared between Tapidor and Westar (Fig. 4). If genes lie on the diagonal line in Fig. 4a,b, it indicates that the same number of gene copies are expressed in both Tapidor and Westar, while deviation from that line shows that more copies are expressed in either Tapidor (below the line) or Westar (above the line). This data is also represented as count data of the number of genes exhibiting copy number expression bias (Fig. 4c,d). For example, a gene with five copies expressed in Tapidor and three copies expressed in Westar would be said to have a two copy bias towards Tapidor. A striking observation, unrelated to copy number expression bias, is that 47.7% of OSR flowering gene homologue families have two or four copies expressed in the apex in both varieties (44.5% of families in the leaf). This is likely due to the hybridisation of the A and C genomes in OSR, resulting in a bias for an even number of expressed copies in both varieties (Fig. 4a,b). In the apex, the same percentage of homologue families exhibit variety-specific bias towards Tapidor and Westar (10.8%; Fig. 4a,c). However, in the leaf there are significantly (p-value = 0.03442; binomial test) more sets of homologue families in Tapidor (18.0%) relative to Westar (10.5%; Fig. 4b,d). Percentages of homologue families exhibiting copy number expression bias (Fig. 4) are higher than the percentages of OSR flowering genes exhibiting variety-specific expression (Fig. 3). For example, 10.5% of homologue families have more genes expressed in the leaf in Westar relative to Tapidor (Fig. 4b), whereas 4% of OSR flowering genes are expressed specifically in Westar (Fig. 3b). This suggests that OSR flowering genes exhibiting variety-specific expression are generally well distributed among different homologue families, rather than the pattern being the result of relatively few families. The same pattern, albeit marginally less strong, is also observed when all 79,014 OSR genes with an Arabidopsis orthologue are compared (Supplementary Fig. 2). A scenario that is not captured by these plots is compensatory orthologue expression. That is, gene copies with Tapidor-specific expression are compensated for by copies with Westar-specific expression, resulting in the gene still being located on the diagonal in Fig. 4 and Supplementary Fig. 2 despite variety-specific expression. While this scenario is observed, the frequency is very low, being observed in just 1.6–2.7% of homologue families, depending on the gene subset and tissue considered (Supplementary Figs. 3, 4).
The majority of Arabidopsis floral genes have the same number of expressed OSR orthologues in each variety. (a, b) OSR homologue families were defined as OSR genes which have the same Arabidopsis gene as their highest scoring BLAST hit. Expressed OSR genes were determined as those that had a maximal expression value above or equal to 2.0 FPKM at one or more time points in the time series. The size and colour of the circles indicate the number of data points at that position. The upper limit of the colour scale is the maximal off-diagonal value. Points on the diagonal, grey line represent OSR homologue families that have equal numbers of homologues expressed in both Tapidor and Westar. The left most percentage within each graph represent the percentage of Arabidopsis genes that have more homologues expressed in Westar, whereas the right most percentage is the corresponding percentage for Tapidor. (c, d) Bar charts sum the number of data points on the diagonals in a and b, with copy number expression bias determined as the absolute difference between the number of expressed genes in a OSR homologue family in both Tapidor and Westar.
Self-organising maps reveal characteristic expression dynamics within the apex and leaf
To assess the temporal dynamics captured by our dataset, we employed self-organising maps (SOMs) to cluster gene expression profiles (Fig. 5 and Supplementary Figs. 5, 6). Neighbouring SOM clusters have similar expression profiles, with clusters at the opposite boundaries being adjacent to prevent boundary effects. In this way, SOMs represent a toroidal landscape of expression profiles onto which genes are mapped. Selecting the clusters with the most genes mapped to them highlights several distinct expression profile types which will be used to comment on the results. “Early/vernalisation responsive” clusters show high expression initially, with expression decreasing during the vernalisation treatment. This name reflects that, given our experimental setup, genes which decrease in expression across development regardless of cold, and genes which decrease in expression due to being vernalisation responsive, will cluster together. “Treatment responsive” clusters exhibit an increase in expression during the vernalisation treatment, while “Late” clusters peak in expression towards the end of the time series. Finally, “Treatment responsive/Late” clusters consist of expression profiles which show increases in expression during the cold treatment and towards the end of the time series.
Self-organising maps based on apex transcriptomes reveal that variety-specific genes tend to be expressed at the end of the time series. SOMs generated using either all OSR with Arabidopsis homologues (a, b) or OSR genes exhibiting variety-specific expression (c, d). (a, b) The representative expression traces of the SOM clusters with the most genes assigned to them are plotted below the SOM. (c, d) The representative expression traces of the SOM clusters most enriched for variety-specific genes are plotted below the SOM.
For the apex data from Tapidor, the clusters with the most genes mapped to them are “Late” and “Treatment responsive” (clusters 46, 88, and 98, which have 2.3%, 2.4%, and 2.3% of the total number of genes mapped to them respectively; Fig. 5a). The “Late” cluster is highly enriched for genes with the GO terms “carpel development”, “gynoecium development”, and “floral whorl development”. The two “Treatment responsive” clusters are both enriched for genes with the GO term “circadian rhythm”. This is likely due to the change in day length during the vernalisation treatment. Interestingly, cluster 88 is also enriched for genes with the GO terms “starch catabolic process” and “glucan catabolic process”, suggesting a change in nutrient usage during the vernalisation period. A similar pattern is observed in the Westar apex data (Fig. 5b), with the most genes mapping to a “Treatment responsive” cluster (cluster 19, 3.2% of genes mapped), enriched for genes with the GO terms “long-day photoperiodism” and “circadian rhythm”, and a “Late” cluster (cluster 56, 2.5% of genes mapped), enriched with the GO terms “carpel development” and “specification of floral organ identity”.
The expression profiles of the SOM clusters with the most genes mapping to them are also somewhat similar in the leaf samples, although different GO terms are enriched (Supplementary Fig. 6). “Treatment responsive” clusters in both varieties (cluster 25 in Tapidor and cluster 99 in Westar, with 3.2% and 2.0% of the total number of genes mapped to them respectively) are strongly enriched for “translation” and “peptide biosynthetic process” GO terms. The GO term “cell wall modification” is also highly enriched in both clusters, suggesting that the large transcriptional changes observed are related to the leaves acclimatising to the colder growth conditions. The “Late” clusters in both varieties (cluster 59, 3.1% of genes, in Tapidor and cluster 19, 2.4% of genes, in Westar) are strongly enriched for stress, cell death, and age-related GO terms. A potential explanation is given by both clusters also being enriched for “leaf senescence”, which is consistent with the first true leaf being sampled throughout development (Fig. 1).
Mapping the 1380 OSR floral genes to the apex SOMs (Supplementary Fig. 5) reveals the same “Late” and “Treatment responsive” clusters as was seen when all genes are considered. However, for the leaf SOMs, floral genes map to different “Treatment responsive” and “Treatment responsive/Late” clusters (clusters 5, 8, 15, and 67 in Tapidor and clusters 55, 66, and 77 in Westar; Supplementary Fig. 6). Cluster 5 in Tapidor and cluster 55 in Westar are both “Treatment responsive” clusters, and are enriched for the GO term “photoperiodism, flowering”, suggesting that floral genes in the leaf are primarily responding to the change in day length during the vernalisation period.
The SOM analysis in the apex in both varieties revealed “Late” SOM clusters as being among the clusters with the most genes assigned to them (cluster 46 in Tapidor, with 2.3% of mapped genes, and cluster 56 in Westar, with 2.5% of mapped genes). Both clusters were enriched for genes with GO terms related to flower development. To analyse this with more detail, we looked at the expression of the ABC floral patterning genes, which have been found to specify floral organ development through overlapping and combinatoric gene expression domains57 (Fig. 6). OSR homologues of APETELA1 (AP1), PISTILLA (PI), and AGAMOUS (AG) all exhibit an expression delay in Tapidor when compared to Westar (Fig. 6). The levels of expression of BnaAP1 (Fig. 6a,b) and BnaPI (Fig. 6c,d) copies in Westar on day 72 of the time series are only reached in Tapidor by the day 83 time point. With the copies of BnaAG, equivalent expression levels to Westar are not obtained in Tapidor during the sampled time series (Fig. 6e,f). These temporal differences closely follow the delayed onset of flowering in Tapidor relative to Westar (Fig. 1).
ABC floral genes show alignment with developmental stage of the plant. Expression traces across the developmental time series in apex tissue for representative copies of BnaAP1 (a, b), BnaPI (c, d), and BnaAG (e, f). The blue segments indicate the time points sampled during the vernalisation treatment.
We can also use the SOM clusters to summarise which genes correlate most with the global transcriptome distance differences observed between varieties in Fig. 2. Calculating Pearson correlation coefficients between the global and gene-based distance matrices, and using an area under the curve metric, we identified the three SOM clusters, for each variety and tissue, with the highest proportion of genes highly correlated with the global pattern (Materials and Methods; Fig. 7). It should be noted that because we have considered the proportion of genes within each SOM cluster, this will not necessarily represent the genes which most contribute to the global transcriptome differences, but those which are most characteristic of it by correlation. In the apex, three “Late” clusters have the highest area under the curve for both varieties, whereas “Early / vernalisation responsive” clusters have the highest area for the leaf in both varieties. This again supports our finding that the apex is driven by developmental progression towards flowering, whereas the leaves are more affected by environmental changes.
“Late” SOM clusters show greatest correlation to the global expression changes in the apex, whereas "early/vernalisation responsive" clusters show greatest correlation in the leaf. Euclidean distances matrices were created for each gene and compared to the global transcriptome response (Fig. 2) by Pearson correlation coefficient. The relationship between the proportion of genes within each SOM cluster with a correlation coefficient greater than a particular threshold is shown. The representative expression traces of the three SOM clusters with the highest area under this curve (areas reported within brackets in figure headers) are shown below the plots.
Tapidor-specific genes in cold-responsive SOM clusters contain known vernalisation genes
To better characterise transcriptome differences between varieties, we tested SOM clusters for enrichment of genes expressed in a variety-specific manner. Fisher tests were used to test each cluster in the SOM for enrichment of genes expressed in a variety-specific manner, relative to all expressed OSR genes in that variety with identified Arabidopsis orthologues (Materials & Methods). The top three clusters showing enrichment, with an adjusted p-value below 10–5, are labelled (Fig. 5c,d; Supplementary Table 3). Across both varieties and tissues, the variety-specific genes tend to be enriched in “Late” clusters (Fig. 5c,d; Supplementary Fig. 6). This suggests that the expression profiles of the varieties diverge towards the final time points. In addition, the genes expressed specifically in Tapidor are also enriched in “Early/vernalisation responsive” clusters in both tissues. This enrichment is not seen in Westar and mimics the expression pattern of documented vernalisation responsive genes, raising the possibility that some of the Tapidor-specific expressed genes mediate, or are downstream of, the vernalisation response in Tapidor.
As differential enrichment of “Early/vernalisation responsive” clusters was observed, we investigated the genes mapped to these clusters in more detail (Supplementary Table 4). In terms of Tapidor-specific expressed genes mapped in the apex (Cluster 1; Fig. 5c) we find two orthologues of FLOWERING LOCUS C (FLC) as the only two annotated flowering time genes (BnaFLC.A10 and BnaFLC.C3c. BnaFLC.C3c is a split model for BnaFLC.C3b45; model details for genes discussed by name are given in Supplementary Table 5). In the Tapidor-specific expressed genes mapped in the leaf (Cluster 61; Supplementary Fig. 6c) we find four orthologues of FLC (BnaFLC.A3b, BnaFLC.A10, BnaFLC.C3a, and BnaFLC.C3c) and one orthologue of MADS AFFECTING FLOWERING 4 (MAF4; BnaMAF4.A2a) as the only annotated flowering genes, all of which are documented vernalisation responsive genes in Arabidopsis and Brassica species58,59,60.
We then expanded our focus beyond genes which are expressed in a variety-specific manner in these clusters but continued to restrict the list of genes to annotated flowering genes. Doing so identifies an additional three BnaFLC genes (BnaFLC.A2, BnaFLC.A3a, and BnaFLC.C3b), two OSR orthologues of AUXIN RESPONSE TRANSCRIPTION FACTOR 3, one OSR orthologue of PSEUDO-RESPONSE REGULATOR 7, and one OSR orthologue of FT-INTERACTING PROTEIN 1. Similarly, in the leaf, an additional three BnaFLC genes are clustered to cluster 61 but do not exhibit variety-specific expression (BnaFLC.A2, BnaFLC.A3a, and BnaFLC.C2). OSR orthologues of PHOSPHOGLUCOSE ISOMERASE 1, EARLY FLOWERING MYB PROTEIN, APETALA 2, and GIBBERELLIN 20-OXIDASE 3 are also annotated as flowering genes and mapped to cluster 61.
BnaFLC gene expression reveals copy-specific differences in expression level between Tapidor and Westar and different reactivation dynamics between apex and leaf
Two copies of BnaFLC were found to be expressed only in Tapidor in the tissues we investigated, and additional copies of BnaFLC are mapped to SOM clusters enriched for genes expressed specifically in Tapidor. Given the key role FLC plays in the vernalisation response in Arabidopsis and Brassica species, we decided to look in more detail at the expression dynamics of the various BnaFLC copies and how they differ between the winter (Tapidor) and spring (Westar) variety. We observed qualitatively similar expression responses for each BnaFLC copy in both the apex and the leaf (Fig. 8 and Supplementary Fig. 7). All copies except BnaFLC.C9b exhibit a decrease in expression during the cold treatment, often in both varieties. Only BnaFLC.A3a has a similar expression profile and expression level in both varieties and is expressed to an appreciable level (Fig. 8b). For other copies (BnaFLC.A2, BnaFLC.A3b, BnaFLC.A10, BnaFLC.C2, and BnaFLC.C3c) the expression of the gene in Westar is considerably less than in Tapidor. This expression difference is most prevalent at the beginning of the time series, due to decreasing gene expression levels during the cold treatment. However, for some copies, a reactivation of the gene is observed. Copies of BnaFLC.A3b (Supplementary Fig. 7), BnaFLC.C2 (Fig. 8d), and BnaFLC.C3c (Supplementary Fig. 7) in the apex all decrease in expression during the cold treatment, but in Tapidor the expression of the genes increase post-cold. The reactivation is not observed in Westar. Differences in reactivation post-cold also define the major differences in expression profiles between apex and leaf, with BnaFLC.A2 (Fig. 8a) and BnaFLC.A10 (Fig. 8c) exhibiting expression reactivation in Tapidor leaf samples yet show more stable repression in the Tapidor apex samples.
BnaFLC.A2 and BnaFLC.A10 genes appear to be the primary copies of BnaFLC mediating the vernalisation response, based on expression in the apex. Expression traces across the developmental time series in apex tissue for selected copies of BnaFLC: BnaFLC.A2 (a), BnaFLC.A3a (b), BnaFLC.A10 (c), and BnaFLC.C2 (d). The blue segments indicate the time points sampled during the vernalisation treatment.
The two copies of BnaFLC on chromosome C9 show higher expression in Westar rather than Tapidor (Supplementary Fig. 7). The expression of BnaFLC.C9b is interesting given that it is highly expressed in both varieties, and counter to the other copies of BnaFLC increases in expression over the time series sampled here. There are an additional two BnaFLC copies on chromosome C3, which are both very lowly expressed in these two varieties and across this period of development (Supplementary Fig. 7).
BnaSOC1 genes exhibit tissue-specific subfunctionalisation in terms of expression
SOC1 is involved with integration of inputs from an array of different flowering time pathways, including vernalisation61. Due to these many functions in the control of flowering, we investigated the expression of OSR SOC1 homologues in both Westar and Tapidor, to try and unpick the effects of the vernalisation pathway on the expression of BnaSOC1 genes. We found that the expression of BnaSOC1 genes was consistently lower in Tapidor compared to Westar across the time series. This is consistent with the repression of BnaSOC1 genes by BnaFLC genes, as in Arabidopsis (Fig. 9). The expression of some copies shows tissue-specificity. Compared to other BnaSOC1 genes expressed in the apex, BnaSOC1.A3 shows very high expression (Fig. 9) and increases in expression concurrently with the homologues of the ABC floral genes (Fig. 6). However, in the leaf this copy is lowly-expressed relative to the other BnaSOC1 genes. Conversely, the A4 copy of BnaSOC1 is among the most highly expressed copies in the leaf, but is lowly expressed in the apex. While the patterns are more subtle, a similar tissue preference can be observed in the A5 and the C4 copies of BnaSOC1. In addition, the copies showing leaf and apex expression preference also exhibit different expression patterns. The genes with a stronger apex preference (BnaSOC1.A3 and BnaSOC1.A5) both reach their highest expression level post-cold and follow the patterns of the ABC floral genes (Fig. 6). However, the genes with a leaf expression preference peak in expression during the vernalisation period. These results all suggest that the BnaSOC1 genes have undergone sub-functionalisation, with some genes retaining apex-specific roles and others leaf-specific functions. This is consistent with the varied functions identified for SOC1 in Arabidopsis10,27,28,29,30,31,32 and might reflect the roles SOC1 plays in the vernalisation, photoperiod, and the intermittent cold-sensing pathways.
BnaSOC genes have undergone subfunctionalisation, resulting in copies which are more prevalent in particular tissues. Expression traces across the developmental time series in apex (a, c, e, g) and leaf (b, d, f, h) tissue for selected copies of BnaSOC1: BnaSOC1.A3 (a, b), BnaSOC1.A4 (c, d), BnaSOC1.A5 (e, f), and BnaSOC1.C4_random (g, h). The blue segments indicate the time points sampled during the vernalisation treatment.
BnaFT genes show large expression differences between varieties, and suggest that flowering in Westar was delayed by the cold treatment
In Arabidopsis, the effect of FLC on flowering time is largely mediated by the FLC protein repressing the expression of FT in the leaves10,11. To determine if a similar mode of action was taking place in OSR, we looked at the expression of BnaFT genes. All four copies are very highly expressed in the leaf in Westar, decreasing to very low expression only during the cold treatment (Fig. 10a–d). This is likely due to the change in day length during the vernalisation period. This effect was seen in the SOM analysis of the leaf (Supplementary Fig. 6) with clusters enriched in the GO term “photoperiodism, flowering” increasing in expression during the cold treatment (Supplementary Table 6). This is expected given the role FT plays in sensing day length in Arabidopsis16,17,18,19. In Tapidor, BnaFT.A2 and BnaFT.C2 are very lowly expressed throughout the time series. BnaFT.A7 and BnaFT.C6 are also lowly expressed, although increase towards the end of the time series, but not to the same extent as the genes in Westar.
BnaFT and BnaTFL1 genes tend to be more highly expressed in Westar relative to Tapidor, although BnaTFL1 genes exhibit qualitatively similar expression patterns in both varieties. Expression traces across the developmental time series in leaf for BnaFT genes (a-d) and apex for BnaTFL1 genes (e–h). The blue segments indicate the time points sampled during the vernalisation treatment.
FT and TFL1 act antagonistically in the gene regulatory network underlying flowering in Arabidopsis26. In our previous work we highlighted the divergence of the BnaTFL1 genes in OSR, which correlated with the presence and absence of regulatory sequences upstream of the gene37. Comparing the expression of the BnaTFL1 genes between varieties (Fig. 10e–h), we see that the most highly expressed copy on C3 shows a very similar expression profile in both varieties in the apex (Fig. 10g). The other copies, while showing similar expression profiles, are more lowly expressed in Tapidor relative to Westar throughout the time series, with the copies on A10 and C2 being below 2.0 FPKM throughout the time series (Fig. 10e,f).
Discussion
The pathways controlling flowering in eudicots have been primarily characterised in Arabidopsis. Our use of both a winter and a spring variety of OSR, and the short-day vernalisation treatment, allowed us to investigate how the flowering pathways function in B. napus. In particular, it seems that the photoperiod flowering pathway was not inhibiting flowering in Westar plants before the cold treatment. This is evidenced most clearly when looking at individual floral genes. At the first time point, the expression of BnaFT genes in the leaves in Westar is high (Fig. 10a–d), yet the expression of BnaAP1 genes, used as an indicator of the floral transition26,62, is low in the apex at the same time (Fig. 6c,d). This is indicative of the plants being grown in a photoperiod conducive to flowering but having not yet transitioned from vegetative to floral growth. The floral transition was then inhibited by the transfer to short-day conditions during the vernalisation treatment, which is again represented by the expression of BnaFT genes decreasing to near-zero in the Westar leaf samples (Fig. 10a–d). This is in line with the photoperiod control of FT in Arabidopsis12,13,14 and the results from our SOM analysis, which indicate that circadian rhythm and photoperiodism genes are enriched in “treatment responsive” clusters (Fig. 5).
Further evidence that the Westar plants were capable of flowering throughout the time series are the flowering gene-specific distance heatmaps. All Westar time points were found to be most similar in expression to Tapidor time points sampled after the vernalisation treatment (Fig. 2c,d). Tapidor has a facultative, rather than obligate, vernalisation requirement, meaning that vernalisation speeds up the onset of flowering in Tapidor, rather than being a prerequisite of flowering. Cold treatment therefore causes the plants to become competent to flower earlier in development than they would have without the treatment41. The observed similarity in Westar samples and Tapidor samples post-cold treatment is therefore consistent with Westar plants being capable of flowering throughout the time series, with Tapidor becoming competent to flower after the cold treatment. Given that the Tapidor plants had visible flower buds at the final time point sampled, it is perhaps surprising that the BnaFT genes in the Tapidor leaf samples do not reach the same expression levels as observed in the Westar leaf samples (Fig. 10a–d). Potentially the low expression of BnaFT genes in the Tapidor leaf samples is a consequence of the age of the leaves, raising the possibility that the ability of leaves to signal within the photoperiod flowering pathway decreases over time. However, a similar transcriptome time series found that expression of the age-dependent pathway factor, SPL5, also increases during vernalisation in a winter variety of OSR, suggesting that this pathway may also be contributing to the floral transition in Tapidor50.
The global gene expression differences between varieties suggest that the apex transcriptome reflects floral development, whereas the leaf transcriptome reflects the age of the plant. The distance heatmaps suggest this, with the leaf transcriptomes between varieties tending to be most similar on the same sampling day (Fig. 2b). While this is true of the beginning of the apex time series, the trajectories diverge after the vernalisation treatment; the final time points, sampled when both varieties had reached BBCH stage 51 (Fig. 1), are most similar despite being sampled on different days (Fig. 2a). This conclusion is supported by the GO term enrichment for the most populated “Late” SOM clusters, with floral development terms enriched in those clusters in the apex (Fig. 5 and Supplementary Table 6) and leaf senescence and age-related terms enriched in the leaf (Supplementary Fig. 6 and Supplementary Table 6). This finding is consistent with the results of a study comparing two rapid-cycling B. rapa varieties, with gene expression in the first true leaf synchronised over time and gene expression in apex tissue following floral development more closely52. The authors suggested that differences in the rate of development between the varieties was likely to be the result of changes at the shoot apex, rather than the leaf, which also seems to be the case with the OSR varieties tested in this study.
Findings from the global level analysis are also observed in the behaviour of key floral genes. The synchronisation of the apex transcriptome with flower development can be seen when looking at copies of the ABC floral genes BnaAP1, BnaPI, and BnaAG (Fig. 6). The expression of these genes in the apex, which can be seen as direct evidence of the floral transition having taken place, begins to increase on day 65 in Westar, whereas expression of these genes is not detected in Tapidor until the final time point at day 83 (Fig. 6). The flowering gene distance heatmaps show that the vernalisation treatment results in the Tapidor and Westar transcriptomes becoming more similar in both tissues (Fig. 2c,d). In Arabidopsis, the expression of FLC is a readout of the vernalisation state of the plant; if FLC is stably silenced after cold treatment then the plant has been vernalised, while rapid-cycling accessions of Arabidopsis have low levels of FLC throughout development primarily due to loss of activity of the FRIGIDA gene8,63. Looking at the BnaFLC genes in OSR, we see that two genes follow this pattern of expression: BnaFLC.A2 and BnaFLC.A10. This is in line with previous findings, with the A2 and A10 homologues of FLC being found multiple times to be associated with flowering time in OSR and B. rapa40,41,42, 44, 50, 64,65,66,67. In Westar, BnaFLC.A10 contains a LINE insertion and BnaFLC.A2 contains a hAT retrotransposon, making both genes non-functional68. Therefore, despite both genes being lowly expressed at the beginning of the time series, the genes are unlikely to be having any repressive effect on flowering. Although the repression of BnaFLC.A2 and BnaFLC.A10 is stable in the apex in Tapidor (Fig. 8a,c), the same is not true in the leaf (Supplementary Fig. 7). This may reflect the number of cells dividing in both tissues. In the meristem of the apex, the more frequent cell divisions may lead to more stable repression compared to the first true leaf69,70. However, this explanation does not fully explain the stability of BnaFLC gene expression, as other gene copies, such as BnaFLC.A3b, exhibit reactivation post-cold in the apex (Supplementary Fig. 7).
Looking to the other copies of BnaFLC, we find BnaFLC.A3b (Supplementary Fig. 7) and BnaFLC.C2 (Fig. 8d) are both more highly expressed in Tapidor at the beginning of the time series, although show an expression reduction during the vernalisation treatment in both varieties. While this expression remains low in Westar, in Tapidor there is reactivation post-cold. Reactivation is observed in some Arabidopsis FLC alleles when not given adequate cold treatment71,73,73. This may suggest that these particular copies have not received adequate cold in our experiment in order to become fully repressed in Tapidor. In B. oleracea, variation in reactivation of BoFLC.C2 contributes to differences in flowering time in different genotypes74. In Arabidopsis, variation in the efficiency of FLC silencing contributes to natural variation in vernalisation response73,75, with different FLC alleles found to have different optimal vernalisation temperatures71,72. If this is true for the different BnaFLC genes in OSR, with different copies having different optimal vernalisation temperatures, it may allow OSR plants to disentangle the length of vernalisation and the temperatures experienced during cold, to better adjust its development. Alternatively, FLC reactivation similar to BnaFLC.A3b and BnaFLC.C2 is observed in the perennial Arabidopsis halleri76,77. It may be the case that these BnaFLC genes exhibiting reactivation retain a perennial mode of action as a result of, or perhaps in spite of, domestication. Another possibility is that the difference between BnaFLC genes is an indicator of the vernalisation requirements of the B. rapa and B. oleracea ancestors that hybridised to form B. napus less than 10,000 years ago33. However, this is difficult to assess as modern-day B. rapa and B. oleracea exhibit considerable diversity in their flowering regimes78,79 and FLC homologue behaviour80,82,82. This is complicated by homologous exchange events, such as the BnaFLC.C2 gene being replaced by the BnaFLC.A2 gene in Tapidor, but not in Westar68. The array of different BnaFLC dynamics observed suggests that these genes have undergone subfunctionalisation. Indeed, in Arabidopsis, the FLC protein was found to bind at hundreds of sites across the genome and, in addition to flowering, potentially have roles in stress and hormone response pathways83,84. This also seems to be the case in other Brassicaceae, such as Arabis alpina85. It is therefore possible that BnaFLC genes which were lowly expressed in both varieties in this time series have roles in pathways other than flowering.
In a previous work we showed that BnaTFL1 genes exhibited very different expression patterns, and this seemed to correspond to differences in the DNA sequence surrounding the gene, which had been found to contain cis-regulatory elements37. Comparing between varieties in this study we find the C3 copy of BnaTFL1 to be highly expressed and have similar expression dynamics in both varieties (Fig. 10g). The other three copies of BnaTLF1, however, are more lowly expressed in Tapidor relative to Westar. Further evidence for sub- or neo-functionalisation of flowering time genes comes from comparing BnaSOC1 genes, which show a clear tissue preference (Fig. 9). This supports other studies which have found SOC1 homologues in Brassica species to have different expression patterns, due to a partitioning of transcription factor binding sites between the different copies48,86, 87. There are also between-variety differences between the copies, with the genes being consistently higher expressed in Westar relative to Tapidor (Fig. 9). This may be due to the higher overall BnaFLC expression in Tapidor, as in Arabidopsis FLC binds within the SOC1 promoter in vitro and is required for SOC1 repression in vivo10,29.
Many gene-level findings here support those of Matar and colleagues, with similar BnaFLC copies exhibiting vernalisation responsiveness and upregulation of BnaSOC1 genes from reference chromosomes A5 and C4_random in the apex50. Where our results diverge, however, are in the expression of BnaAP1 and BnaPI genes, which are expressed during vernalisation in the previous study but expressed after vernalisation in this study (Fig. 6). This may well be due to the longer vernalisation period used (9 weeks compared to 6 weeks) or may represent a physiological difference between both Tapidor and Westar and Express617. Matar et al. also found no significant difference between BnaSOC1 genes between apex and leaf samples. This is likely due to overall transcript levels being assessed, which would mask the tissue subfunctionalisation we observe here (Fig. 9). Other transcriptomic studies for OSR also exist, investigating differential gene expression between different developmental and vernalisation states49,51. Although all these studies focus on OSR homologues of known flowering genes, they often lack more general analysis of global transcriptome dynamics. In this work we show that by calculating Euclidean distances between transcriptomes, and using SOMs, we can observe global dynamics which suggest the apex and first true leaf transcriptomes are both strongly affected by the cold treatment, which appears to be primarily due to the change in day length. This allows gene-level results, such as the downregulation of BnaFT genes (Fig. 10) to be considered in context with what is happening at the global level.
In this work we have compared the transcriptomes of two varieties of OSR during pre-floral development. Taking the whole transcriptome into consideration, we find that the apex transcriptome seems to represent the developmental state of the plant, while the first true leaf transcriptome reflects the age of the plant. Within each tissue, however, the transcriptomes between varieties were found to be largely similar. Differences became apparent between varieties when focussing on floral genes, and in particular, BnaFLC genes. Additionally, BnaSOC1 genes appear to have undergone tissue-specific subfunctionalisation. The transcriptomes presented here provide an important resource for OSR researchers investigating flowering related genes, and as such, we have made the data available in a web based expression browser54 (order.jic.ac.uk).
Materials and methods
Plant growth and sampling
Brassica napus cv. Westar and Brassica napus cv. Tapidor plants were grown and sampled as previously described37. Plants were sown on 7th May 2014 in cereals mix and grown in unlit glasshouses in Norwich, UK, with temperatures set at 18 °C during the day and 15 °C at night. The day length was approximately 16 h. On day 22 plants were transferred to a 5 °C controlled environment room with 8 h days. After 6 weeks at 5 °C, plants were transferred back to the unlit glasshouses and grown until the plants flowered.
The first true leaf and shoot apex of each plant was sampled at 22, 43, 64, 65, 67, 69, 72, and 83 days after sowing (between BBCH stages 13 and 52). To ensure that this developmental range was aligned between varieties, towards the end of the time series varieties were samples on different days from each other (Table 1). The first true leaves were cut and immediately frozen in liquid nitrogen. The shoot apices were dissected on a chilled tile before being transferred to liquid nitrogen.
Samples were pooled and ground in preparation for RNA extraction. For apex tissue, 0.1 g of apices were ground. For leaf samples, between 6 and 10 leaf samples from separate plants were pooled and ground. RNA extraction and DNase treatment were performed following the method provided with the E.Z.N.A® Plant RNA Kit (Omega Bio-tek Inc., www.omegabiotek.com).
Library preparation and RNA sequencing were carried out by the Earlham Institute, Norwich, UK (www.earlham.ac.uk). RNA sequencing (RNA-Seq) was performed on an Illumina HiSeq2500, generating 100-bp single-end reads. The number of biological pools sequenced for each time point, for each tissue, is indicated in Table 1. Gene expression levels were quantified using the previously published approach. Briefly, the Darmor-bzh reference genome sequence88 was used to align reads to. AUGUSTUS89 (version 3.2.2) was used to perform gene model prediction, using the aligned reads to aid intron detection. The Tuxedo suite of software was used to align reads and quantify gene expression levels90. As in our previous work, genes were defined as expressed if their maximal expression level across the transcriptomic time series was greater than, or equal to, 2.0 FPKM.
Floral gene annotation
OSR gene models were assigned homologous Arabidopsis gene names based on a blastn91 (version 2.2.30 +91) approach already published37. OSR floral genes are defined as those gene models with homologous Arabidopsis gene models which are in the FLOR-ID database of flowering genes56.
Euclidean distance calculation
The dist function in the R statistical programming language was used to calculate the Euclidean distance between transcriptomes. Either all genes were used, or just the OSR flowering time genes as defined above.
UpSet plots of expressed genes
UpSet plots were plotted using the UpSetR package92 (version 1.4.0).
Self-organising map analysis
The self-organising map (SOM) analysis was carried out as previously reported37, with SOM ratio set to the ratio of eigenvalues of the first two principal components and the size determined using a sum of squared distance metric. For each tissue and variety combination, all 155,240 OSR genes were filtered to remove any genes not detected in half of time points, any genes not expressed above 2.0 FPKM at any point in the time series, and any genes which did not have an Arabidopsis homologue, in that order. The number of genes removed by each filtering step, and the number of genes clustered for each SOM, is shown in Table 2. The kohonen package93 (version 2.0.19) was used to calculate and plot the SOMs.
Enrichment of variety specific genes within SOM clusters
For each SOM cluster, a Fisher test was conducted using the fisher.test function in the R statistical programming language to test if variety specific genes were enriched in that cluster relative to the total number of genes mapped to that cluster. The p-values from these tests were False Discovery Rate94 adjusted using the p.adjust function.
Gene ontology analysis
Gene ontology (GO) terms were assigned to OSR gene models based on Arabidopsis GO term assignments. The Arabidopsis gene with the highest BLAST bit score to each OSR gene model was taken, and the Arabidopsis GO assignments for that gene, from the org.At.tair.db package95 were assigned to the OSR gene. GO enrichment analysis was performed using the topGO package96.
SOM cluster correlation
For each SOM cluster, between-variety Euclidean distance matrices, such as those in Fig. 2, were calculated for each individual gene. A tissue-specific Pearson correlation coefficient was calculated for each gene by calculating the coefficient between each gene-specific distance matrix and the global distance matrix by flattening the matrices into a vector. Using this method, genes with a high correlation coefficient have a similar normalised distance matrix to the normalised global matrix. The cumulative proportion of genes within each SOM cluster below a certain correlation threshold was determined (Fig. 7), and the area under these curves was used to identify the three SOM clusters with the highest proportion of highly correlated genes.
Plant research statement
Both Tapidor and Westar, as Brassica napus cultivars, are covered by Annex 1 of the International Treaty on Plant Genetic Resources for Food and Agriculture. The authors assert that all institutional, national, and international guidelines and legislation were followed during study. Seeds for Tapidor and Westar can be obtained by contacting Dr. Rachel Wells (rachel.wells@jic.ac.uk).
Data availability
The raw reads have been deposited in the NCBI Sequence Read Archive under the BioProject numbers PRJNA398789 and PRJNA565743.
References
Tardieu, F. Plant response to environmental conditions: Assessing potential production, water demand, and negative effects of water deficit. Front. Physiol. 4, 17 (2013).
Friedman, J. The evolution of annual and perennial plant life histories: Ecological correlates and genetic mechanisms. Annu. Rev. Ecol. Evol. Syst. 51, 461–481 (2020).
Friedt, W. & Snowdon, R. Oilseed rape. In Oil Crops (eds Vollmann, J. & Rajcan, I.) 91–126 (Springer, 2010). https://doi.org/10.1007/978-0-387-77594-4_4.
Brown, J. K. M., Beeby, R. & Penfield, S. Yield instability of winter oilseed rape modulated by early winter temperature. Sci. Rep. 9, 6953 (2019).
Tun, W., Yoon, J., Jeon, J.-S. & An, G. Influence of climate change on flowering time. J. Plant Biol. 64, 193–203 (2021).
Andrés, F. & Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639 (2012).
Xu, S. & Chong, K. Remembering winter through vernalisation. Nat. Plants 4, 997–1009 (2018).
Michaels, S. D. & Amasino, R. M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 (1999).
Sheldon, C. C. et al. The FLF MADS box gene: A repressor of flowering in arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458 (1999).
Helliwell, C. A., Wood, C. C., Robertson, M., James Peacock, W. & Dennis, E. S. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46, 183–192 (2006).
Searle, I. et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912 (2006).
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M. & Araki, T. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962 (1999).
Samach, A. et al. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616 (2000).
Yoo, S. K. et al. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139, 770–778 (2005).
Tiwari, S. B. et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187, 57–66 (2010).
Corbesier, L. et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033 (2007).
Jaeger, K. E. & Wigge, P. A. FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17, 1050–1054 (2007).
Mathieu, J., Warthmann, N., Küttner, F. & Schmid, M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17, 1055–1060 (2007).
Notaguchi, M. et al. Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 49, 1645–1658 (2008).
Alvarez, J., Guli, C. L., Yu, X.-H. & Smyth, D. R. terminal flower: A gene affecting inflorescence development in Arabidopsis thaliana. Plant J. 2, 103–116 (1992).
Shannon, S. & Meeks-Wagner, D. R. A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell Online 3, 877–892 (1991).
Bradley, D., Ratcliffe, O., Vincent, C., Carpenter, R. & Coen, E. Inflorescence commitment and architecture in Arabidopsis. Science 275, 80–83 (1997).
Ho, W. W. H. & Weigel, D. Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell Online 26, 552–564 (2014).
Wigge, P. A. et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059 (2005).
Abe, M. et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056 (2005).
Jaeger, K. E., Pullen, N., Lamzin, S., Morris, R. J. & Wigge, P. A. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell Online 25, 820–833 (2013).
Lee, H. et al. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376 (2000).
Onouchi, H., Igeño, M. I., Périlleux, C., Graves, K. & Coupland, G. Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12, 885–900 (2000).
Hepworth, S. R., Valverde, F., Ravenscroft, D., Mouradov, A. & Coupland, G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21, 4327–4337 (2002).
Moon, J. et al. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35, 613–623 (2003).
Seo, E. et al. Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and Its upstream negative regulator FLC. Plant Cell 21, 3185–3197 (2009).
Wang, J.-W., Czech, B. & Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749 (2009).
Rana, D. et al. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 40, 725–733 (2004).
Allender, C. J. & King, G. J. Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 10, 54 (2010).
He, Z. et al. Genome structural evolution in Brassica crops. Nat. Plants 7, 757–765 (2021).
Schiessl, S., Samans, B., Hüttel, B., Reinhard, R. & Snowdon, R. J. Capturing sequence variation among flowering-time regulatory gene homologs in the allopolyploid crop species Brassica napus. Front. Plant Sci. 5, 404 (2014).
Jones, D. M. et al. Spatio-temporal expression dynamics differ between homologues of flowering time genes in the allopolyploid Brassica napus. Plant J. 96, 103–118 (2018).
Jones, D. M. & Vandepoele, K. Identification and evolution of gene regulatory networks: insights from comparative studies in plants. Curr. Opin. Plant Biol. 54, 42–48 (2020).
Schiessl, S. V., Huettel, B., Kuehn, D., Reinhardt, R. & Snowdon, R. J. Flowering time gene variation in Brassica species shows evolutionary principles. Front. Plant Sci. 8, 1742 (2017).
Osborn, T. C. et al. Comparison of flowering time genes in Brassica rapa, B. napus and Arabidopsis thaliana. Genetics 146, 1123–1129 (1997).
Long, Y. et al. Flowering time quantitative trait loci analysis of oilseed Brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics 177, 2433–2444 (2007).
Zou, X. et al. Comparative analysis of FLC homologues in Brassicaceae provides insight into their role in the evolution of oilseed rape. PLoS ONE 7, e45751 (2012).
Schranz, M. E. et al. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162, 1457–1468 (2002).
Tudor, E. H. et al. QTL-seq identifies BnaFT.A02 and BnaFLC.A02 as candidates for variation in vernalization requirement and response in winter oilseed rape (Brassica napus). Plant Biotechnol. J. 18, 2466 (2020).
Calderwood, A. et al. Total FLC transcript dynamics from divergent paralogue expression explains flowering diversity in Brassica napus. New Phytol. 229, 3534–3548 (2021).
Wang, J. et al. Promoter variation and transcript divergence in Brassicaceae lineages of FLOWERING LOCUS T. PLoS ONE 7, e47127 (2012).
Guo, Y., Hans, H., Christian, J. & Molina, C. Mutations in single FT- and TFL1-paralogs of rapeseed (Brassica napus L.) and their impact on flowering time and yield components. Front. Plant Sci. 5, 282 (2014).
Matar, S. & Melzer, S. A 598-bp InDel variation in the promoter region of Bna.SOC1.A05 is predominantly present in winter type rapeseeds. Front. Plant Sci. 12, 547 (2021).
Schiessl, S. V., Quezada-Martinez, D., Tebartz, E., Snowdon, R. J. & Qian, L. The vernalisation regulator FLOWERING LOCUS C is differentially expressed in biennial and annual Brassica napus. Sci. Rep. 9, 1–15 (2019).
Matar, S., Kumar, A., Holtgräwe, D., Weisshaar, B. & Melzer, S. The transition to flowering in winter rapeseed during vernalization. Plant Cell Environ. 44, 506–518 (2021).
Shah, S. et al. Whole-transcriptome analysis reveals genetic factors underlying flowering time regulation in rapeseed (Brassica napus L.). Plant Cell Environ. 41, 1935–1947 (2018).
Calderwood, A. et al. Comparative transcriptomics reveals desynchronisation of gene expression during the floral transition between Arabidopsis and Brassica rapa cultivars. Quant. Plant Biol. 2, e4 (2021).
Murphy, L. A. & Scarth, R. Vernalization response in spring oilseed rape (Brassica napus L.) cultivars. Can. J. Plant Sci. 74, 275–277 (1994).
Jones, D. M. et al. The oilseed rape developmental expression resource: a resource for the investigation of gene expression dynamics during the floral transition in oilseed rape. BMC Plant Biol. 20, 344 (2020).
Lancashire, P. D. et al. A uniform decimal code for growth stages of crops and weeds. Ann. Appl. Biol. 119, 561–601 (1991).
Bouché, F., Lobet, G., Tocquin, P. & Périlleux, C. FLOR-ID: An interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 44, D1167–D1171 (2016).
Irish, V. The ABC model of floral development. Curr. Biol. 27, R887–R890 (2017).
Ratcliffe, O. J., Kumimoto, R. W., Wong, B. J. & Riechmann, J. L. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169 (2003).
Huang, F., Liu, T. & Hou, X. Isolation and functional characterization of a floral repressor, BcMAF1, From Pak-choi (Brassica rapa ssp. Chinensis). Front. Plant Sci. 9, 290 (2018).
Akter, A. et al. The histone modification H3 lysine 27 tri-methylation has conserved gene regulatory roles in the triplicated genome of Brassica rapa L.. DNA Res. 26, 433–443 (2019).
Lee, J. & Lee, I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254 (2010).
Pullen, N., Jaeger, K. E., Wigge, P. A. & Morris, R. J. Simple network motifs can capture key characteristics of the floral transition in Arabidopsis. Plant Signal. Behav. 8, 820 (2013).
Johanson, U. et al. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347 (2000).
Kole, C., Quijada, P., Michaels, S. D., Amasino, R. M. & Osborn, T. C. Evidence for homology of flowering-time genes VFR2 from Brassica rapa and FLC from Arabidopsis thaliana. Theor. Appl. Genet. 102, 425–430 (2001).
Lou, P. et al. Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. J. Exp. Bot. 58, 4005–4016 (2007).
Xiao, D. et al. The Brassica rapa FLC homologue FLC2 is a key regulator of flowering time, identified through transcriptional co-expression networks. J. Exp. Bot. https://doi.org/10.1093/jxb/ert264 (2013).
Zhao, J. et al. BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J. Exp. Bot. 61, 1817–1825 (2010).
Song, J.-M. et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 6, 34–45 (2020).
Finnegan, E. J. & Dennis, E. S. Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr. Biol. 17, 1978–1983 (2007).
Wellensiek, S. J. Dividing cells as the prerequisite for vernalization. Plant Physiol. 39, 832–835 (1964).
Coustham, V. et al. Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science 337, 584–587 (2012).
Duncan, S. et al. Seasonal shift in timing of vernalization as an adaptation to extreme winter. eLife 4, e06620 (2015).
Shindo, C., Lister, C., Crevillen, P., Nordborg, M. & Dean, C. Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev. 20, 3079–3083 (2006).
Irwin, J. A. et al. Nucleotide polymorphism affecting FLC expression underpins heading date variation in horticultural brassicas. Plant J. 87, 597–605 (2016).
Hepworth, J. et al. Natural variation in autumn expression is the major adaptive determinant distinguishing Arabidopsis FLC haplotypes. eLife 9, e57671 (2020).
Satake, A. et al. Forecasting flowering phenology under climate warming by modelling the regulatory dynamics of flowering-time genes. Nat. Commun. 4, 2303 (2013).
Nishio, H., Iwayama, K. & Kudoh, H. Duration of cold exposure defines the rate of reactivation of a perennial FLC orthologue via H3K27me3 accumulation. Sci. Rep. 10, 16056 (2020).
Guo, Y., Chen, S., Li, Z. & Cowling, W. A. Center of origin and centers of diversity in an ancient crop, Brassica rapa (Turnip Rape). J. Hered. 105, 555–565 (2014).
Woodhouse, S. et al. Validation of a novel associative transcriptomics pipeline in Brassica oleracea: Identifying candidates for vernalisation response. BMC Genomics 22, 539 (2021).
Akter, A. et al. Genome triplication leads to transcriptional divergence of FLOWERING LOCUS C genes during vernalization in the genus Brassica. Front. Plant Sci. 11, 619417 (2021).
Ridge, S., Brown, P. H., Hecht, V., Driessen, R. G. & Weller, J. L. The role of BoFLC2 in cauliflower (Brassica oleracea var. botrytis L.) reproductive development. J. Exp. Bot. 66, 125–135 (2015).
Takada, S. et al. The role of FRIGIDA and FLOWERING LOCUS C genes in flowering time of Brassica rapa leafy vegetables. Sci. Rep. 9, 13843 (2019).
Deng, W. et al. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. 108, 6680–6685 (2011).
Mateos, J. L. et al. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 16, 31 (2015).
Mateos, J. L. et al. Divergence of regulatory networks governed by the orthologous transcription factors FLC and PEP1 in Brassicaceae species. Proc. Natl. Acad. Sci. 114, E11037–E11046 (2017).
Sri, T., Mayee, P. & Singh, A. Sequence and expression variation in SUPPRESSOR of OVEREXPRESSION of CONSTANS 1 (SOC1): Homeolog evolution in Indian Brassicas. Dev. Genes Evol. 225, 287–303 (2015).
Sri, T., Gupta, B., Tyagi, S. & Singh, A. Homeologs of Brassica SOC1, a central regulator of flowering time, are differentially regulated due to partitioning of evolutionarily conserved transcription factor binding sites in promoters. Mol. Phylogenet. Evol. 147, 106777 (2020).
Chalhoub, B. et al. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345, 950–953 (2014).
Stanke, M., Diekhans, M., Baertsch, R. & Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644 (2008).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Camacho, C. et al. BLAST+: Architecture and applications. BMC Bioinformatics 10, 421 (2009).
Conway, J. R., Lex, A. & Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017).
Wehrens, R. & Buydens, L. M. C. Self- and super-organising maps in R: The kohonen package. J Stat. Softw. 21, 1–19. https://doi.org/10.18637/jss.v021.i05 (2007).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 (1995).
Carlson, M. org.At.tair.db: Genome wide annotation for Arabidopsis. (2020).
Alexa, A. & Rahnenfuhrer, J. topGO: Enrichment analysis for gene ontology. (2020).
Acknowledgements
Many thanks to Dr. Judith Irwin for her important contributions to the project and experimental design.
Funding
MJ acknowledges support from the John Innes Foundation for the JIC Rotation PhD Programme and from the Global Challenges Research Fund (BBS/OS/GC/200013B). RW and MT are grateful for support from BBSRC's Institute Strategic Programme on Growth and Development (BB/J004588/1) and Genes in the Environment (BB/P013511/1), RM, JH and RW acknowledge support from the BBSRC sLoLa ‘Brassica Rapeseed and Vegetable Optimisation’ (BB/P003095/1). RM acknowledges support from BBSRC's Institute Strategic Programme on Biotic Interactions underpinning Crop Productivity (BB/J004553/1) and Plant Health (BB/P012574/1). RM and RW are supported by the BBSRC funded Institute Strategic Programme Building Robustness in Crops (BRiC) (BB/X01102X/1).
Author information
Authors and Affiliations
Contributions
R.M. conceived the project. M.J., N.P., R.W., M.T., and R.M. designed the experiments that were carried out by R.W. with support from N.P. and M.J. The sequence analysis was carried out by M.J. with help from M.T. M.J. performed all transcriptomic time-series analyses and produced all the figures. M.J., J.H., and R.M. drafted the manuscript. All authors contributed to editing and proofreading the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jones, D.M., Hepworth, J., Wells, R. et al. A transcriptomic time-series reveals differing trajectories during pre-floral development in the apex and leaf in winter and spring varieties of Brassica napus. Sci Rep 14, 3538 (2024). https://doi.org/10.1038/s41598-024-53526-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-53526-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.