Abstract
Safety concerns, stemming from the presence of complex and unpredictable adulterants, permeate the entire industrial chain of traditional Chinese medicines (TCMs). The Notopterygii Rhizoma et Radix (NReR) from the Apiaceae family, commonly known as “Qiang-huo”, is a widely used herbal medicine. The recent surge in its demand has given rise to a proliferation of counterfeit and substituted products in the market. Traditional identification presents inherent limitations, while DNA mini-barcoding, reliant on sequencing a short-standardized region, has received considerable attention as a new potential means to identify processed medicinal materials. In this study, we constructed a comprehensive Internal Transcribed Spacer 2 (ITS2) matrix encompassing genuine NReR and their commonly found adulterants for the first time. Leveraging this matrix, we conducted a thorough assessment of the genetic profiles and sources of NReR available in the Chinese herbal medicine market. Following established DNA barcoding protocols, the intra-specific genetic divergences within NReR species were found to be lower than the inter-specific genetic divergences from other species. Among the 120 samples that were successfully amplified, ITS2 exhibits an outstanding species-level identification efficiency of 100% when evaluated using both the BLASTN and neighbor-joining (NJ) tree methods. We concluded that ITS2 is a mini-barcode that has shown its potential and may become a universal mini-barcode for the quality control of “Qiang-huo”, thereby ensuring the safety of clinical medication.
Similar content being viewed by others
Introduction
Traditional Chinese medicine (TCM), encompassing Chinese herbal medicine, continues to gain international recognition. According to the National Bureau of Statistics of China, the turnover of the Chinese herbal medicine market in 2019 reached 165.3 billion yuan for the domestic market and $6.175 billion for the international side1. Apiaceae, a family of flowering plants, is recognized as a significant resource for TCM, comprising 65 genera and 262 species2. Among them, Notopterygii Rhizoma et Radix (NReR), known as "Qiang-huo" in Chinese, holds a long history dating back to the Han Dynasty, approximately 2000 years ago3. According to the Pharmacopoeia of the People's Republic of China, NReR is derived from the roots and rhizomes of Notopterygium incisum Ting ex H. T. Chang or N. franchetii H. de Boissieu4. It encompasses a complex array of chemical constituents, including volatile oils and terpenes, coumarins, sugars, glycosides, phenolic acids, polyalkynes, and alkaloids5. Modern pharmacological studies have demonstrated its anti-inflammatory, antibacterial, antioxidant, antiarrhythmic, anticancer, antipyretic, and analgesic activities6. Currently, NReR serves as a raw material for over two hundred types of Chinese (Tibetan) patent medicines.
Notopterygium incisum and N. franchetii were listed as national third-class protected plants as early as 1987, and they were successively included as "Near-Threatened" species in China's Red List of Biodiversity and China Species Red List. In the past few decades, excessive excavation and habitat destruction lead the wild resources of NReR to be drastically reduced7,8,9. The scarcity of resources and the increase of market demand have driven the price up, which motivated adulteration intentionally. Reports indicate that Angelica sylvestris L., Pleurospermum rivulorum (Diels) Hiroe, Polygonum cuspidatum Sieb et Zucc, and Sanguisorba officinalis L. are frequently sold as NReR in the medicinal market10,11,12. These species share similar organoleptic characteristics but differ in chemical constituents compared to NReR13,14,15,16,17. Traditional methods used to authenticate NReR and its adulterants, such as macroscopy11,18,19, microscopy20, and chemical profiles21,22, can provide certain recognition and differentiation to some extent. But, these methods are prone to geographical variations, growth stages, and storage conditions, which may affect identification accuracy23. Therefore, it is necessary to establish a simple and accurate identification method to distinguish NReR from adulterations.
DNA barcode technology is currently used as an effective tool to identify species. This method provides a large amount of genetic information with high accuracy and objectivity, and it can standardize and automate the identification process, establishing an easy-to-use application system in a short time23. One potential DNA barcode for identifying medicinal plants and their close relatives is the internal transcribed spacer 2 (ITS2), which has attracted attention due to its unique advantages such as being short and conducive to amplifying degraded samples24,25. While a few studies have examined the molecular identification of NReR and its adulterants using DNA barcoding26,27,28, the composition of commercial NReR in the Chinese medicinal market needs to be sorted out to ensure the subsequent formulation of quality control standards and clinic safety for NReR.
Thus, the aim of this study is to investigate whether ITS2 is a valuable marker for identifying genuine NReR from its adulterants and to gain insight into the composition of commercial NReR in China.
Materials and methods
Plant materials
Eighteen ITS sequences available for genuine NReR (N. incisum and N. franchetii) were first downloaded and screened from NCBI GenBank as reference. Moreover, 168 commercial crude drug samples under the name of NReR were collected from herbal markets, pharmacies, and online shops in 23 provinces and municipalities of China. Voucher specimens are deposited in Herbarium of Kunming Medical University. Detailed information is presented in the Table S1. All methods of experimental research on plants were performed in accordance with the relevant institutional, national, and international guidelines and legislation.
DNA extraction, polymerase chain reaction (PCR) amplification, and sequencing
The surface of all herbal materials was cleaned with 75% ethanol to avoid fungal DNA contamination. About 50 mg of the materials were cut into pieces, added with 10% polyvinylpyrrolidone (PVP), and then ground with a FastPrep bead mill (Retsch MM400, Germany). Total genomic DNA was extracted using the modified CTAB procedure of Doyle and Doyle29 or using the Plant Genomic DNA Kit (Tiangen Biotech, Beijing, China). Agarose gel electrophoresis showed slight smearing in some DNA samples, indicating partial degradation. The universal primers ITS-S2F (5′-ATGCGATACTTGGTGTGAAT-3′) and ITS-S3R (5′-GACGCTTCTCCAGACTACAAT-3′) were used to amplify the complete ITS2 region24. The PCR reaction conditions were the same as described previously30. PCR products purifying and sequencing were completed by Tsingke Biotechnology Co., Ltd (Beijing, China).
Data analysis
The sequences of genuine and commercial NReR were assembled using the MAFFT v.731, and manually adjusted where necessary using the BioEdit32. The assembled sequences were annotated and trimmed to obtain the complete ITS2 region based on a Hidden Markov Model (HMM)33. The genetic distances were calculated using MEGA v.734 according to Kimura 2-parameter (K2P) model34. Barcoding gaps comparing the distributions of the pairwise intra- and inter-specific distances with distance intervals of 0.002 were estimated in Microsoft Excel 2016. The true NReR presenting a minimum inter-specific distance value higher than their maximum intra-specific distance were considered successfully discriminated from potential adulterant plant species35. Wilcoxon two-sample tests were performed as described previously24,36. Haplotype matrix was generated by DNAsp v.637. BLASTN and the nearest distance methods were both used to evaluate the species authentication efficacy38. Sequences were uploaded onto NCBI database with a minimum identity cut of 99% for a top match according to the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Neighbor-joining (NJ) tree was constructed based on haplotypes, performing 1,000 bootstrap replicates in MEGA v.734.
Results
Amplification, sequencing and sequence characteristics
Genomic DNA was extracted from a total of 168 commercial "Qiang-huo" products, out of which 48 samples failed to amplify due to severe DNA degradation. The length of 138 combined sequences ranges from 226 to 256 bp. The GC content of the sequences shows a mean value of 58.3% with a range of 53.2% to 68.2%. The aligned length of 263 bp exhibits 143 variable sites, a rate of 54.4% (Table 1). These findings suggest that sequences for the sampled "Qiang-huo" were relatively variable.
Assessment of barcoding gap
The average interspecific distance between N. incisum and N. franchetii was 0.039. The interspecific distance between N. incisum and the adulterant species ranges from 0.037 (N. oviforme) to 0.659 (Broussonetia papyrifera). Notopterygium franchetii shows a similar interspecific distance with the adulterants, with the maximum interspecific distance being 0.699 from B. papyrifera and the minimum being 0.036 from N. oviforme. The intraspecific genetic distance within N. incisum (0.005) and N. franchetii (0.005) was both smaller than interspecific distance between NReR and adulterants (Fig. 1). Our results show that the intra- and inter-specific variation of ITS2 had distinct gaps (Fig. 2). Additionally, Wilcoxon’s two-sample tests reveals that the mean of the inter-specific divergences was significantly higher than that of the corresponding intra-specific variations (p < 0.001, Table 2).
Evaluation of species authentication capability of ITS2
The BLASTN method exhibits a 100% success rate in identifying the tested commercial samples (Table 3). These samples consist of nine species, namely N. incisum, N. franchetii, N. oviforme, Levisticum officinale, A. amurensis, Ostericum scaberulum, B. papyrifera, Haplosphaera himalayensis, and Heracleum fargesii. Each identification result was supported by best hit of accessions obtained from the NCBI database (Table S1). A few samples initially identified as O. scaberulum, B. papyrifera, Ha. himalayensis and He. fargesii were subsequently confirmed through additional sampling and sequencing.
A total of 25 haplotypes were generated from the ITS2 sequences of genuine NReR and 120 commercial samples (Fig. S1). Combining these haplotypes with the BLASTN results, N. incisum was assigned to Haps_1-7, N. franchetii to Haps_8-15, L. officinale to Hap_17, A. amurensis to Hap_18, O. scaberulum to Haps_19-20, Ha. himalayensis to Haps_21-22, O. scaberulum to Hap_23, He. fargesii to Hap_24, and B. papyrifera to Hap_25 (Table 4). With the exception of N. incisum, the NJ tree analysis reveals that haplotypes representing different species formed isolated clades. While the haplotypes representing N. incisum does not form a monophyletic group on the NJ tree, the sequences to be identified as potential authentic species were clustered together with haplotypes representing genuine species (Fig. 3). Hence, the NJ tree method also exhibits a 100% success rate for NReR identification (Table 3).
Neighbor joining (NJ) tree of NReR and its adulterants constructed based on the haplotypes. Detailed information about the haplotypes associated with each species is shown in Table 4. Bootstrap values are shown (≥ 50%) next to the branch.
Survey of commercial NReR in the Chinese medicine markets
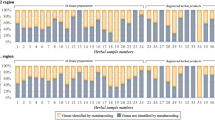
This study represents the most comprehensive nationwide sampling of commercial “Qiang-huo” to date, comprising a total of 168 samples obtained from 23 provinces. Based on their external morphology and odor characteristics, these samples proved challenging to distinguish from one another (Fig. 4A). Of 168 samples, except 48 failed to be amplified, molecular identification results show that 65 samples (54.2%) were identified as authentic “Qiang-huo”, while 55 samples (45.8%) were identified as adulterants (Fig. 4B). Further identification using the BLASTN method reveals that the adulterants belonged to seven different species, namely L. officinale (17 samples), A. amurensis (13 samples), O. scaberulum (five samples), B. papyrifera (L.) Vent. (two samples), Ha. himalayensis (two samples) and He. fargesii (one sample) (Table S1). Levisticum officinale, the most widely sold adulterant, was found in medicinal markets of nine provinces, followed by N. oviforme, A. amurensis and O. scaberulum found in eight, six and two provinces, respectively. Notably, B. papyrifera and Ha. himalayensis were only detected in Yunnan and Xizang, respectively. Regarding spatial distribution, samples from Guizhou, Hunan, Jiangsu, Qinghai, and Zhejiang were all confirmed as genuine NReR, while in Chongqing, Liaoning, Inner Mongolia, Shaanxi, and Xizang, no authentic NReR was detected. In ten provinces, including Hubei, Jilin, Jiangxi, and others, only one adulterant was discovered. Similarly, within the six provinces, such as Sichuan, Guangxi, Gansu, and others, two distinct types of adulterants were observed. The scenario in Yunnan and Anhui is more complicated, as three distinct types of adulterants were found (Tables S1, S2).
(A) Morphology of commercial “Qiang-huo” (SC06: Notopterygium incisum, AH08: N. franchetii, AH16: Angelica amurensis, AH18: Levisticum officinale, AH19: N. oviforme, SC16: Heracleum fargesii, XZ02: Haplosphaera himalayensis, YN06: Ostericum scaberulum, YN14: Broussonetia papyrifera), (B) The composition and proportion of the commercial “Qiang-huo” products identified by barcode ITS2.
Discussion
In recent years, DNA molecular identification technology has emerged as a robust tool for TCMs identification. This technology stands out for its ease of operation, cost-effectiveness, and high accuracy. In 2009 at the 3rd World DNA Barcode Conference, it was announced that the matK and rbcL markers are the core sequences of plant DNA barcodes, with ITS and trnH-psbA as complementary sequences39. The longer length of these markers has shown some weakness in amplification, sequencing and alignment, which is exacerbated if the materials are highly processed and have degraded DNAs40,41. Unlike Western herbs, most Chinese medicinal herbs are subjected to traditional processing procedures to increase their potency, minimize negative effects, and change their medicinal properties for a particular clinical use before they are released into dispensaries, practitioners, and the market42. According to the Chinese pharmacopoeia, the medicinal herbs are typically cleaned, cut, dried, and then processed, including stir-frying, charring, steaming, boiling, and calcining4,43. Raw materials processing methods can cause DNA degradation, posing challenges in obtaining standard DNA barcode sequences for samples. The use of mini-DNA barcoding technology, which focuses on shorter yet more efficiently amplified sequences through PCR, can partially overcome this limitation24,41,44,45.
ITS2, a mini-barcode spanning 160 to 330 bp in length, has emerged as the predominant marker for identifying plant medicinal materials24. Its growing popularity can be attributed to its ease of amplification and remarkable discriminatory capabilities across different taxonomic levels24. In this study, ITS2 performs well, with a higher amplification rate of 71.4%. Unsuccessfully PCR amplified NReR samples could be attributed to the high temperature drying process, causing DNA degradation. Both nucleotide signature (NS) and genome skimming metagenomics (GSM) emerge as promising solutions for fragmented and degraded plant materials identification46. NS consists of distinct nucleotide sequences that are exclusive to a specific taxonomic group. Previous studies, such as those on American Ginseng47, Cistanches Herba48, and Pinelliae Rhizoma49, have successfully demonstrated the efficacy of nucleotide signatures in identifying medicinal materials. GSM is the low-coverage shotgun sequencing of total DNA. When this approach is applied on herbal products, sequencing library is built without PCR amplification of barcode regions, circumventing the limitations of PCR in conventional DNA barcoding, such as DNA degradation during product manufacturing and PCR bias because of primer mismatch, etc. GSM produces millions of reads in a single run. After quality control, reads could then be clustered into operational taxonomic units (OUTs) based on similarity at defined threshold (usually 99–100%). Representative consensus sequences from each cluster would then be subject to taxonomic assignment, usually by alignment-based software like BLAST or k-mer based methods like Kraken46,50. As its sequencing cost is decreasing year by year, GSM technology will be a prospective method for the identification of TCMs herbs.
The validity of DNA barcoding relies heavily on the availability of a precise reference database, as it serves as the cornerstone of DNA barcoding. In this study, we constructed a comprehensive ITS2 matrix encompassing genuine NReR and their commonly found adulterants for the first time. This matrix will play a crucial role in both monitoring NReR and exploring potential substitute sources. The matrix includes two authentic species of NReR (N. incisum and N. franchetii, comprising 16 haplotypes) and seven confused species (nine haplotypes) (Table 4; Fig. S1). Out of the 120 samples sold as "Qiang-huo", only 54.2% were identified as authentic NReR, while the rest were identified as adulterants, including L. officinale, N. oviforme, A. amurensis, O. scaberulum, He. fargesii, Ha. himalayensis, and B. papyrifera (Fig. 4B). These findings highlight the complexity of the "Qiang-huo" market, with the presence of previously unreported species. According to the Chinese Pharmacopeia, only N. incisum and N. franchetii are listed as sources of NReR, and the former one exhibits superior quality and efficacy51. Our analysis revealed that N. incisum accounted for two-thirds of the authentic products, further indicating a preference for N. incisum in the market (Fig. 4B; Table S1). The NJ tree showed that adulterants of NReR were distantly related to N. incisum and N. franchetii (Fig. 3). Levisticum officinale was introduced to China in 1957, and used as a substitute for the traditional Chinese medicine “dang gui”, the roots of A. amurensis, He. fargesii and O. scaberulum52. The chemical and pharmacological analysis results of L. officinale and A. amurensis dramatically differs from those of NReR53,54,55,56,57,58. Notopterygium oviforme and Ha. himalayensis were grouped together in a strongly supported clade (bootstrap support = 99), closely related to authentic NReR, exhibiting genetic distances of 0.059–0.084 and 0.036–0.037, respectively (Figs. 1, 3). While N. oviforme has been traditionally regarded as a regional substitute to NReR, albeit with inferior quality59, additional research is urgently needed to explore the chemical constituents and pharmacological efficacy of both N. oviforme and Ha. himalayensis. This investigation aims to ascertain their potential as viable substitutes for NReR. Broussonetia papyrifera may represent a contaminant in the Yunnan samples, as its morphology significantly differs from that of NReR, and we also detected genuine NReR in these samples.
Conclusions
In this study, the origin plants of commercial “Qiang-huo” in the market was clarified, and the reference matrix of NReR and its adulterants was successfully established. This achievement is crucial for the future industrial development of NReR. However, it is important to acknowledge that the amplification efficiency of the ITS2 region is not always optimal, which is a challenge encountered in DNA barcoding identification of many medicinal materials. Therefore, alternative approaches such as NS and GSM appear to be promising solutions for overcoming this issue. These cutting-edge methodologies have the potential to revolutionize the field, providing more comprehensive and accurate identification results for medicinal materials.
Data availability
New sequenced and other published ITS2 sequences can be found in GenBank (https://www.ncbi.nlm.nih.gov/genbank/), and the accession numbers showed in Table S1.
References
Zhu, S., Liu, Q. Z., Qiu, S. M., Dai, J. P. & Gao, X. X. DNA barcoding: An efficient technology to authenticate plant species of traditional Chinese medicine and recent advances. Chin. Med. 17, 112 (2022).
Wei, J., Gao, Y. Z., Zhou, J. & Liu, Z. W. Collection and sorting of medicinal plants in Chinese Apiaceae (Umbelliferae). Chin. J. Chin. Mater. Med. 44, 5329–5335 (2019).
Sun, X. Y. & Sun, F. J. Shennong Materia Medica (The People’s Health Publishing House, 1982).
Chinese Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China Vol. 1 (People’s Medical Publishing House, 2020).
Ma, L. M. & Yang, J. L. Research progress on chemical constituents and pharmacological activities of Notopterygii Rhizoma et Radix. Chin. Tradit. Herbal. Drugs. 52, 6111–6120 (2021).
Azietaku, J. T. et al. A review of the ethnopharmacology, phytochemistry and pharmacology of Notopterygium incisum. J. Ethnopharmacol. 202, 241–255 (2017).
Sun, H. et al. Studies on habitats suitability of endangered medicinal plant Notopterygium incisum. Chin. J. Chin. Mater. Med. 34, 535–538 (2009).
Zhou, Yi. et al. Resources crisis and protective measures on Rhizoma et Radix Notopterygii. Chin. Tradit. Herbal Drugs. 34, 12–14 (2003).
Jiang, S. Y. et al. Industrialization condition and development strategy of Notopterygii Rhizoma et Radix. Chin. J. Chin. Mater. Med. 42, 2627–2632 (2017).
Xu, G. J., Xu, L. S. & Wang, Z. T. Variety Arrangement and Quality Evaluation of Common Chinese Herbal Medicines Vol. 3 (Fujian Science and Technology Press, 1995).
Wu, X. M. & Yuan, W. S. The identification of Notopterygii Rhizoma et Radix and its adulterants. Chin. Healthc. Innov. 1, 113 (2007) (in Chinese).
Wang, Z., Chen, S. L., Huang, L. F., Song, J. Y. & Du, J. Advances in quality control of Notopterygii Rhizoma et Radix. Chin. J. Mod. Appl. Pharm. 29, 209–214 (2012).
Wang, S. & Wang, T. Z. Qualitative analysis of chemical constituents of the essential olis from Xinjiangqianghuo (Angelica silvestris L.). West Chin. J. Pharmaceut. Sci. 12, 92–93 (1997).
Xiao, Y. Q. et al. Studies on chemical components of Pleurospermum rivulorum (Diels) (I). China J. Chin. Materia Medica 20, 324–424 (1995).
Cui, S. L., Liu, X. H., Yang, L. X., Liu, D. & Xiao, Y. Q. Studies on chemical components of Pleurospermum rivulorum (Diels) (II). Chin. J. Chin. Mater. Med. 10, 743–744 (1995).
Wang, L. N. et al. Hydrosoluble chemical constituents in Sanguisorba officinalis (II). Chin. Tradit. Herbal Drugs 50, 3017–3023 (2019).
Zuo, W. P., Cai, Y., Liu, W. Y. & Wang, D. G. Chemical constituents from Polygonum cuspidatum. Chin. Pharmaceut. J. 55, 189–193 (2020).
Liu, W. L. & Zhou, J. Identification of Notopterygium and Angelica silvestris L.. Shandong Pharma. Industry 17, 15–16 (1998).
Gu, Q. & Zhang, Y. F. Morphological identification of Notopterygii Rhizoma et Radix and its adulterants. Lishizhen Med. Mate. Med. Res. 10, 947 (2001).
Zhou, X. T. et al. Study on characteristics and microscopic identification of fruits of Notopterygium franchetii and N. forrestii. Chin. J. Chin. Mater. Med. 43, 3466–3470 (2018).
Guo, H. Q., Li, Y. Q., Wang, Z. X., Zhang, Z. K. & Ma, C. H. HPLC fingerprint analysis of Notopterygii Rhizoma et Radix with different commercial specifications. Chin. J. Exp. Tradit. Med. Formul. 25, 184–188 (2019).
Jin, H. Y., Chen, X. H., Zhang, S. T., Li, Q. & Bi, K. S. Establishment of chromatographic fingerprint and identification of Rhizoma et Radix Notopterygii and Radix Angelicae Pubescentis by GC. J. Shenyang Pharmaceut. Univ. 26, 369–375 (2009).
Yuan, Q. J. et al. On the methods and principles of molecular identification of Chinese herbs. Plant Divers. 34, 607–613 (2012).
Chen, S. L. et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 5, e8613 (2010).
China Plant BOL Group et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. PNAS 108, 19641–19646 (2011).
Xin, T. Y. et al. Stability and accuracy of the identification of Notopterygii Rhizoma et Radix using the ITS/ITS2 barcodes. Acta. Pharm. Sin. 47, 1098–1105 (2012).
Sun, Z. Y., Chen, S. L., Yao, H. & Song, J. Y. Identification of Notopterygii Rhizoma et Radix and its adulterants using DNA barcoding method based on ITS2 sequence. Chin. Tradit. Herbal Drugs. 43, 568–571 (2012).
Zheng, M. D., Sun, M. M., He, Z. H. & Li, H. Z. Molecular authentication of the medicinal species of Rhizoma et Radix Heraclei, Radix Angelicae Sinensis, Radix Angelicae Pubescentis and Rhizoma et Radix Notopterygii by integrating ITS2 and its secondary structure. Acta. Pharm. Sin. 12, 2289–2294 (2021).
Doyle, J. J. & Doyle, J. L. A rapid DNA isolation procedure for small quantities of fresh leaf issue. Phytochem. Bull. 19, 11–15 (1987).
Liu, Z. W., Gao, Y. Z. & Zhou, J. Molecular authentication of the medicinal species of Ligusticum (Ligustici Rhizoma et Radix, ‘Gao-ben’) by integrating non-coding internal transcribed spacer 2 (ITS2) and its secondary structure. Front. Plant Sci. 10, 429 (2019).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Hall, T. BioEdit: biological sequence alignment editor for Win95/98/NT/2K/XP version 6.0.7. (1999).
Keller, A. et al. 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene 430, 50–57 (2009).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007).
Meier, R., Zhang, G. Y. & Ali, F. The use of mean instead of smallest interspecific distances exaggerates the size of the ‘barcoding gap’ and leads to misidentification. Syst. Biol. 57, 809–813 (2008).
Kress, W. J. & Erickson, D. L. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2, e508 (2007).
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302 (2017).
Ross, H. A., Murugan, S. & Li, W. L. S. Testing the reliability of genetic methods of species identification via simulation. Syst. Biol. 57, 216–230 (2008).
CBOL. A DNA barcode for land plants. PNAS 106, 12794–12797 (2009).
Chen, S. Y., Xia, T., Wang, Y. J., Liu, J. Q. & Chen, S. L. Molecular systematics and biogeography of Crawfurdia, Metagentiana and Tripterospermum (Gentianaceae) based on nuclear ribosomal and plastid DNA sequences. Ann. Bot. 96, 413–424 (2005).
Taberlet, P. et al. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35, e14 (2007).
Zhao, Z. Z. et al. A unique issue in the standardization of Chinese Materia Medica: Processing. Planta Med. 76, 1975–1986 (2010).
Zhao, Z. Z. et al. Authentication is fundamental for standardization of Chinese medicines. Planta Med. 72, 865–874 (2006).
Valentini, A. et al. New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: The trnL approach. Mol. Ecol. Resour. 9, 51–60 (2009).
Meusnier, I. et al. A universal DNA mini-barcode for biodiversity analysis. BMC Genomics 9, 214 (2008).
Wu, H. Y. & Shaw, P. C. Strategies for molecular authentication of herbal products: From experimental design to data analysis. Chin. Med. 17, 38 (2022).
Liu, Y. et al. A nucleotide signature for the identification of American ginseng and its products. Front Plant Sci 7, 319 (2016).
Wang, X. Y. et al. Detection of Cistanches Herba (Rou Cong Rong) medicinal products using species-specific nucleotide signatures. Front. Plant Sci. 9, 1643 (2018).
Zhang, T. Y. et al. A nucleotide signature for the identification of Pinelliae Rhizoma (Banxia) and its products. Mol. Biol. Rep. 49, 7753–7763 (2022).
Handy, S. M. et al. HPLC-UV, metabarcoding and genome skims of botanical dietary supplements: A Case Study in Echinacea. Planta Med. 87, 314–324 (2021).
Peng, R. Study on the Chemical Constituents and Quality Evaluate Method of the Notopterygium incisum (Qiang-huo) (Nanjing University of Chinese Medicine, 2021).
Sheh, M. L. & Watson, M. F. Apiaceae lindley. In Flora of China (eds Wu, Z. Y. & Raven, P. H.) (Science Press, 2005).
Sattari, R., Khayati, G. R. & Hoshyar, R. Biosynthesis of silver-silver chloride nanoparticles using fruit extract of Levisticum Officinale: Characterization and anticancer activity against MDA-MB-468 cell lines. J. Clust. Sci. 32, 593–599 (2021).
Zhang, L. Y. Research on Chinese medicine resources and evaluation of pharmacological activity of Anglica L. in the northeast three provinces (Changchun University of Chinese Medicine, 2020).
Le, W., Wu, Y. L. & Qiu, R. L. Comparative study on Angelica sinensis and Levisticum officinale based on HPLC and chemometrics. J. Chin. Med. Mater. 41, 1846–1850 (2018).
Sertel, S., Eichhorn, T., Plinkert, P. K. & Efferth, T. Chemical composition and antiproliferative activity of essential oil from the leaves of a medicinal herb, Levisticum officinale, against UMSCC1 head and neck squamous carcinoma cells. Anticancer Res. 31, 185–191 (2011).
Abd El-Hamid, S. R. & Abeer, Y. I. Anti-inflammatory, antioxidant, anti-tumor and physiological studies on Levisticum officinale-Koch plant. Planta Med. 75, PE62 (2009).
Lv, R. M., Lin, M., Liu, T. C. & Fang, Q. C. The chemical constituents of Levisticum officinale. Chin. Tradit. Herbal Drugs. 12, 5–6 (1981).
Zhang, H. K. Summary of Chinese Traditional Medicine Resources (Science Press, 1994).
Funding
This work was supported by the National Natural Science Foundation of China (no.31960048), the Foundation of Yunnan Science and Technology Department (no. 202201AT070118), the Ten Thousand Talents Program of Yunnan (YNWRQNBJ-2019–208) and the Hundred-Talent Program of Kunming Medical University (no.60118260127).
Author information
Authors and Affiliations
Contributions
Z.-W.L and J.Z conceived and designed the research protocols; Z.-W.L did most of the analysis, creation of figures, and manuscript writing; Z.-W.L and J.Z edited the final manuscript. J.Z supervised the work. All the authors contributed to the revision and publication processes.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, ZW., Zhou, J. DNA barcoding of Notopterygii Rhizoma et Radix (Qiang-huo) and identification of adulteration in its medicinal services. Sci Rep 14, 2879 (2024). https://doi.org/10.1038/s41598-024-53008-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-53008-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.