Abstract
Acinetobacter baumannii is a Gram-negative bacterium considered an emerging multi-drug-resistant pathogen. Furthermore, this bacterium can survive in extreme environmental conditions, which makes it a frequent cause of nosocomial infection outbreaks. Gene expression analyses by Reverse Transcription Quantitative real-time PCR (RT-qPCR) depend on a reference gene, also called an endogenous gene, which is used to normalize the generated data and thus ensure an accurate analysis with minimal errors. Currently, gene expression analyses in A. baumannii are compromised, as there are no reports in the literature describing the identification of validated reference genes for use in RT-qPCR analyses. For this reason, we selected twelve candidate reference genes of A. baumannii and assessed their expression profile under different experimental and culture conditions. The expression stability of the candidate genes was evaluated by using statistical algorithms such as BestKeeper, geNorm, NormFinder, Delta CT, and RefFinder, in order to identify the most suitable candidate reference genes for RT-qPCR analyses. The statistical analyses indicated rpoB, rpoD, and fabD genes as the most adequate to ensure accurate normalization of RT-qPCR data in A. baumannii. The accuracy of the proposed reference genes was validated by using them to normalize the expression of the ompA gene, encoding the outer membrane protein A, in A. baumannii sensible and resistant to the antibiotic polymyxin. The present work provides suitable reference genes for precise RT-qPCR data normalization on future gene expression studies with A. baumannii.
Similar content being viewed by others
Introduction
Acinetobacter baumannii is an opportunistic pathogen, primarily associated with hospital-acquired infections, and which has proved to be an eminently problematic pathogen globally. A. baumannii needs minimal nutritional requirements, which makes it able to survive in extreme environmental conditions and last for long periods on surfaces. Since they are ubiquitous in nature, Acinetobacter species are intrinsically resistant to most currently available antibiotics. In addition, A. baumannii has a great ability to adapt to the environment, being able to acquire virulence and resistance genes from other pathogens, which makes them even more capable of spreading in hospital environments and infecting vulnerable patients, particularly in intensive care units (ICUs). The alarming spread of A. baumannii was even more evident during the COVID-19 pandemic, when multidrug-resistant A. baumannii strains have figured among the most common secondary bacterial infections in patients with COVID-19, often with a fatal outcome1,2.
Carbapenems remain effective for the treatment of most hospital-acquired critical infections caused by multidrug-resistant Gram-negative bacteria, specially A. baumannii. However, the emergence of carbapenem-resistant Acinetobacter baumannii (CRAB) over the last decades has become a serious problem of healthcare-associated infections worldwide due to the limited options of antimicrobials for treatment3,4,5. In fact, the WHO has appointed CRAB among the top-priority bacteria that deserve support for the research and development of new effective drugs6. Currently, polymyxin antibiotics are recognized as last-resort antibiotics against CRAB treatment, although A. baumannii may develop polymyxin resistance and their use is limited due to nephrotoxicity and neurotoxicity4,5,7,8,9.
Among the main virulence factors that allow A. baumannii to acquire resistance, the porin OmpA stands out as an outer membrane protein that plays key roles in the pathogenicity of A. baumannii by regulating the adhesion, biofilm formation, and the immune response of the host10,11,12. In addition, OmpA plays an important role in resistance to carbapenems13,14. Therefore, the expression level of OmpA is shown to be a diagnostic index for A. baumannii resistant to antibiotics15, either through the reduction of its expression when resistant or by joining the efflux pump mechanism to eject the antibiotic present in the periplasmic space out of the bacterial cell11,16. Thus, the characterization of porin OmpA is important to support studies that seek new therapeutic strategies against antibiotic resistance mechanisms.
Considering the current relevance of multidrug resistant strains of A. baumannii, it is essential to characterize the virulence genes of this bacterium and how these genes are expressed during the colonization of a host. Different methods of gene expression analysis are available and their choice varies according to budget, laboratory facilities, cost-effectiveness, availability of hardware and software, the purpose of the study, and the number of genes of interest17.
Currently, gene expression analyses in A. baumannii have been performed by comparative transcriptomic profiling18,19,20,21, PCR array22, reporter vector systems23, and mostly by Reverse Transcription Quantitative real-time Polymerase Chain Reaction (RT-qPCR)7,18,19,24,25,26,27,28,29,30.
Since its first proposal, RT-qPCR has remained the most versatile and ubiquitous gene expression analysis technique due to its practicality and accessibility. The introduction of fluorescent markers not only reduced the risks of contamination but also eliminated the need for post-PCR reaction processing, such as agarose gel electrophoresis31. However, a crucial step in RT-qPCR analysis is the normalization of the raw data by an internal control, also called the reference gene or an endogenous gene. The use of a reference gene is essential to achieve reliable results, as in addition to establishing itself as a normalizer for the other genes under study, it will also correct possible errors in pipetting, RNA extraction, or cDNA synthesis32. A suitable reference gene for RT-qPCR reactions must have unchanged expression, regardless of the bacterial culture condition33. In addition, to be used as an internal control, the candidate reference gene must be experimentally validated, following the Minimum Information for Publication Quantitative Real-Time PCR Experiments (MIQE) guidelines31.
Unfortunately, there are no reports of a comprehensive evaluation of experimentally validated reference genes to be used in RT-qPCR in A. baumannii. Therefore, we describe here the investigation of twelve candidate genes as potential internal controls in RT-qPCR analyses with A. baumannii. The expression pattern of the candidate genes was measured in A. baumannii subjected to different experimental conditions, and the stability of their expression were statistically evaluated using Bestkeeper, NormFinder, geNorm, Delta CT, and RefFinder programs. Statistical analyzes ranked rpoB, rpoD, and fabD as the most suitable reference genes, and these genes were used to normalize the expression of the ompA gene in clinical samples of A. baumannii sensitive and resistant to the antibiotic polymyxin. The contribution of the present study was to provide suitable reference genes for precise RT-qPCR data normalization so that future analysis of gene expression on A. baumannii be precisely performed.
Results
Selection of candidate reference genes, PCR amplification efficiencies, and expression profile under different experimental conditions
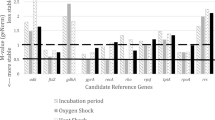
In this study, the following 12 genes were selected and evaluated as potential candidate reference genes for RT-qPCR in A. baumannii: 16S, atpD, era, fabD, ftsZ, gapA, groEL, gyrA, proC, rho, rpoB, and rpoD. The efficiency of PCR amplification of primer pairs for each gene was calculated from standard curves constructed from RT-qPCR reactions performed with serial dilutions of chromosomal DNA from the bacteria. The expression profile of the candidate genes was evaluated from A. baumannii subjected to four different culture conditions, namely: (1) cells isolated at different growth phases (lag, mid-log, and stationary phases); (2) cells subjected to pH conditions above and below the ideal for their growth (pH 9.0 and 5.0, respectively); (3) cells submitted to thermal shock at temperatures also above and below the ideal for their growth (42 °C and 28 °C, respectively), and (4) cells grown in solid LB medium. Supplementary Fig. S1 shows the growth patterns of A. baumannii grown in LB broth and under stressful cultivation conditions.
Table 1 shows the candidate genes selected in this study, the product they encode, the nucleotide sequences of the primer pairs for each gene, and the amplicon size. The PCR amplification specificity of each primer pair was confirmed by the presence of a single PCR product of the expected size on agarose gel electrophoresis (Supplementary Fig. S2). Furthermore, all 12 candidate genes presented a single peak in the dissociation-curve analyses, indicating the absence of primer-dimers and/or non-specific PCR products (Supplementary Fig. S3).
Table 1 also displays the calculations for the PCR amplification efficiencies (E) for each pair of primers, which were calculated based on standard curves generated for each gene, and the cycle threshold (CT) values. The PCR efficiency of era and ftsZ genes, of 122% and 121% respectively, were above the acceptable range and, therefore, were excluded from the subsequent analyses. The PCR efficiency of the other genes ranged from 102% for groEL to 115% for fabD, which were within the acceptable range.
The CT values ranged from 16.03 to 24.79, with most genes having average CT values of 22.06. The 16S ribosomal gene presented the lowest CT value (16.03 ± 1.71, mean CT ± standard deviation) and, therefore, was the most abundantly expressed gene across all samples. On the other hand, the proC gene was considered the least expressed gene since it had the highest CT (24.79 ± 0.98, CT mean ± standard deviation). The 16S and atpD genes presented the largest standard deviation of the mean CT values (standard deviation of 1.71 and 1.1, respectively), whereas rpoD and fabD genes showed the least variation on the standard variation (0.36 and 0.49, respectively).
Expression stability analyses of candidate reference genes
The stability of transcription levels of candidate genes under the different experimental conditions was evaluated using the statistical algorithms BestKeeper, geNorm, NormFinder, Delta CT, and RefFinder; the latter program integrates all four algorithms to elaborate a comprehensive analysis. Table 2 summarizes the ranking of the genes with the most stable expression across all experimental conditions tested and calculated by all five statistical programs.
BestKeeper ranks the most stable gene based on the correlation coefficient (r), the coefficient of variation of the CTs (CV values), and the standard deviation (SD values). Genes with the lowest SD values and highest coefficient of correlation are considered the most stable. BestKeeper ranked fabD and rpoD with the lowest SD values (0.484 and 0.522, respectively) and, therefore, the genes with the most stable expression under the tested experimental conditions (Table 2). According to BestKeeper, genes with SD values greater than 1 are considered inappropriate as reference genes. Among all tested genes, only 16S presented SD value greater than 1 (1.534). This result corroborates the result presented in Table 1, which showed 16S as the gene with the greatest variation in the CT number among all the genes analyzed.
geNorm determines the expression stability of each gene by calculating an expression stability M value. The program recommends a cut-off of M < 1.5: the lower the value, more stable is the expression of the gene. geNorm indicated the rpoB, rpoD, and fabD genes with the lowest M values (0.519, 0.519, and 0.556, respectively) and, therefore, with the most stable expression (Table 2). On the other hand, the program determined 16S with the highest M value (1.166) and, thus, the least stable gene.
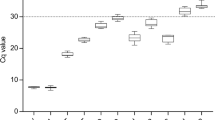
geNorm also employs pairwise variation (Vn/Vn+1) analyses to generate a V value that indicates the optimal number of reference genes needed for the reliable normalization of RT-qPCR data. The program recommends a V value threshold of 0.15, below which the addition of more reference genes is not required. As displayed in Fig. 1, the pairwise variation of V2/3, considering the two most stable genes (rpoB and rpoD), was below the cut-off of 0.15 (0.1479), indicating that the addition of a third gene is not necessary for an accurate normalization.
geNorm analysis indicates the optimal number of reference genes for normalization of RT-qPCR data. geNorm calculates the pairwise variation (Vn/Vn+1, V value) to determine the minimum number of reference genes required for accurate normalization. The program suggests a cut-off threshold of 0.15 (indicated by the red dashed line), below which the inclusion of an additional reference gene is not necessary. The V2/3 value of 0.1479 (red arrow) indicates that the inclusion of the third most stable reference gene (fabD) would have no significant contribution to the accuracy of the data normalization.
NormFinder combines intra- and inter-group variations to calculate a stability (S) value: genes with the lowest S values present the most stable expression. As shown in Table 2, NormFinder indicated rpoB, rpoD, and fabD as the top ranked genes with S values of 0.315, 0.430, and 0.448, respectively. On the other hand, NormFinder indicated 16S (S value of 1.711) and groEL (S value of 1.069) with the highest S values and, therefore, with the least stable expression. The stability ranking performed by the Delta CT method was similar to the one generated by NormFinder, indicating rpoB, fabD, and rpoD as the top ranked genes with the lowest average of the standard deviation (SD of 0.923, 0.943, and 0.968, respectively).
Finally, Table 2 also shows the ranking of the most stable genes generated by RefFinder. This is a web-based program that generates a comprehensive ranking based on the geometric mean of the results obtained by the other statistical analysis softwares (geNorm, NormFinder, BestKeeper, and Delta CT methods). This comprehensive ranking generated by RefFinder indicated rpoB, rpoD, and fabD as the three top-ranked genes with the lowest geometric mean (geomean) of ranking values (1.41, 1.86, and 2.06, respectively).
The proposed reference genes are highly conserved among different strains of A. baumannii and other species of the genus Acinetobacter. The primer sequences for each gene on A. baumannii strains AB736 and AB5075-UW and on other species of the genus Acinetobacter, such as A. calcoaceticus, A. haemolyticus, A. johnsonii, A. junii, A. lwoffii, A. nosocomialis, A. pittii, and A. schindleri, are displayed on Supplementary Tables S1–S11.
Validation of selected reference genes by expression analyses of ompA gene
To evaluate the reliability of the selected reference genes, the three genes with the most stable expression in this study—rpoB, rpoD, and fabD—were used to normalize the expression of ompA gene in a clinical isolate of A. baumannii sensitive to the antibiotic polymyxin B and after acquiring resistant to this antibiotic. To compare the normalization using the most stable genes with the least stable, the 16S gene was also used to normalize the RT-qPCR data, since it is commonly used as a reference gene for A. baumannii and presented the least stable expression in the present study.
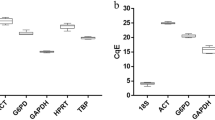
As shown in Fig. 2, the normalization of the RT-qPCR data using the three most stable reference genes alone or as a geometric mean revealed statistically significant down-regulation of ompA gene at the isolate resistant to polymyxin when compared to the isolate sensitive to the antibiotic. On the other hand, normalization with the 16S gene rendered results similar to the one obtained with the most stable genes, but the results were sub-estimated and not statistically significant.
Relative expression of the ompA gene in a clinical isolate of A. baumannii sensitive to polymyxin and after having acquired resistance to this antibiotic. The expression data were normalized by the three most stable genes—fabD, rpoB, and rpoD—individually and with a geometric mean across the three genes (geomean). As a comparison, ompA expression was normalized with 16S, the least stable gene. *p < 0.05, **p ≤ 0.01, ***p ≤ 0.001, and n.s.: not significant (p ≥ 0.05).
Discussion
A. baumannii is currently considered a pathogenic bacterium of clinical relevance, mainly due to the emergence of strains resistant to multiple drugs and associated with infections related to health care34,35. In this scenario, genetic tools that allow the study of genes that encode virulence factors and confer resistance to antimicrobials is of paramount importance for a better understanding of the pathogenicity of this bacterium36.
The RT-qPCR technique is considered the gold standard method in gene expression analysis37. One of the most critical steps in this technique is the use of reliable reference genes, which act as internal controls of the RTq-PCR reactions and are fundamental to ensure a proper normalization of the obtained results. The choice of a reference gene for RT-qPCR data normalizations should involve careful selection, as an adequate endogenous gene should preferably have a stable expression, regardless of the conditions in which the bacterium is subjected31,38. Most of the time, the genes that normally show a stable expression pattern are the constitutive genes, which encode proteins of the bacteria's basal metabolism33.
Unfortunately, there is no description in the literature of properly validated reference genes for RT-qPCR for A. baumannii, despite the clinical significance of this bacterium. Although their stable expressions have not been experimentally validated, several genes have been used in RT-qPCR analysis in A. baumannii, such as groEL22, gyrB18, gyrA30, recA22, rpoB22, rpoD29, and the ribosomal RNA coding genes 50S39 and mostly 16S7,19,24,25,26,27,28. However, the use of a gene encoding a ribosomal RNA as an internal control in RT-qPCR reactions may be challenging, as its high expression hinders its use in mRNA transcript normalizations33.
In response to the lack of suitable reference genes for gene expression analysis in A. baumannii, a systematic approach was performed in this study to identify and validate ideal reference genes to be used in RT-qPCR analyses in A. baumannii. Twelve candidate reference genes (16S, atpD, era, fabD, gapA, groEL, gyrA, proC, rho, rpoB, and rpoD) commonly used in other bacterial species were selected and evaluated, some of them already used in A. baumannii despite of not having their stable expressions validated. The set of primers designed for the candidate genes yielded amplicon sizes ranging from 97 to 118 bp, which is within the range recommended for SYBR Green-based assays40,41.
The expression profile of the selected genes was evaluated in A. baumannii cells submitted to different experimental conditions and then the stability of the expression of the candidate genes was checked using the statistical algorithms Bestkeeper, geNorm, NormFinder, Delta CT, and RefFinder. Except for the Bestkeeper analysis, all other statistical algorithms (geNorm, NormFinder, Delta CT, and RefFinder) recommended rpoB, rpoD, and fabD as the top three most stable reference genes across all the experimental conditions, with only a slight variation in the ranking order by the Delta CT method. BestKeeper's analysis presented the greatest difference in the ranking of the most stable candidate genes. Although Bestkeeper also ranked rpoD and fabD among the three most stable genes, the program ranked groEL as the third and rpoB as the fourth most stable genes. However, groEL was not considered a stable gene by the other statistical programs. On the contrary, most algorithms ranked the groEL gene among the genes with the least stable expression. Thus, we suggest that the groEL gene be disregarded as a stable gene for RT-qPCR data normalizations in A. baumannii. Unanimously, all five statistical programs indicated the 16S gene as the one with the least stable expression and, therefore, the least suitable for normalizations of RT-qPCR data in A. baumannii.
Based on the evaluation by all statistical programs, this study suggests the rpoB, rpoD, and fabD genes as the most suitable reference genes for normalizing RT-qPCR data in A. baumannii. Congruent with our study, these genes were also indicated as internal controls for analysis of RT-qPCR data in other bacteria, such as in Pseudomonas brassicacearum GS2042, Staphylococcus aureus43,44, Saccharopolyspora spinosa45, and also for Aeromonas salmonicida46.
In order to test the legitimacy of the proposed genes as normalizers of RT-qPCR reactions in A. baumannii, we used the expression of fabD, rpoB, and rpoD to normalize the expression of the ompA gene in a strain of A. baumannii sensitive to the polymyxin B antibiotic and after the same strain acquires resistance to this antibiotic. As a comparison, we also used the least stable gene expression, 16S, as a normalizer of ompA expression. We chose to normalize ompA expression in these conditions because polymyxin B is the main therapeutic option available to treat infections caused by A. baumannii strains resistant to multiple drugs. The ompA gene encodes the outer membrane protein OmpA, which is considered not only a virulence factor, as it acts as an adhesin in the bacteria's adhesion processes, but it is also of great importance for the genus Acinetobacter because it is involved in resistance to antibiotics of the class of beta-lactams13.
In the present study, the normalization of ompA expression using the three recommended genes (rpoB, rpoD, and fabD) generated the same expression pattern, either performing the normalization with the genes individually or using a geometric mean between them. In contrast, normalization with the 16S gene generated underestimated and statistically non-significant data, despite resulting in an expression pattern similar to that obtained with the recommended reference genes. Therefore, despite the 16S gene being used as a reference gene for the bacteria A. baumannii in some studies, caution should be taken when using this gene on RT-qPCR normalizations in A. baumannii.
In a study that evaluated the mechanisms of antimicrobial resistance in clinical isolates of A. baumannii initially susceptible to polymyxin B and after acquiring resistance to this antibiotic, the authors showed changes in the surface of A. baumannii strains submitted to cultivation in the presence of polymyxin B, in addition to changes in the expression of genes that encode outer membrane proteins, such as porins and efflux pumps7. The data obtained in the present study show that while the expression of the ompA gene is induced in polymyxin-sensitive A. baumannii, the expression of the gene is reduced when the same strain becomes resistant to polymyxin. This result suggests that resistance to polymyxin acquired by bacteria does not seem to involve the expression of the outer membrane protein OmpA. Interestingly, this result goes against a study that showed that the resistance of A. baumannii to polymyxin is due to the presence of OmpA in the bacteria, as the knockout of the ompA gene makes the bacteria more sensitive to the antibiotic13,14. However, this reduction in polymyxin resistance is much more likely due to the loss of cell wall integrity in a mutant strain lacking the OmpA protein.
A limitation of the present study is that, except for the static culture in a solid LB medium, the expression stability of the proposed reference genes was defined from cultivation and experimental conditions that involved the planktonic mode of growth. It is plausible to state that these genes may not be equally suitable under other experimental conditions. For this reason, it is strongly advised that the expression stability of the proposed reference genes be checked in any physiological or experimental condition to be investigated, to guarantee the reliability of the normalization of RT-qPCR data with these genes. For instance, biofilm formation has recently become a major research field in pathogenicity studies with A. baumannii18,26,47,48. Before using the proposed endogenous genes in gene expression analysis during biofilm growth by A. baumannii, it is advisable to test the stability of their expression in this specific condition. Likewise, although the proposed genes are highly homologous between different strains of A. baumannii and even between other species of the genus Acinetobacter, it is worth testing the expression stability of the proposed reference genes and their suitability on RT-qPCR data normalization in a given strain or Acinetobacter species under study. As there is no ideal reference gene with stable expression regardless of physiological state and experimental conditions, the stable expression of any potential reference gene must be validated under all physiological conditions or experimental treatment under study before its use in RT-qPCR normalizations33.
In summary, a reliable normalizing of RT-qPCR data relies on the use of stable reference genes, which demands testing the candidate reference genes at the most different experimental conditions. In the present study, rpoB, rpoD, and fabD were recommended as reliable reference genes for RT-qPCR data normalization in A. baumannii under the chosen experimental conditions. It is advisable that the expression stability of the proposed reference genes be assessed in a given experimental treatment under study, prior to their use on the normalization of gene expression by RT-qPCR analysis in this bacterium. To our knowledge, this is the first study aimed to identify reference genes for RT-qPCR analysis in A. baumannii.
Methods
Selection of candidate reference genes for A. baumannii
The selection of endogenous candidate genes was performed on the annotated genome of A. baumannii ATCC 19606 (NCBI reference sequence NZ_CP045110.1, available at https://www.ncbi.nlm.nih.gov/nuccore/NZ_CP045110.1?report=gbwithparts&log$=seqview). Special attention must be given to the choice of candidate genes to be evaluated, which must involve a rigorous gene selection procedure and taking into account the specific conditions under which the study will be conducted49,50. The criterion used in the present study for selecting candidate genes was to choose internal controls commonly used in other bacteria in the most diverse cultivation and experimental conditions as described in the literature. Among the selected genes are 16S (16S ribosomal RNA)7,19,24,25,26,27,28,51, atpD (ATP synthase β subunit)52, era (era GTPase)53, fabD (ACP S-malonyltransferase)54, ftsZ (cell division protein FtsZ)42,55,56, gapA (glyceraldehyde-3-phosphate dehydrogenase A)57, groEL (GroEL chaperonin)22,58,59, gyrA (DNA gyrase subunit A)30,42,60, proC (pyrroline-5-carboxylate reductase)33,61, rho (transcription termination factor Rho)33,42,45, rpoB (β subunit of RNA polymerase)22,44,45,62 and rpoD (RNA polymerase Sigma factor RpoD)33,42,63. In order to minimize the chance of coregulation of the selected genes, the candidate genes were selected in order to belong to different functional categories such as cellular metabolism (atpD, era, fabD, gapA, and proC), protein synthesis (groEL and 16S), cell division (ftsZ) and DNA metabolism (gyrA, rho, rpoB, and rpoD).
BLAST multiple alignments were performed to confer the homology of the proposed genes from A. baumannii ATCC 19606 with the homologous genes from other strains of A. baumannii and other species of the genus Acinetobacter.
Primers design and PCR amplification efficiencies
Pairs of primers specific for each selected gene were designed with the web-based program Primer3 v. 0.4.064, from the nucleotide sequence of the coding region of each gene. Primer pairs were designed in order to present an annealing temperature of 60 °C, GC content of 40–60%, and yield product size of 95–120 bp.
Primers were initially tested by conventional PCR, using genomic DNA from A. baumannii ATCC 19606, extracted using the Wizard® Genomic DNA Purification kit (Promega), and the Platinum™ PCR SuperMix kit (Invitrogen™), following the instructions of the manufacturers. The amplicons were analyzed by agarose gel electrophoresis to confirm the specificity of the PCR amplification. The images were captured with a digital Gel Doc™ XR photodocumentation system (Biorad) and analyzed with the Image Lab™ software version 5.0 (Biorad).
To determine the PCR amplification efficiency and the regression coefficient (R2) of each pair of primers, standard curves for each selected candidate gene were constructed from real-time PCR reactions performed with increasing concentrations of genomic DNA extracted from the bacteria. The reactions were performed in a 7300 Real-Time PCR System equipment (Applied Biosystems™) with the Platinum™ SYBR™ Green qPCR SuperMix-UDG w/ROX kit (Invitrogen™), following the manufacturer's instructions. At the end, an additional dissociation step was performed (40 cycles with a decrease of 1 °C every 15 s, starting at 95 °C) to generate a dissociation (melting) curve. The analysis of this curve made it possible to verify the specificity of the reaction, that is, whether there were nonspecific bands and/or primer-dimers formation. The analysis of the generated data was performed using the 7300 Real-Time PCR System Sequence Detection software v1.4.1 (Applied Biosystems™).
Standard curves were constructed for each candidate gene by plotting the mean CT numbers versus the logarithmic amount of the serially diluted template DNA. The formula E (%) = [10(− 1/slope) − 1] × 100 was used to calculate the PCR amplification efficiency (E) of each pair of primers, and the efficiency of each gene was considered in the subsequent statistical analyses. In this process, the era and ftsZ genes were eliminated on further analyses, as they presented a slope value outside the optimal range.
Bacterial strain and culture conditions
A. baumannii strain ATCC 19606 was used in this study. Bacteria were cultured in LB broth at 37 °C and under 250 rpm of agitation. Bacterial growth was monitored by reading the optical density in a spectrophotometer at a wavelength of 600 nanometers (D.O.600 nm), using the GeneQuant spectrophotometer (GE Healthcare).
The expression of the selected genes was measured from A. baumannii cultivated under eight different experimental conditions, namely: cultivation of the bacteria in LB broth until the growth phases lag (D.O.600 nm = 0.2), exponential (D.O.600 nm = 0.6), and stationary (D.O.600 nm = 1.0); growth of the bacterial cells until the exponential phase (D.O.600 nm of around 0.5) under the thermal stress of 28 °C and 42 °C; growth of the bacterial cells until the exponential phase (D.O.600 nm of around 0.5) under the pH stress of 5.0 and 9.0, and cultivation of A. baumannii in solid LB medium.
To validate the reference genes recommended by the statistical programs, the expression pattern of the ompA gene was analyzed in an A. baumannii strain sensitive to the antibiotic polymyxin B isolated from bloodstream infection. The expression pattern of ompA was compared in the strain initially susceptible to polymixin B and after it had acquired resistance to this antibiotic. Antibiotic resistance was induced by culturing the clinical isolate in increasing concentrations of polymyxin B sulfate (Sigma-Aldrich) on 10 consecutive days, starting with a concentration of 0.125 µg/mL. Subcultures were carried out in solid LB medium until a minimum inhibitory concentration (MIC) of 4 µg/mL was obtained, which corresponds to polymyxin B resistance according to the Clinical and Laboratory Standards Institute (CLSI) guidelines65. The expression of the ompA gene was investigated in polymyxin B sensitive and resistant strains grown in TSB medium (Himedia) up to the exponential phase of growth. The resistant strain was grown in the presence of 4 µg/mL of polymyxin B in order to maintain resistance.
Bacterial cells cultured under the abovementioned conditions were harvested by centrifugation, and the cell pellets were kept in RNAprotect® Bacteria Reagent (Qiagen) for RNA stabilization until the total RNA extraction procedures.
RNA extraction, cDNA synthesis and real-time PCR reactions
Total bacterial RNA and cDNA synthesis were performed following protocols published elsewhere66,67. Total RNA was extracted from the A. baumannii cells cultured under the abovementioned conditions by using the TRIzol™ Plus RNA Purification kit and Phasemaker™ Tubes Complete System (Invitrogen™), following the manufacturer’s instructions. To ensure a DNA-free total RNA, on-column DNase treatments were performed using PureLink™ DNase (Invitrogen™), according to the manufacturer’s recommendations. The integrity of the extracted RNA was analyzed by agarose gel electrophoresis and its purity and concentration were calculated through the optical density of the samples at 260 and 280 nm using the NanoDrop 2000 spectrophotometer (ThermoFisher).
For the cDNA synthesis, 1 μg of total RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems™), as recommended by the manufacturer’s protocol. The real-time PCR reactions were conducted with cDNA samples diluted with nuclease-free water to a final concentration of 100 ηg/uL. Reactions were done in triplicates using the Platinum™ SYBR™ Green qPCR SuperMix-UDG w/ROX kit on the 7300 Real-Time PCR System equipment, following the manufacturer’s recommendations. The data obtained were analyzed by the 7300 Real-Time PCR System Sequence Detection software v1.4.1, which generated the raw cycle threshold (CT) values for each reaction. The means of the CT values of each candidate reference gene were used on the subsequent statistical analyses to determine the expression stability of the candidate genes.
Statistical analyses of the expression stability of the reference genes
To identify reference genes with stable and reliable expression for normalization of RT-qPCR data, the expression stability of the candidate genes was evaluated on A. baumannii cultured under the abovementioned conditions, using the statistical algorithms BestKeeper, geNorm, NormFinder, Delta CT, and RefFinder.
Bestkeeper uses raw CT values to calculate the standard deviation (SD) of the CT values (SD values) and the coefficient of variation (CV [% CT])68. Reference genes with the lowest SD values are considered the most stable genes, and genes with SD values greater than 1 are considered inconsistent by Bestkeeper.
To evaluate the expression stability of candidate genes, geNorm requires relative data which is obtained by transforming the raw CT values into 2−ΔCT values where ΔCT is the difference between each CT value and the lowest CT value across all samples. geNorm ranks the most stable genes by calculation an expression stability value M, which represents the average pairwise variation of a specific reference gene with all other genes tested49. Following a stringent analysis33, M value ≤ 1.0 indicates the most stable expression gene and suitable for RT-qPCR data normalization. geNorm also indicates the optimal number of reference genes for accurate RT-qPCR normalizations by calculating the pairwise variation (Vn/Vn+1) between sequential normalization factors (NFn/NFn+1) across all experimental conditions. The program recommends a cut-off threshold of 0.15, bellow each the inclusion of additional reference genes is not necessary to improve the normalization accuracy49.
NormFinder also utilizes relative data calculated from the raw CT values to identify the genes with the most stable expression. The program estimates the intra- and inter-group expression variations for a given set of experiments to produce a Stability value for each candidate gene50. Genes with the smallest Stability value are ranked the most stable according to NormFinder.
Delta CT method compares the relative expression of “pairs of genes” within each sample to identify suitable reference genes for accurate RT-qPCR normalization69. According to the method, the smaller the average of the standard deviations (Average of SD), the smaller the variability in gene expression.
Finally, the expression stability of the candidate reference genes was also analyzed by RefFinder, a web-based program that integrates the abovementioned algorithms (BestKeepr, geNorm, NormFinder, and the Delta CT method)70. By using raw CT values, RefFinder generates a comprehensive ranking based on the geometric mean of the ranking values calculate by the other algorithms.
Validation of the recommended reference genes
To validate the recommended reference genes, the expression of ompA on polymyxin- resistant and -sensitive A. baumannii strains were normalized using the three most stable reference genes—rpoB, rpoD, and fabD—individually and as a geometric mean of the three genes. Data were also normalized using the 16S, since this gene was considered the least stable and it is commonly used as internal control in A. baumannii RT-qPCR assays. The relative expression levels of the ompA gene were calculated using the comparative critical threshold 2−ΔΔCT method71. The differences on the expression levels were evaluated by Student’s t test with correction for multiple tests using the GraphPad Prism 7.00 software. Differences were considered statistically significant with p-values ≤ 0.05.
Data availability
All data supporting the findings of this study are included within the published article and in its Supplementary Material files.
References
Shafran, N. et al. Secondary bacterial infection in COVID-19 patients is a stronger predictor for death compared to influenza patients. Sci. Rep. 11, 12703. https://doi.org/10.1038/s41598-021-92220-0 (2021).
Ellis, R. C., Roberts, E. K., Grier, J. T. & Fiester, S. E. Acinetobacter baumannii infections that are resistant to treatment: Warning signs from the COVID-19 pandemic. Future Microbiol. 17, 1345–1347. https://doi.org/10.2217/fmb-2022-0153 (2022).
Kyriakidis, I., Vasileiou, E., Pana, Z. D. & Tragiannidis, A. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens 10, 373. https://doi.org/10.3390/pathogens10030373 (2021).
Nguyen, M. & Joshi, S. G. Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital-acquired infections: A scientific review. J. Appl. Microbiol. 131, 2715–2738. https://doi.org/10.1111/jam.15130 (2021).
Jiang, Y. et al. Carbapenem-resistant Acinetobacter baumannii: A challenge in the intensive care unit. Front. Microbiol. 13, 1045206. https://doi.org/10.3389/fmicb.2022.1045206 (2022).
Tacconelli, E. et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. https://doi.org/10.1016/S1473-3099(17)30753-3 (2018).
Girardello, R. et al. Diversity of polymyxin resistance mechanisms among Acinetobacter baumannii clinical isolates. Diagn. Microbiol. Infect. Dis. 87, 37–44. https://doi.org/10.1016/j.diagmicrobio.2016.10.011 (2017).
Piperaki, E.-T., Tzouvelekis, L. S., Miriagou, V. & Daikos, G. L. Carbapenem-resistant Acinetobacter baumannii: In pursuit of an effective treatment. Clin. Microbiol. Infect. 25, 951–957. https://doi.org/10.1016/j.cmi.2019.03.014 (2019).
Sheu, C.-C., Chang, Y.-T., Lin, S.-Y., Chen, Y.-H. & Hsueh, P.-R. Infections caused by carbapenem-resistant Enterobacteriaceae: An update on therapeutic options. Front. Microbiol. 10, 80. https://doi.org/10.3389/fmicb.2019.00080 (2019).
Nie, D. et al. Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection. J. Biomed. Sci. 27, 26. https://doi.org/10.1186/s12929-020-0617-7 (2020).
Uppalapati, S. R., Sett, A. & Pathania, R. The outer membrane proteins OmpA, CarO, and OprD of Acinetobacter baumannii confer a two-pronged defense in facilitating its success as a potent human pathogen. Front. Microbiol. 11, 589234. https://doi.org/10.3389/fmicb.2020.589234 (2020).
Viale, A. M. & Evans, B. A. Microevolution in the major outer membrane protein Ompa of Acinetobacter baumannii. Microb. Genomics 6, e000381. https://doi.org/10.1099/mgen.0.000381 (2020).
Kwon, H. I. et al. Outer membrane protein A contributes to antimicrobial resistance of Acinetobacter baumannii through the OmpA-like domain. J. Antimicrob. Chemother. 72, 3012–3015. https://doi.org/10.1093/jac/dkx257 (2017).
Kwon, H. I., Kim, S., Oh, M. H., Shin, M., & Lee, J. C. Distinct role of outer membrane protein A in the intrinsic resistance of Acinetobacter baumannii and Acinetobacter nosocomialis. Infect. Genet. Evol. 67, 33–37. https://doi.org/10.1016/j.meegid.2018.10.022 (2019).
Martín-Peña, R., Domínguez-Herrera, J., Pachón, J. & McConnell, M. J. Rapid detection of antibiotic resistance in Acinetobacter baumannii using quantitative real-time PCR. J. Antimicrob. Chemother. 68, 1572–1575. https://doi.org/10.1093/jac/dkt057 (2013).
Lee, C.-R. et al. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol. 7, 55. https://doi.org/10.3389/fcimb.2017.00055 (2017).
Moustafa, K. & Cross, J. M. Genetic approaches to study plant responses to environmental stresses: an overview. Biology (Basel) 5, 20; https://doi.org/10.3390/biology5020020 (2016).
Rumbo-Feal, S. et al. Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS ONE 8, e72968. https://doi.org/10.1371/journal.pone.0072968 (2013).
Chang, K.-C. et al. Transcriptome profiling in imipenem-selected Acinetobacter baumannii. BMC Genomics 15, 815. https://doi.org/10.1186/1471-2164-15-815 (2014).
Murray, G. L. et al. Global gene expression profile of Acinetobacter baumannii during bacteremia. J. Infect. Dis. 215, S52–S57. https://doi.org/10.1093/infdis/jiw529 (2017).
Qin, H. et al. Comparative transcriptomics of multidrug-resistant Acinetobacter baumannii in response to antibiotic treatments. Sci. Rep. 8, 3515. https://doi.org/10.1038/s41598-018-21841-9 (2018).
Lannan, F. M. et al. Evaluation of virulence gene expression patterns in Acinetobacter baumannii using quantitative real-time polymerase chain reaction array. Mil. Med. 181, 1108–1113. https://doi.org/10.7205/MILMED-D-15-00437 (2016).
Lucidi, M. et al. New shuttle vectors for Real-Time gene expression analysis in multidrug-resistant Acinetobacter species: in vitro and in vivo responses to environmental stressors. Appl. Environ. Microbiol. 85, e01334–e1419. https://doi.org/10.1128/AEM.01334-19 (2019).
Sahu, P. K., Iyer, P. S., Barage, S. H., Sonawane, K. D. & Chopade, B. A. Characterization of the algC gene expression pattern in the multidrug resistant Acinetobacter baumannii AIIMS 7 and correlation with biofilm development on abiotic surface. ScientificWorldJournal 2014, 593546. https://doi.org/10.1155/2014/593546 (2014).
Geisinger, E., Mortman, N. J., Vargas-Cuebas, G., Tai, A. K. & Isberg, R. R. A global regulatory system links virulence and antibiotic resistance to envelope homeostasis in Acinetobacter baumannii. PLoS Pathog. 14, e1007030. https://doi.org/10.1371/journal.ppat.1007030 (2018).
Amin, M. et al. Genes related to biofilm formation and biofilm-specific resistance of Acinetobacter baumannii strains isolated from burn infection in Ahvaz. Iran. Infect. Drug. Resist. 12, 3867–3881. https://doi.org/10.2147/IDR.S228981 (2019).
Na, S.-H. et al. Therapeutic effects of inhibitor of ompA expression against carbapenem-resistant Acinetobacter baumannii strains. Int. J. Mol. Sci. 22, 12257. https://doi.org/10.3390/ijms222212257 (2021).
Kashyap, S., Sharma, P. & Capalash, N. Tobramycin stress induced differential gene expression in Acinetobacter baumannii. Curr. Microbiol. 79, 88. https://doi.org/10.1007/s00284-022-02788-7 (2022).
AlQumaizi, K. I., Kumar, S., Anwer, R. & Mustafa, S. Differential gene expression of efflux pumps and porins in clinical isolates of MDR Acinetobacter baumannii. Life 12, 419. https://doi.org/10.3390/life12030419 (2022).
Shakib, P., Halimi, S., Rezaei, F. & Delfani, S. Expression of bap gene in clinical Acinetobacter baumannii isolates in Khorramabad. Iran. Gene Cell. Tissue 10, e124061. https://doi.org/10.5812/gct-124061 (2023).
Bustin, S. A. & Wittwer, C. T. MIQE: A step toward more robust and reproducible quantitative PCR. Clin. Chem. 63, 1537–1538. https://doi.org/10.1373/clinchem.2016.268953 (2017).
Bustin, S. A., Benes, V., Nolan, T. & Pfaffl, M. W. Quantitative real-time RT-PCR—a perspective. J. Mol. Endocrinol. 34, 597–601. https://doi.org/10.1677/jme.1.01755 (2005).
Gomes, A. É. I. et al. Selection and validation of reference genes for gene expression studies in Klebsiella pneumoniae using reverse transcription quantitative real-time PCR. Sci. Rep. 8, 9001. https://doi.org/10.1038/s41598-018-27420-2 (2018).
Tu, Q. et al. Acinetobacter baumannii phages: past, present and future. Viruses 15, 673. https://doi.org/10.3390/v15030673 (2023).
Wu, H.-J. et al. Drug-resistant Acinetobacter baumannii: from molecular mechanisms to potential therapeutics (Review). Exp. Ther. Med. 25, 209. https://doi.org/10.3892/etm.2023.11908 (2023).
Sykes, E. M. E., Deo, S. & Kumar, A. Recent advances in genetic tools for Acinetobacter baumannii. Front. Genet. 11, 601380. https://doi.org/10.3389/fgene.2020.601380 (2020).
Aviv, G. & Gal-Mor, O. Real-time reverse transcription PCR as a tool to study virulence gene regulation in bacterial pathogens. Methods Mol. Biol. 1734, 23–32. https://doi.org/10.1007/978-1-4939-7604-1_3 (2018).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. https://doi.org/10.1373/clinchem.2008.112797 (2009).
Abirami, G., Durgadevi, R., Velmurugan, P. & Ravi, A. V. Gene expressing analysis indicates the role of Pyrogallol as a novel antibioflm and antivirulence agent against Acinetobacter baumannii. Arch. Microbiol. 203, 251–260. https://doi.org/10.1007/s00203-020-02026-3 (2020).
Bustin, S. & Huggett, J. qPCR primer design revisited. Biomol. Detect. Quantif. 14, 19–28. https://doi.org/10.1016/j.bdq.2017.11.001 (2017).
Debode, F., Marien, A., Janssen, E., Bragard, C. & Berben G. The influence of amplicon length on real-time PCR results. Biotechnol. Agron. Soc. Environ. 21, 3–11. https://doi.org/10.25518/1780-4507.13461 (2017).
Bai, B., Ren, J., Bai, F. & Hao, L. Selection and validation of reference genes for gene expression studies in Pseudomonas brassicacearum GS20 using real-time quantitative reverse transcription PCR. PLoS One 15, 1e0227927. https://doi.org/10.1371/journal.pone.0227927 (2020).
Theis, T., Skurray, R. A. & Brown, M. H. Identification of suitable internal controls to study expression of a Staphylococcus aureus multidrug resistance system by quantitative real-time PCR. J. Microbiol. Methods 70, 355–362. https://doi.org/10.1016/j.mimet.2007.05.011 (2007).
Sihto, H. M., Tasara, T., Stephan, R. & Johler, S. Validation of reference genes for normalization of qPCR mRNA expression levels in Staphylococcus aureus exposed to osmotic and lactic acid stress conditions encountered during food production and preservation. FEMS Microbiol. Lett. 356, 134–140. https://doi.org/10.1111/1574-6968.12491 (2014).
Liu, X. et al. rpoB and efp are stable candidate reference genes for quantitative real-time PCR analysis in Saccharopolyspora spinosa. Biotechnol. Biotechnol. Equip. 35, 619–632. https://doi.org/10.1080/13102818.2021.1899852 (2021).
Connors, E. et al. Identification and validation of reliable Aeromonas salmonicida subspecies salmonicida reference genes for differential gene expression analyses. Infect. Genet. Evol. 73, 314–321. https://doi.org/10.1016/j.meegid.2019.05.011 (2019).
Azizi, O. et al. Molecular analysis and expression of bap gene in biofilm-forming multi-drug-resistant Acinetobacter baumannii. Rep. Biochem. Mol. Biol. 5, 62–72. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5214686/ (2016).
Eze, E. C., Chenia, H. Y. & El Zowalaty, M. E. Acinetobacter baumannii biofilms: effects of physicochemical factors, virulence, antibiotic resistance determinants, gene regulation, and future antimicrobial treatments. Infect. Drug. Resist. 11, 2277–2299. https://doi.org/10.2147/IDR.S169894 (2018).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 34. https://doi.org/10.1186/gb-2002-3-7-research0034 (2002).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. https://doi.org/10.1158/0008-5472.CAN-04-0496 (2004).
Ogonowska, P. & Nakonieczna, J. Validation of stable reference genes in Staphylococcus aureus to study gene expression under photodynamic treatment: a case study of SEB virulence factor analysis. Sci. Rep. 10, 16354. https://doi.org/10.1038/s41598-020-73409-1 (2020).
Ludwig, W. et al. Phylogenetic relationships of Bacteria based on comparative sequence analysis of elongation factor Tu and ATP-synthase β-subunit genes. Antonie Van Leeuwenhoek 64, 285–305 (1993).
Inoue, K., Alsina, J., Chen, J. & Inouye, M. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol. Microbiol. 48, 1005–1016. https://doi.org/10.1046/j.1365-2958.2003.03475.x (2003).
Kutchma, A. J., Hoang, T. T. & Schweizer, H. P. Characterization of a Pseudomonas aeruginosa fatty acid biosynthetic gene cluster: purification of acyl carrier protein (ACP) and malonyl-coenzyme A:ACP transacylase (FabD). J. Bacteriol. 181, 5498–5504. https://doi.org/10.1128/JB.181.17.5498-5504.1999 (1999).
Rocha, D. J. P., Santos, C. S. & Pacheco, L. G. C. Bacterial reference genes for gene expression studies by RT-qPCR: Survey and analysis. Antonie Van Leeuwenhoek 108, 685–693. https://doi.org/10.1007/s10482-015-0524-1 (2015).
Williams, M. L. & Ghanem, M. Evaluation of candidate reference genes stability for gene expression analysis by reverse transcription qPCR in Clostridium perfringens. Sci. Rep. 12, 19434. https://doi.org/10.1038/s41598-022-23804-7 (2022).
Toyoda, K., Teramoto, H., Inui, M. & Yukawa, H. Involvement of the LuxR-type transcriptional regulator RamA in regulation of expression of the gapA gene, encoding glyceraldehyde-3-phosphate dehydrogenase of Corynebacterium glutamicum. J. Bacteriol. 191, 968–977. https://doi.org/10.1128/JB.01425-08 (2009).
Florindo, C. et al. Selection of reference genes for real-time expression studies in Streptococcus agalactiae. J. Microbiol. Methods 90, 220–227. https://doi.org/10.1016/j.mimet.2012.05.011 (2012).
Krzyżanowska, D. M., Supernat, A., Maciąg, T., Matuszewska, M. & Jafra, S. Selection of reference genes for measuring the expression of aiiO in Ochrobactrum quorumnocens A44 using RT-qPCR. Sci. Rep. 9, 13129. https://doi.org/10.1038/s41598-019-49474-6 (2019).
Zhao, P., Xu, L., Zhang, A., Zhu, B. & Shao, Z. Evolutionary analysis of gyrA gene from Neisseria meningitidis bacterial strains of clonal complex 4821 collected in China between 1978 and 2016. BMC Microbiol. 20, 71. https://doi.org/10.1186/s12866-020-01751-5 (2020).
Takle, G. W., Toth, I. K. & Brurberg, M. B. Evaluation of reference genes for real-time RT-PCR expression studies in the plant pathogen Pectobacterium atrosepticum. BMC Plant. Biol. 7, 50. https://doi.org/10.1186/1471-2229-7-50 (2007).
Wen, S., Chen, X., Xu, F. & Sun, H. Validation of reference genes for real-time quantitative PCR (qPCR) analysis of Avibacterium paragallinarum. PLoS One 11, e0167736. https://doi.org/10.1371/journal.pone.0167736 (2016).
McMillan, M. & Pereg, L. Evaluation of reference genes for gene expression analysis using quantitative RT-PCR in Azospirillum brasilense. PLoS One 9, e98162. https://doi.org/10.1371/journal.pone.0098162 (2014).
Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. https://doi.org/10.1385/1-59259-192-2:365 (2000).
CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed. CLSI supplement M100 (Wayne, Pennsylvania: Clinical and Laboratory Standards Institute, 2020).
Gomes, A. É. I. et al. Functional insights from KpfR, a new transcriptional regulator of fimbrial expression that is crucial for Klebsiella pneumoniae pathogenicity. Front. Microbiol. 11, 601921. https://doi.org/10.3389/fmicb.2020.601921 (2021).
Pacheco, T. et al. SdiA, a quorum-sensing regulator, suppresses fimbriae expression, biofilm formation, and quorum-sensing signaling molecules production in Klebsiella pneumoniae. Front. Microbiol. 12, 597735. https://doi.org/10.3389/fmicb.2021.597735 (2021).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. https://doi.org/10.1023/b:bile.0000019559.84305.47 (2004).
Silver, N., Best, S., Jiang, J. & Thein, S. L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7, 33. https://doi.org/10.1186/1471-2199-7-33 (2006).
Xie, F., Xiao, P., Chen, D., Xu, L. & Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80, 75–84. https://doi.org/10.1007/s11103-012-9885-2 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using Real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. https://doi.org/10.1006/meth.2001.1262 (2001).
Acknowledgements
This study was funded by the São Paulo Research Foundation (FAPESP), #2018/26203-9 to LFCF. PAAO and ASFM were supported by scholarships from the Coordination for the Improvement of Higher Education Personnel (CAPES, Ministry of Education of Brazil). COAR received a scholarship from Casa de Nossa Senhora da Paz—Ação Social Franciscana (CNSP-ASF). JB was supported by the Institutional Scientific Initiation Scholarship Program (PIBIC in Portuguese) from the National Council for Scientific and Technological Development (CNPq). The authors gratefully acknowledge Casa de Nossa Senhora da Paz - Ação Social Franciscana (CNSP-ASF) for the financial support for the publication fees of this study.
Author information
Authors and Affiliations
Contributions
L.F.C.F., R.G., and P.A.A.O. conceived and designed the experiments. P.A.A.O., J.B., C.O.A.R., and A.S.F.M. conducted bacterial culture experiments and nucleic acids analyses. P.A.A.O., J.B., C.O.A.R., and A.S.F.M. conducted bacterial culture experiments and nucleic acids analyses. P.A.A.O. and C.O.A.R. conducted the RT-qPCR analyses, and L.F.C.F., P.A.A.O., and A.P. performed the statistical analysis. L.F.C.F. wrote the manuscript and coordinated its revision. R.G. and A.P. assisted with critical revision of the manuscript. All authors participated in data analysis, contributed to the manuscript revision and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
de Oliveira, P.A.A., Baboghlian, J., Ramos, C.O.A. et al. Selection and validation of reference genes suitable for gene expression analysis by Reverse Transcription Quantitative real-time PCR in Acinetobacter baumannii. Sci Rep 14, 3830 (2024). https://doi.org/10.1038/s41598-024-51499-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-51499-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.