Abstract
Anterior Visual Pathway (aVP) damage may be linked to diverse inflammatory, degenerative and/or vascular conditions. Currently however, a standardized methodological framework for extracting MRI biomarkers of the aVP is not available. We used high-resolution, 3-D MRI data to generate a probabilistic anatomical atlas of the normal aVP and its intraorbital (iOrb), intracanalicular (iCan), intracranial (iCran), optic chiasm (OC), and tract (OT) subdivisions. We acquired 0.6 mm3 steady-state free-precession images from 24 healthy participants using a 3 T scanner. aVP masks were obtained by manual segmentation of each aVP subdivision. Mask straightening and normalization with cross-sectional area (CSA) preservation were obtained using scripts developed in-house. A probabilistic atlas (“aVP-24”) was generated by averaging left and right sides of all subjects. Leave-one-out cross-validation with respect to interindividual variability was performed employing the Dice Similarity Index (DSI). Spatially normalized representations of the aVP subdivisions were generated. Overlapping CSA values before and after normalization demonstrate preservation of the aVP cross-section. Volume, length, CSA, and ellipticity index (ε) biometrics were extracted. The aVP-24 morphology followed previous descriptions from the gross anatomy. Atlas spatial validation DSI scores of 0.85 in 50% and 0.77 in 95% of participants indicated good generalizability across the subjects. The proposed MRI standardization framework allows for previously unavailable, geometrically unbiased biometric data of the entire aVP and provides the base for future spatial-resolved, group-level investigations.
Similar content being viewed by others
Introduction
The normal structure of the anterior Visual Pathway (aVP) may be affected by a variety of acquired conditions, such as glaucoma1,2, inflammatory3,4, post-traumatic5, ischemic6, radiation-induced damage7, as well as inheritable diseases8. Despite several previous efforts9,10,11,12,13, there is currently no dedicated methodological framework for the morphometric evaluation of the entire aVP. Optic computed tomography (OCT) allows only for a quantitative assessment of the optic head retinal nerve fiber layer (RNFL), but provides no direct information on the remaining aVP. The very thin and tortuous morphology of the aVP and motion artefacts from eyeball movement pose specific technical challenges for MRI assessment making classical imaging protocols, typically employing two-dimensional (2-D) supra-millimetre slices, suboptimal for a geometrically unbiased assessment.
To address this situation, we developed a dedicated and relatively simple framework for the standardized high-resolution MRI assessment of the entire aVP and its intraorbital (iOrb), intracanalicular (iCan), intracranial (iCran), optic chiasm (OC), and optic tract (OT) anatomical subdivisions. We used three-dimensional (3-D) 0.6 mm3 images from a group of 24 healthy participants to develop an image normalization pipeline based on manually segmented data of the normal aVP, in order to: (1) allow for standardized structural analyses, (2) extract salient biometrics, and (3) generate a probabilistic atlas providing statistical knowledge of the aVP morphology.
Methods
Study participants
This study was approved by the local ethics committee of Canton Ticino, Bellinzona, Switzerland, (REF: 2017-00814; CE3224). Written informed consent was obtained from all participants before taking part in the study. The study was conducted according to ethical principles laid down by the latest version of the Declaration of Helsinki. Participants were recruited and underwent the MRI acquisition at the Neurocenter of Southern Switzerland between November 2017 and March 2019 as part of a prospective investigation14. Inclusion criteria were: (1) healthy male or female, (2) age ≥ 18 and < 65. Exclusion criteria were: (1) history of neurological or (2) ophthalmological diseases (including multiple sclerosis, tumors, glaucoma, retinopathy, maculopathy), (3) spherical refractive errors worse than − 6D15, (4) tumors, (5) severe eye and/or head trauma, (6) pregnancy, (7) claustrophobia. One female subject was excluded from the study due to high myopia (− 8.5D), 24 participants (10 females, 14 males; mean age 35.8 ± 9 years) were included and their scans analyzed.
Subject preparation and MRI data acquisition
Images were acquired on a single 3 T MRI “Skyra” scanner (Siemens, Erlangen, Germany) using a 64-channel head coil. Several previously proposed procedures were applied to minimize voluntary and/or spontaneous gaze and saccades movements, that have potential to cause image quality degradation14. If needed, MRI-compatible eyeglasses were provided to correct for refractive defects. Dimmed room lights optimized visual comfort for the participants. They were asked to lie still during image acquisition and to focus their gaze on a target placed on the scanner gantry in front of their eyes to maintain a straight gaze, or to keep their eyes closed if/when feeling unable to maintain fixation.
Imaging data for segmentation consisted of “constructive interference in steady state” (CISS) images covering the entire aVP with the following parameters: TR = 8.1 ms, TE = 3.74 ms, FoV = 150 mm2, matrix = 245 × 245, slice thickness = 0.6 mm, acquired voxel size = 0.6 × 0.6 × 0.6 mm3, number of slices = 72, scan duration = 3′11″. Images were transferred to a PC workstation running Linux and were checked for major artefacts by an experienced neuroradiologist (E.P.).
aVP extraction
To establish reproducible and MRI-suitable landmarks, we adapted the aVP anatomical subdivision criteria14, that were based on several sources of information from previous post-mortem descriptions16,17,18,19, as illustrated in Fig. 1. Specifically, the subdivisions were:
Illustration of the aVP subdivision boundaries applied for segmentation, as seen in one subject on the CISS images. The left panel shows a curved reconstruction of the aVP along its longitudinal axis, with red bars marking the boundaries (a). The iOrb/iCan boundary is located at the end of the intraorbital fat (f) and corresponds to the orbital apex at the level of the annulus of Zinn envisioned in (b). The transition from iCan to iCran is marked by the cisternal CSF (*) surrounding the nerve as seen in (c). The OC is the segment where the left and right aVPs merge perpendicular to the midsagittal plane (dashed line in d). The hypothalamus (h), representing the medial OT boundary, is highlighted in (a).
iOrb: from the optic nerve (ON) head to the last section where the orbital fat can be seen surrounding the nerve sheath.
iCan: the ON segment coursing within the optic canal, defined as the segment without fat and without cisternal CSF surrounding the nerve.
iCran: from the iCan segment end to the anterior OC boundary.
OC: the aVP section where the left and right iCran and OT segments merge into a single trunk, and the corresponding sides cannot be separated. For the present study, the OC was split according to the midsagittal plane into the left and right hemi-trunks.
OT: from the posterior OC boundary to the lateral geniculate nucleus, defined as the section where no CSF surrounding the nerve can be seen.
Using ITK-SNAP (v.3.6.0 freely available at www.itksnap.org), a neuroradiologist in training (A.D.) labelled each aVP subdivision on the CISS images according to the anatomical landmarks described above under the constant supervision of an experienced neuroradiologist (E.P.). Readers had the post-mortem normal anatomy descriptions constantly on hand for reference. Accuracy was optimized by iteratively revising segmentation until agreement with the senior neuroradiologist was achieved. The labels were saved as separate binary masks for each subdivision and side.
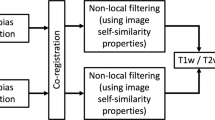
Image normalization pipeline
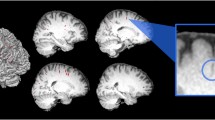
The image normalization pipeline steps are indicated in gray in Fig. 2. Its first step consisted of aligning the centroids of the label masks in the coronal plane, with the effect of straightening the ON (Fig. 3). Each aVP segment mask and side were processed separately in an identical manner. For each ki labelled slice orthogonal to the y axis (representing the image anterior–posterior axis orthogonal to the coronal plane), the centroids and the corresponding Di, i+1 distance between them in the ki, ki+1 slices were calculated. Then, the segments length (TL) was calculated as:
Diagram illustrating the aVP straightening process. Following manual segmentation in the native space (a), the slice centroids and the corresponding between-centroid distance were calculated (red markers in b). Then, slices were linearly shifted on the (x, z) transverse plane to the image centerline (red line in c), with preservation of the original between-centroid distance and CSA. The resulting gaps between shifted slices were filled by slice-wise interpolation, preserving the average CSA (light-blue slices in d). Finally, the straightened iOrb, iCan, iCran, OC and OT sections were concatenated along the image centerline (e). The (b–d) sample sketches are simplified for illustrative purposes.
Each ki slice was linearly shifted to the image C(x, z) center, so that:
where Ni corresponds to the mask centroid and Ci the image center for the j = x-z-axis. Following shifting, if \(\left(|\Delta x|,|\Delta z|\right)>0\), the straightened distance between consecutive centroids is smaller than Di, i+1. To preserve aVP length, we performed slice-wise interpolation, by ten-fold replication (in effect creating a 10−1 (y) coronal slice thickness (i.e. 0.06 mm)) slice, and introducing sufficient slices by nearest neighbor replication to counter the reduction of straightened distance. This procedure was repeated for all slices, ensuring the same TL for the straightened aVP subdivisions.
After assembling all the aVP subdivisions (iOrb, iCan, iCran, OC, OT) of the same side, linear normalization with respect to the y-axis was performed to rescale each aVP length to the average TL (61.1 mm) across all subjects and sides. For each subject and aVP side, the total length (TLi) was temporarily increased to a nominal length by linearly increasing the Di, i+1 and maintaining proportionality of each subdivision, applying the same interpolation procedure as for straightening (see above). Importantly, the CSA of each slice generated from an original slice, was preserved. After the TL was adapted, the final voxel size resampled to an isotropic 0.6 mm3 resolution.
All procedures were conducted on the manually segmented label masks using scripts developed in-house with Matlab (v.R2016a) and its Image Processing toolbox (MathWorks, Natick, USA, https://www.mathworks.com), as well as fslutils (v6.0.5, Analysis Group, FMRIB, Oxford, UK, https://fsl.fmrib.ox.ac.uk), to allow for a slice-wise comparison of the cross-sectional area (CSA) between subjects.
Probabilistic atlas creation
The probabilistic atlas was created by taking the voxel-wise average of all the binary normalized aVP masks, i.e. all the L and R-to-L flipped masks, for a total of N = 48 aVP masks and multiplying by 100 to obtain percentage values in the range of 0–100. Therefore, each resulting voxel intensity represented the overlap across the 48 aVP of the 24 participants in terms of probability of the atlas voxels to belong to all participants’ aVP. The image origin was set at the centerline of the most anterior iOrb section corresponding to the nerve head. Individual atlases for each corresponding segment of the iOrb, iCan, iCran, OC, and OT subdivisions were also generated applying the same procedure (Fig. 2). To estimate the impact of interindividual anatomical variability on the atlas morphology, and thus to estimate the atlas generalizability, we used a leave-one-out approach by testing the similarity between each subject’s normalized aVP segmentation and the atlas with all subjects’ aVP averaged except for that particular one to be tested. Similarity was estimated with the Dice Similarity Index (DSI) defined as
where A represents the individual subject aVP, B the atlas’s aVP with threshold at 50% probability, and ∩ their intersection.
aVP biometry
Four biometric variables were extracted: volume, length, mean cross-sectional area, and ellipticity index (ε), separately for the left and right side. (1) Volume was directly obtained from the CISS images label masks (see the “aVP extraction” section above). (2) Length was obtained by estimating the TL distance between all aVP slice centroid, (see “Image Normalization pipeline” section above). (3) mCSA was calculated adapting a method described for the spinal cord20,21 by dividing each segment volume by its length. (4) ε estimates the degree to which the nerve section approaches an elliptical configuration applying the formula:
where h1 > h2 represents the two orthogonal cross-sectional hemi-axes, as reported from post-mortem studies17,22,23. Values close to 0 indicate a circular shape, whereas values close to 1 an elliptical configuration.
Statistical analysis
Descriptive statistics (mean with standard deviation, minimum and maximum, and 95% confidence interval for the variables with Gaussian distribution; median with the first and third quartile for the corresponding variables with non-Gaussian distribution) were calculated for continuous variables. For categorical variables, counts and percentages were provided. Left side vs Right side measurements were compared with paired t-tests. Linear Mixed-Effects Models were applied to investigate associations between the measurements (dependent variable) and the demographic characteristics (i.e., age and sex) while accounting for intra-subject variability. Mixed models allowed estimation of the between-subject proportion of the measurement variance. The coefficient of variation, describing the normalized between-subject standard deviation relative to the mean, was used to compare inter-subject segment variability. Between-subject variability was assessed by defining variables obtained from the absolute raw measurements by standardizing the volume and the length of each segment on the total aVP values (i.e., the proportion of aVP located in the segment), while mCSA and ε were standardized on the corresponding mean aVP values (i.e., the variation of each segment from a hypothetical nerve with same volume, the same length but a constant mCSA/ε). P-values < 0.05 of two-tailed tests were considered statistically significant.
Results
Standardization pipeline
For each subject, aVP was deformed in the y-axis to match the same length corresponding to the average of group and side (i.e. 61.1 mm). The “spaghetti” diagrams in Fig. 4 show how normalization was applied to the y-axis without affecting the CSA throughout the nerve (Fig. 4A), while preserving each subdivision’s proportional length (Fig. 4B).
Results of the normalization procedure. (A) The CSA for each subject before (upper) and after normalization (lower), as a function of the distance (mm) from the brain showed different length values between subjects, due to the expected anatomical interindividual variability. The corresponding median values are presented in red and cyan colors and demonstrate a good agreement indicating CSA preservation. (B) The deformations due to the normalization pipeline, as a function of position/subdivision along the y-axis for each section, comparing the length before (red) and after (cyan) normalization of each subject’s values. The number of individual lengths are shown as bands with fixed transparency levels; areas with higher transparency represent fewer number of subjects.
aVP biometry
Table 1 displays the aVP volume, length, mCSA and ε biometric data. Concurring with pathology studies17, there was no statistically significant difference between the left and right side for each parameter (P range 0.249–0.931). Therefore, side was not included in the subsequent analyses, and all subsequent metrics represent averaged left, right values. The mCSA was larger in the iOrb (7.67 mm2) and iCran (6.92 mm2) than in the iCan (5.62 mm2) subdivision (P < 0.001 for both). The OC, split into left and right hemi-trunks, presented the largest mCSA (14.37 mm2) subdivision, being almost twice as big than the contiguous iCran (6.92 mm2 P = 0.018) subdivision. ε was significantly different across all segments: (P < 0.001). The lowest index was found in iOrb (0.15) indicating an almost circular cross-section, and the highest in OC (0.57), corresponding to an elliptical cross-section.
Between-subject variability estimates for aVP volume, length, mCSA, and ε are reported in Table 2. The relative mCSA had the lowest between-subject coefficient of variation (CV) (average CV per segment 9.5%, range 6.9– − 10.7%), suggesting that this metric was the least influenced by between-subject anatomical differences in our population. Relatively higher CV values were obtained for a VP volume (average CV per sector 11.1%, range 9.9–12.8%), length (average CV per segment 10.0%, range 7.1–12.3%) and ε (average CV per segment 13.7%, range 7.9–24.1%).
aVP-24 probabilistic atlas
The results of the normalization procedure and probabilistic atlas generation for the aVP and its subdivisions constitute the anterior Visual Pathway-24 (aVP-24) atlas and are depicted in Fig. 5 in color scale. Each color represents the probability that the voxel belongs to the aVP of all participants. The leave-one-out cross-validation yielded DSI scores of 0.85 in 50% and 0.77 in 95% of the participants (Supplementary Figure S1) and thus shows a good capacity to represent the aVP general anatomy, as the scores obtained correspond to good levels of similarity24. At the same time, the 0.77 score provides an estimate of the maximum allowed spatial level that two different populations may share, to permit still the detection of differences when compared.
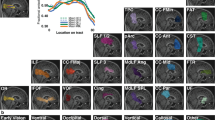
The aVP-24 probabilistic atlas with its iOrb, iCan, iCran, OC and OT anatomical subdivisions seen as coronal (leftmost) and long-axis sections. The millimetre scale indicates the distance from the optic nerve head (which was set as the atlas origin), the color scale the spatial overlap between the 48 nerves: cooler colors indicate lower overlap while warmer colors greater overlap. White arrows highlight the aVP on illustrative coronal oblique CISS images for each corresponding subdivision of the aVP from one participant. The dashed line on the OC coronal image indicates the midline plane employed for splitting. Orientation marker abbreviations: I = inferior, N = nasal, S = superior, T = temporal.
Consistent with the metrics reported for the anatomical subdivisions in Table 1, the atlas further illustrates the smooth decrease of CSA from the retrobulbar iOrb to the iCan section and a slight increase in the iCran section, to raise sharply at the iCran-OC transition and to fall sharply at the OC-OT transition. The overall shape as estimated by ε is almost circular in the extra-cranial segments but becomes increasingly elliptical in the intracranial segments, mostly in the OC.
Following OC split and centerline registration, the OC is represented collinearly with respect to the adjacent iCran and OT segments.
The atlas also provides information about the probability of anatomical subdivision boundaries, highlighting the underlying interindividual differences. The color-coded boundary distribution spread (Fig. 5) illustrates that the iCan segment exhibits the largest boundary uncertainty, being consistent with the known interindividual optic canal length variability17.
Discussion
We developed a dedicated framework for high-resolution and geometrically unbiased evaluation of images of the entire aVP, acquired by MRI. Three main results emerged. First, a relatively simple pipeline including aVP straightening and normalization allows MRI data preparation for standardized group-level analyses. Second, the salient biometry characteristics of the normal aVP and its subdivisions could be extracted. Third, the aVP-24 atlas was developed to provide researchers with probabilistic knowledge of the general aVP MRI morphology of healthy people.
This is a preparatory work to be later expanded to quantitative, standardized investigations about aVP pathological conditions, for example the degree of modification damage related to different optic neuropathies such as CSA degree and its spatial distribution.
The peculiar aVP anatomy, characterized by sharp bends and varying perioptic tissues along its course, hinders any straightforward application of the methodologies currently employed for the brain and/or spinal cord. To the best of our knowledge, the present pipeline is the first one especially dedicated to the assessment of the entire aVP morphometry. One strength is the employment of a specific MRI acquisition technique that yields optimal nerve-to-CSF contrast, high-resolution, and geometrically unbiased tissue sampling in a reasonable scan time (about three minutes). The employed CISS images are based on the “steady-state free-precession” technique, which provides optimal signal contrast at the interface between the cranial nerves and the surrounding cerebrospinal fluid (CSF)25. Another strength is the relatively simple normalization pipeline preserving the cross-section aVP topology, thus, allowing spatially standardized comparisons between groups.
Previous morphometric MRI studies were mostly limited to the ON and/or provided 1-D or 2-D metrics, and/or employed non-dedicated imaging protocols (Table 3). In two studies including healthy subjects, dedicated ultrafast 2D MRI “HASTE” images with 3 mm and 2 mm slice thickness were employed to obtain 1-D and CSA measurements of the iOrb at different arbitrary points9,12. In a small group of 8 healthy subjects a semiquantitative segmentation of 2D T2-TSE images were acquired using a multidynamic scheme to obtain measurements limited to 8–10 slices of the intraorbital ON13. In another clinical study comparing 23 healthy subjects with a group of patients with idiopathic intracranial hypertension, volume calculations of the intraorbital ON was based on 2D T2-weighted 2 mm-thick images11. Finally, in a clinical study including a small group of 12 healthy subjects and 8 patients with glaucoma, an automated volumetry analysis of the aVP using standard 3-D-MPRAGE images and a non-dedicated processing pipeline was performed10. Contrarily to the previous MRI reports and pathology studies17, however, a substantial volume difference between the L and R ON and OT in healthy subjects was reported, which is suggestive for underlying biases related to the non-dedicated pipeline.
We can provide a systematic description of the salient morphometry features of the entire aVP for the first time. These data are in line with the descriptions available from pathology studies17,26,27. In particular, the average ON length estimated in our study is quite close to that reported by Radunovic et al.17 and the iOrb mCSA diameter is consistent with reported average cross-section diameters26. However, whereas the iOrb mCSA (L/R average = 7.67 mm2) diameter approaches the surface area measured at the level of the orbital foramen (L/R average = 6.97 mm2)17, our estimate of the iCan was substantially smaller (L/R average = 5.62 mm2) when compared to the cranial foramen measurements in cadavers (8.76 mm2)17. We speculate that such discrepancies might be related, at least in part, to (1) limitations in the iCan contouring accuracy because of the reduced signal contrast gradient related to the perioptic CSF scarcity at this level and (2) methodological differences in the determination of the iCan boundaries. Supporting such hypotheses, our iCan MRI volume estimates (L/R average = 38.95 mm3) are substantially smaller compared to the pathology data (50.25 mm3)17.
The description of the aVP cross-section ellipticity is another novelty of the present study is. ε increases from an almost circular to an elliptical configuration from the intraorbital to the intracranial subdivisions. This is in line with post-mortem reports17,22, and parallels the configuration of the osseous optic canal17. Another interesting finding was that the OC exhibited the largest mCSA (L/R average = 14.37 mm2) and the highest ε (L/R average = 0.57). This is consistent with the general appearance of the OC on standard MRI as well as with reports from gross anatomy17, and is reasonably linked to the arrangement of the fibers hemidecussation28.
The aVP-24 atlas provides probabilistic knowledge of the general aVP morphology and its anatomical subdivisions and might allow localization of differences in the morphology, trophism and/or lesion distribution between healthy and/or diseased populations in a future extension of this study. Spatial localization of the differences may be obtained (1) in terms of distance in mm from the atlas origin, which was set at the optic nerve head, or (2) in terms of categorization within the different aVP subdivisions. The atlas additionally discloses the aVP general morphology visible in vivo on MRI being characterized by CSA and ellipticity variation, consistent with post-mortem findings17,27. Remarkably, boundary uncertainty was highest for the iCan segment, corresponding to a low spatial localization reliability when studying a group. Two assumptions underlying the aVP-24 atlas creation must be addressed. First, the complex OC geometry was assimilated to that of the remaining aVP by splitting the OC into hemi-trunks. This allowed for methodology harmonization and simplification but led to the artefactual “banjo-like” shape related to the shift of the relatively larger OC slices to the centerline (Fig. 5). The other assumption concerning the left- and right-side symmetry was based on the lack of significant differences in the biometry data detected in our population. These assumptions led to the proposed unpaired atlas generated by straightening and averaging the L and right aVP sides together.
Several limitations need to be acknowledged. First, segmentation was performed manually being time consuming and limiting potential reproducibility. Future development of automated segmentation would be a desirable direction of development. Second, our data was obtained from a relatively small number of healthy adult subjects. Larger studies are warranted to provide age and sex specific normative data. Third, ε might have been overestimated particularly in sharp-angled segments, because a centerline slice shift was conducted according to the image coronal plane upon nerve straightening. Finally, the unpaired atlas configuration implies the loss of the visual field topology information, limiting correlations with functional deficits.
Data availability
The aVP-24 atlas and the original software developed in this study are made freely available online as part of the 'aVP-Toolbox' project at https://github.com/EmanuelePravata/aVP-Toolbox.
References
Quigley, H. A., Miller, N. R. & George, T. Clinical evaluation of nerve fiber layer atrophy as an indicator of glaucomatous optic nerve damage. Arch. Ophthalmol. 98, 1564–1571. https://doi.org/10.1001/archopht.1980.01020040416003 (1980).
Smith, C. A., Vianna, J. R. & Chauhan, B. C. Assessing retinal ganglion cell damage. Eye 31, 209–217. https://doi.org/10.1038/eye.2016.295 (2017).
Dutra, B. G., da Rocha, A. J., Nunes, R. H. & Maia, A. C. M. J. Neuromyelitis optica spectrum disorders: Spectrum of MR imaging findings and their differential diagnosis. Radiographics 38, 169–193. https://doi.org/10.1148/rg.2018170141 (2018).
Harrigan, R. L. et al. Quantitative characterization of optic nerve atrophy in patients with multiple sclerosis. Mult. Scler. J.- Exp., Transl. Clin. 3, 2055217317730097. https://doi.org/10.1177/2055217317730097 (2017).
Sarkies, N. Traumatic optic neuropathy. Eye 18, 1122–1125. https://doi.org/10.1038/sj.eye.6701571 (2004).
Biousse, V. & Newman, N. J. Ischemic optic neuropathies. N. Engl. J. Med. 372, 2428–2436. https://doi.org/10.1056/NEJMra1413352 (2015).
Danesh-Meyer, H. V. Radiation-induced optic neuropathy. J. Clin. Neurosci. 15, 95–100. https://doi.org/10.1016/j.jocn.2007.09.004 (2008).
Newman, N. J. & Biousse, V. Hereditary optic neuropathies. Eye 18, 1144–1160. https://doi.org/10.1038/sj.eye.6701591 (2004).
Furlanetto, R. L. et al. Structural and functional analyses of the optic nerve and lateral geniculate nucleus in glaucoma. PloS one 13, e0194038. https://doi.org/10.1371/journal.pone.0194038 (2018).
Hernowo, A. T., Boucard, C. C., Jansonius, N. M., Hooymans, J. M. & Cornelissen, F. W. Automated morphometry of the visual pathway in primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci. 52, 2758–2766. https://doi.org/10.1167/iovs.10-5682 (2011).
Hoffmann, J. et al. Volumetric assessment of optic nerve sheath and hypophysis in idiopathic intracranial hypertension. AJNR Am. J. Neuroradiol. 35, 513–518. https://doi.org/10.3174/ajnr.A3694 (2014).
Lagrèze, W. A. et al. Morphometry of the retrobulbar human optic nerve: comparison between conventional sonography and ultrafast magnetic resonance sequences. Invest. Ophthalmol. Vis. Sci. 48, 1913–1917. https://doi.org/10.1167/iovs.06-1075 (2007).
Yiannakas, M. C. et al. MRI acquisition and analysis protocol for in vivo intraorbital optic nerve segmentation at 3T. Invest. Ophthalmol. Vis. Sci. 54, 4235–4240. https://doi.org/10.1167/iovs.13-12357 (2013).
Pravatà, E. et al. Dedicated 3D-T2-STIR-ZOOMit imaging improves demyelinating lesion detection in the anterior visual pathways of patients with multiple sclerosis. AJNR Am. J. Neuroradiol. 42, 1061–1068. https://doi.org/10.3174/ajnr.A7082 (2021).
Flitcroft, D. I. et al. IMI-defining and classifying myopia: A proposed set of standards for clinical and epidemiologic studies. Invest. Ophthalmol. Vis. Sci. 60, M20-m30. https://doi.org/10.1167/iovs.18-25957 (2019).
Rabrenović, M. et al. Neurotoxic effects of oxygen in hyperbaric environment: A case report. Vojnosanit. Pregl. 72, 827–830. https://doi.org/10.2298/vsp140312059r (2015).
Radunovic, M. et al. Morphometric characteristics of the optic canal and the optic nerve. Folia Morphol. 78, 39–46. https://doi.org/10.5603/FM.a2018.0065 (2019).
Saeki, N. et al. Histologic characteristics of normal perivascular spaces along the optic tract: New pathogenetic mechanism for edema in tumors in the pituitary region. AJNR Am. J. Neuroradiol. 25, 1218–1222 (2004).
Selhorst, J. B. & Chen, Y. The optic nerve. Semin. Neurol. 29, 29–35. https://doi.org/10.1055/s-0028-1124020 (2009).
Horsfield, M. A. et al. Rapid semi-automatic segmentation of the spinal cord from magnetic resonance images: Application in multiple sclerosis. NeuroImage 50, 446–455. https://doi.org/10.1016/j.neuroimage.2009.12.121 (2010).
Pravatà, E. et al. Influence of CNS T2-focal lesions on cervical cord atrophy and disability in multiple sclerosis. Mult. Scler. J. 26, 1402–1409. https://doi.org/10.1177/1352458519865989 (2020).
Jeffery, G. et al. The human optic nerve: Fascicular organisation and connective tissue types along the extra-fascicular matrix. Anat. Embryol. 191, 491–502. https://doi.org/10.1007/bf00186739 (1995).
Maniscalco, J. E. & Habal, M. B. Microanatomy of the optic canal. J. Neurosurg. 48, 402–406. https://doi.org/10.3171/jns.1978.48.3.0402 (1978).
Molloi, S., Zhou, Y. & Kassab, G. S. Regional volumetric coronary blood flow measurement by digital angiography: In vivo validation. Acad. Radiol. 11, 757–766. https://doi.org/10.1016/j.acra.2004.04.002 (2004).
Sheth, S., Branstetter, B. F. T. & Escott, E. J. Appearance of normal cranial nerves on steady-state free precession MR images. Radiographics 29, 1045–1055. https://doi.org/10.1148/rg.294085743 (2009).
Jonas, J. B., Schmidt, A. M., Müller-Bergh, J. A. & Naumann, G. O. Optic nerve fiber count and diameter of the retrobulbar optic nerve in normal and glaucomatous eyes. Graefes Arch. Clin. Exp. Ophthalmol. 233, 421–424. https://doi.org/10.1007/bf00180945 (1995).
Karim, S., Clark, R. A., Poukens, V. & Demer, J. L. Demonstration of systematic variation in human intraorbital optic nerve size by quantitative magnetic resonance imaging and histology. Invest. Ophthalmol. Vis. Sci. 45, 1047–1051. https://doi.org/10.1167/iovs.03-1246 (2004).
Jain, N. S. et al. Visualization of nerve fiber orientations in the human optic chiasm using photomicrographic image analysis. Invest. Ophthalmol. Vis. Sci. 56, 6734–6739. https://doi.org/10.1167/iovs.15-17443 (2015).
Bäuerle, J. et al. Reproducibility and accuracy of optic nerve sheath diameter assessment using ultrasound compared to magnetic resonance imaging. BMC Neurol. 13, 187. https://doi.org/10.1186/1471-2377-13-187 (2013).
Kim, D. H., Jun, J. S. & Kim, R. Measurement of the optic nerve sheath diameter with magnetic resonance imaging and its association with eyeball diameter in healthy adults. J. Clin. Neurol. 14, 345–350. https://doi.org/10.3988/jcn.2018.14.3.345 (2018).
Geeraerts, T. et al. Use of T2-weighted magnetic resonance imaging of the optic nerve sheath to detect raised intracranial pressure. Crit. Care 12, R114. https://doi.org/10.1186/cc7006 (2008).
Kashiwagi, K., Okubo, T. & Tsukahara, S. Association of magnetic resonance imaging of anterior optic pathway with glaucomatous visual field damage and optic disc cupping. J. Glaucoma 13, 189–195. https://doi.org/10.1097/00061198-200406000-00003 (2004).
Liu, C. et al. Optic nerve sheath diameter measured using magnetic resonance imaging and factors that influence results in healthy Chinese adults: A cross-sectional study. Chin. Med. J. 134, 2263–2265. https://doi.org/10.1097/cm9.0000000000001353 (2021).
Votruba, M. et al. MRI of the intraorbital optic nerve in patients with autosomal dominant optic atrophy. Neuroradiology 42, 180–183. https://doi.org/10.1007/s002340050041 (2000).
Seitz, J. et al. Magnetic resonance imaging in patients diagnosed with papilledema: a comparison of 6 different high-resolution T1- and T2(*)-weighted 3-dimensional and 2-dimensional sequences. J. Neuroimaging 12, 164–171. https://doi.org/10.1111/j.1552-6569.2002.tb00115.x (2002).
Weigel, M., Lagrèze, W. A., Lazzaro, A., Hennig, J. & Bley, T. A. Fast and quantitative high-resolution magnetic resonance imaging of the optic nerve at 3.0 tesla. Invest. Radiol. 41, 83–86. https://doi.org/10.1097/01.rli.0000195820.98062.c5 (2006).
Zhang, Y. Q. et al. Anterior visual pathway assessment by magnetic resonance imaging in normal-pressure glaucoma. Acta Ophthalmol. 90, e295-302. https://doi.org/10.1111/j.1755-3768.2011.02346.x (2012).
Yiannakas, M. C. et al. A method for measuring the cross sectional area of the anterior portion of the optic nerve in vivo using a fast 3D MRI sequence. J. Magn. Reson. Imaging 31, 1486–1491. https://doi.org/10.1002/jmri.22202 (2010).
Ramli, N. M. et al. Novel use of 3T MRI in assessment of optic nerve volume in glaucoma. Graefes Arch. Clin. Exp. Ophthalmol. 252, 995–1000. https://doi.org/10.1007/s00417-014-2622-6 (2014).
Acknowledgements
This work was supported by the Velux Stiftung, Swiss Society of Multiple Sclerosis (SSMS), Advisory board for research Ente Ospedaliero Cantonale (ABREOC), and the Italian Ministry of Health “Ricerca corrente 2022” grants. The Authors are grateful to Liliane Petrini for her help with manuscript editing, and to Dr. Paul Summers, PhD, for his help in preparing the scripts and atlas for their open online publication.
Funding
For conducting this project, E.P. received funding from the Velux Stiftung, Swiss MS Society and Scientific Research Advisory Board (ABREOC); R.N. received funding from the Scientific Research Advisory Board (ABREOC); L.R. and M.P.S. received funding from the Velux Stiftung and from the Italian Ministry of Health (Ricerca Corrente). The remaining Authors did not receive specific funding for this project.
Author information
Authors and Affiliations
Contributions
E.P.: conception, design, acquisition and interpretation of data, manuscript draft. A.D.: data analysis and interpretation, manuscript draft. R.N.: software creation. L.C., M.P.S.: data statistics and interpretation. L.R.: data interpretation, manuscript revision. A.C.: data interpretation, manuscript revision. A.O., A.C., A.C., A.K.: manuscript revision. C.G., C.Z.: data interpretation, manuscript draft, data acquisition. All authors have approved the submitted manuscript version, agreed both to be personally accountable for the author's own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated, resolved, and the resolution documented in the literature.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pravatà, E., Diociasi, A., Navarra, R. et al. Biometry extraction and probabilistic anatomical atlas of the anterior Visual Pathway using dedicated high-resolution 3-D MRI. Sci Rep 14, 453 (2024). https://doi.org/10.1038/s41598-023-50980-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-50980-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.