Abstract
Hepatocellular carcinoma (HCC) is a lethal malignancy worldwide with an increasing number of new cases each year. Apolipoprotein (APOL) isoforms have been explored for their associations with HCC.The GSE14520 cohort was used for training data; The Cancer Genome Atlas (TCGA) database was used for validated data. Diagnostic, prognostic significance and mechanisms were explored using these cohorts. Risk score models and nomograms were constructed using prognosis-related isoforms and clinical factors for survival prediction. Oncomine and HCCDB databases were further used for validation of diagnostic, prognostic significance. APOL1, 3, and 6 were differentially expressed in two cohorts (all P ≤ 0.05). APOL1 and APOL6 had diagnostic capacity whereas APOL3 and APOL6 had prognostic capacity in two cohorts (areas under curves [AUCs] > 0.7, P ≤ 0.05). Mechanism studies demonstrated that APOL3 and APOL6 might be involved in humoral chemokine signaling pathways (all P ≤ 0.05). Risk score models and nomograms were constructed and validated for survival prediction of HCC. Moreover, diagnostic values of APOL1 and weak APOL6 were validated in Oncomine database (AUC > 0.700, 0.694); prognostic values of APOL3 and APOL6 were validated in HCCDB database (all P < 0.05). Differentially expressed APOL1 and APOL6 might be diagnostic biomarkers; APOL3 and APOL6 might be prognostic biomarkers of RFS and OS for HCC via chemokine signaling pathways.
Similar content being viewed by others
Introduction
Global cancer statistics in 2018 indicated that liver cancer in both sexes ranked seventh in morbidity as approximately 4.7% of 36 cancer types and 841,080 new diagnosis in 185 countries. Liver cancer is third in mortality of approximately 8.2% of 36 cancer types with 781,631 cancer deaths in 185 countries1. Higher incidence and mortality by roughly 2- to 3-times are seen in males compared to females in most world regions1. Accounting for most types of primary liver cancer were hepatocellular carcinoma (HCC) at 75–85% and intrahepatic cholangiocarcinoma at 10–15%1. Factors such as chronic hepatitis B virus (HBV) or hepatitis C virus infection, food contamination with aflatoxin, high alcohol consumption, cirrhosis, male sex, smoking and HCC family history are risk elements for HCC tumorigenesis and progression2, 3. In addition to external risk factors, genetic gene alterations are recognized etiological constituents of HCC initiation and progression4. Although advances have occurred in surgical resection, which is the best approach for HCC treatment, the prognosis of HCC remains poor with an approximately 30% 5-year survival rate5. Furthermore, more than 70% of developed tumor recurrences occur at 5 years6, 7. Some potential biomarkers have been identified for HCC diagnosis and prognosis8, 9. However, these biomarkers need further validation in varied populations. Therefore, identification of new biomarkers for HCC early diagnosis and prognosis is crucial for better patient survival.
The apolipoprotein (APO) superfamily contains 10 subfamilies: APOA, APOB, APOC, APOD, APOE, APOF, APOH, APOL, APOM, and APOO (https://www.genenames.org/data/genegroup/#!/group/405). The APOL subfamily has six isoforms: APOL1, APOL2, APOL3, APOL4, APOL5, and APOL610. APOs are critical for the development of high-density lipoprotein (HDL) and low-density lipoprotein complexes11. APOL1 binds to HDL12. As a major source of HDL production and circulating APOs, the liver is pivotal for the circulating pool of APOL113, 14. In addition, APOL1 induces autophagy-mediated cell death independent of caspase-mediated apoptosis and can be a general autophagy mediator15. APOL2 has an anti-apoptotic function in interferon-γ-induced cytotoxicity in human bronchial epithelial cells16. The APOL3 region on chromosome 22q12 was a risk locus in a family-based association analysis of 42 families with hereditary prostate cancer17. Single nucleotide polymorphisms and haplotypes in APOL1, 2 and 4, located on chromosome 22q12.3–13.1, are associated with schizophrenia in African-American, European-American, Chinese and Japanese populations18. APOL5 is reported rare with disease. Overexpression of wild-type APOL6 leads to mitochondrial-mediated apoptosis in DLD-1 cells, characterized by release of cytochrome c and Smac/DIABLO from mitochondria and activation of caspase 919. APOL family isoforms transport HDL in cell membranes are important for the development and maintenance of membrane structure and function20, 21. Therefore, we hypothesized that aberrant expression of APOLs may be associated with HCC tumorigenesis and progression because of APOL isoforms involvement in membrane structure and function and as a major source of HDL production and circulation of APOs in the liver. We conducted this study to explore the potential roles of APOL isoforms in HCC.
Material and methods
Patient data and ethical approval
Gene profile data of GSE14520 was used for a training cohort. To avoid batch effects, only the platform of GPL3921 in this dataset, including 212 HBV-HCC patients, was used for analysis (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14520)22, 23. A total of 370 HCC patients from The Cancer Genome Atlas (TCGA) database were used for validation (https://cancergenome.nih.gov/). GSE14520 dataset had its features with HBV-related, most cirrhosis background, and Asian race, while TCGA dataset had its features with most patients over 40 years old, Asian race with less than 45%, roughly 50% patients with BMI ≥ 24.
Analysis of diagnostic and prognostic significance
Expression of APOL1-6 mRNA was used for diagnostic and prognostic analysis. Expression of APOL1-6 mRNA in HCC and nontumor tissues was used for diagnostic capacity assessment in the GSE14520 and TCGA cohorts. Expression of APOL1-6 in HCC tissues was used for prognosis assessment in the GSE14520 and TCGA cohorts. Expression of APOL1-6 was divided into low and high by median expression levels. Prognosis-related genes were combined for joint-effect analysis for overall survival (OS) and recurrence-free survival (RFS).
Mechanism exploration of prognosis-related genes in genome-wide
Identified prognosis-related genes were explored for potential mechanisms in HCC with genome-wide gene set enrichment analysis (GSEA). GSEA was performed using gene ontologies (GO) of biological processes (BP, c5.bp.b6.1.symbols.gmt), cellular component (CC, c5.cc.v6.1.symbols.gmt), molecular function (MF, c5.mf.v6.1.symbols.gmt), and Kyoto Encyclopedia of Genes and Genomes pathways (KEGG, c2.cp.kegg.v6.1.symbols.gmt). P values ≤ 0.05 and false discovery rate ≤ 0.25 were considered significant.
Risk score model and nomogram construction
Risk score models were used for HCC prognosis predictions by gene expression. A risk score model was constructed using coefficients and expressions of different genes using the formula: risk scores = expression of gene1 x β1 (coefficient) + expression of gene2 x β2 (coefficient) + … + expression of genen x βn (coefficient)24,25,26. Risk score models included risk score rankings, survival status, expression heatmaps, Kaplan–Meier plots and time-dependent receiver operating characteristic curves.
Nomograms were constructed for HCC prediction of clinical factors and gene expression. Prognosis-related genes and prognosis-related clinical factors were used in nomograms. Different expression levels and factors indicated different points and total points equal to the sum of all points.
Co-expression matrixes and interaction networks
Co-expression matrixes of APOL1-6 were constructed using mRNA expression in the GSE14520 and TCGA cohorts. Co-expression networks of gene–gene interaction (GGI) of APOL1-6 were constructed using the geneMANIA plugin of Cytoscape software27, 28. Chemical association networks were constructed using APOL1-6 and visualized for APOL1-6 and chemicals using the STITCH website (http://stitch.embl.de/)29. Visualized GO term interaction networks were constructed using the BinGO plugin of Cytoscape software30.
Validation of diagnostic analysis and prognosis significance by Oncomine and HCCDB databases
Differential expressions and diagnostic significance of APOL isoforms were further validated using Wurmbach dataset in Oncomine database (https://www.oncomine.org/resource/main.html)31. Furthermore, prognosis-related APOL isoforms, including OS and RFS, in TCGA and GSE14520 datasets were further validated in HCCDB database (http://lifeome.net/database/hccdb/home.html)32.
Statistical analysis
Survival analysis was by SPSS software version 24 (IBM, Chicago, IL). Scatter plots and the Kaplan–Meier method were generated using GraphPad 7.0. Calculations of 95% confidence intervals and hazard ratios were by univariate and multivariate Cox regression models. Median survival time and log-rank P value were calculated by the Kaplan–Meier method. RT-PCR was validated using paired t-tests. A P value ≤ 0.05 was considered significant.
Results
Demographic characteristics and mRNA expression analysis
The GSE14520 cohort contained 212 patients with HBV-related HCC. The TCGA cohort contained 370 patients with HCC. Characteristics of the cohorts are in our previous report33.
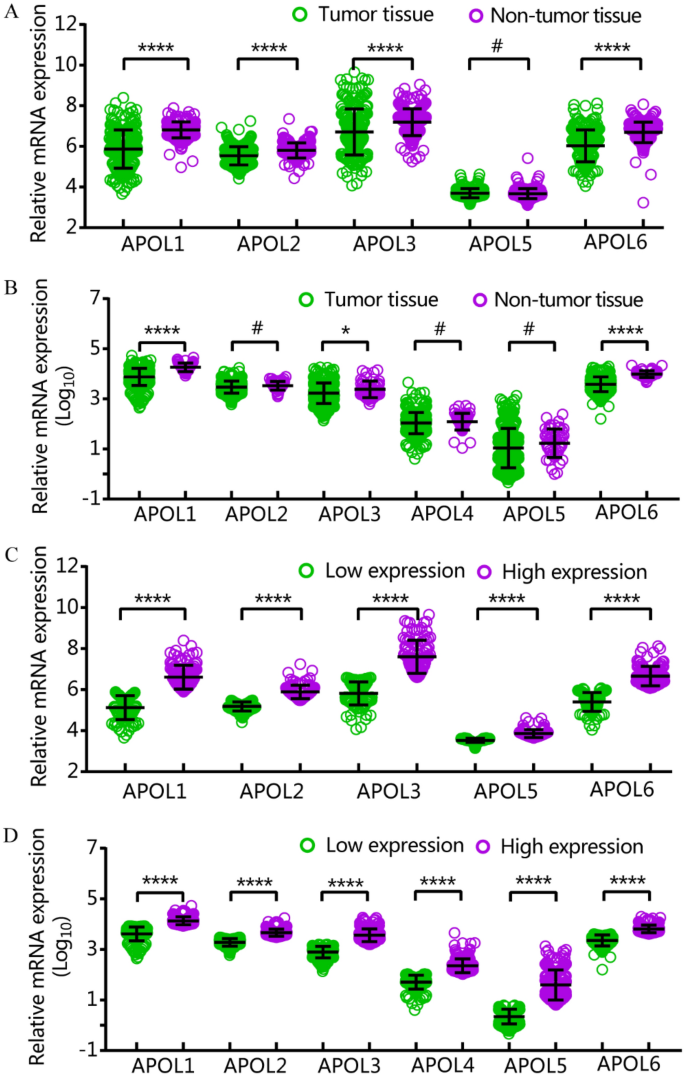
APOL1, 2, 3, and 6 were differentially expressed in tumor and non-tumor tissues in the GSE14520 cohort. APOL1, 3 and 6 were differentially expressed in tumor and non-tumor tissues in the TCGA cohort (Fig. 1A,B). All APOL isoforms were differentially expressed in low and high expression groups in both cohorts (Fig. 1C,D). APOL4 was not included in the GSE14520 cohort.
Diagnostic capacity and prognostic significance analysis
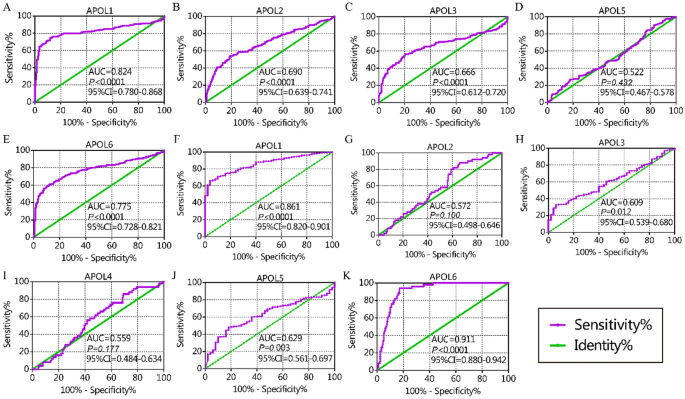
From the diagnostic capacity analysis, in the GSE14520 cohort, APOL1 and APOL6 had diagnostic significance for HCC (APOL1: area under curve [AUC] 0.824, P < 0.0001; APOL6: AUC 0.775, P < 0.0001, Fig. 2A,E). In the TCGA cohort, APOL1 and APOL6 had diagnostic significance for HCC (APOL1: AUC 0.824, P < 0.0001; APOL6: AUC 0.911, P < 0.0001, Fig. 2F,K). Others showed no or weak diagnostic capacity for HCC (Fig. 2).
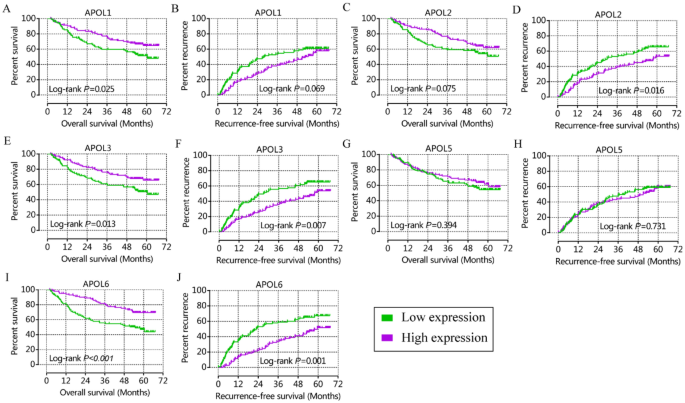
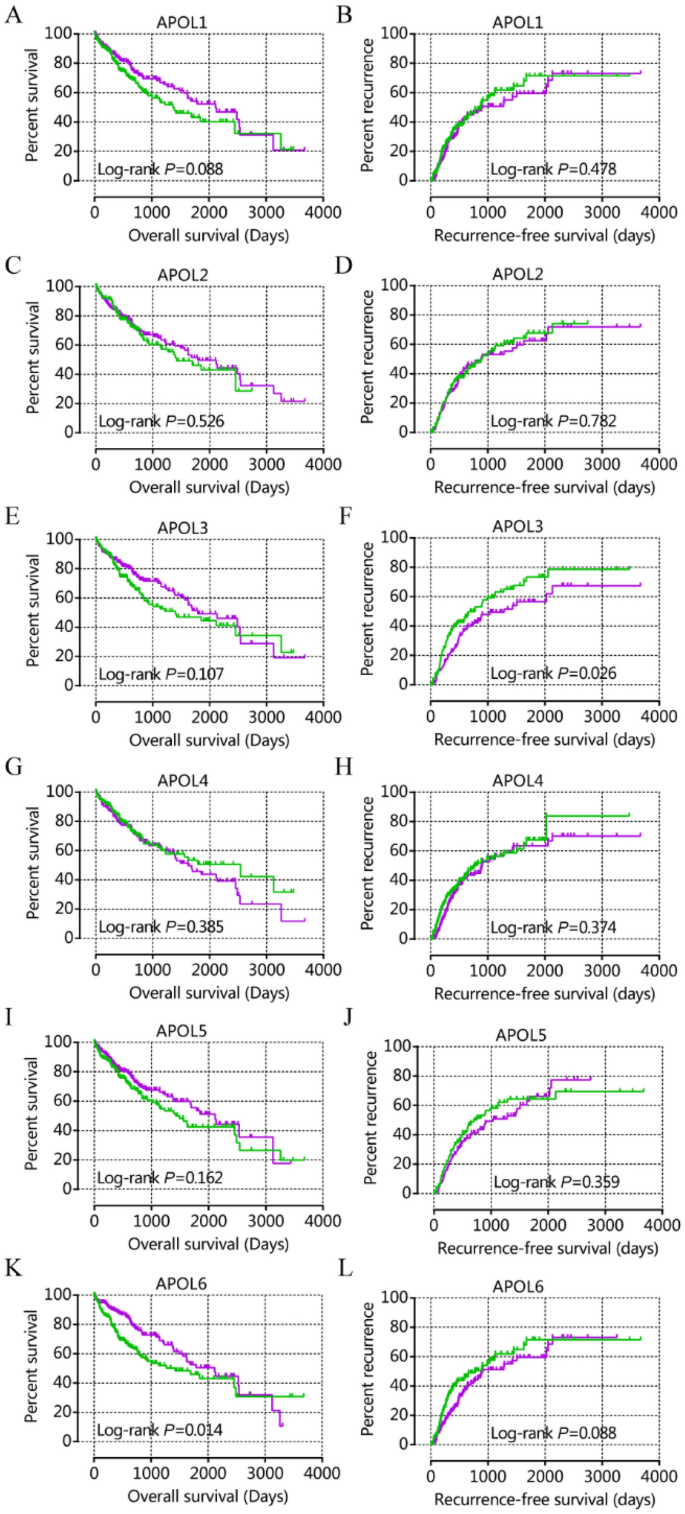
From prognostic significance analysis, in the GSE14520 cohort, APOL1, APOL3 and APOL6 showed prognostic value by univariate analysis. APOL3 and APOL6 showed prognostic value for OS by multivariate analysis (Table 1, Fig. 3). APOL2, APOL3 and APOL6 showed prognostic value for RFS in both univariate and multivariate analysis (Table 1, Fig. 3). In the TCGA cohort, only APOL6 showed prognostic value for OS in univariate and multivariate analysis (Table 2, Fig. 4). APOL3 and APOL4 had prognostic value for RFS in multivariate analysis (Table 2).
Analysis of combined prognosis-related genes
Prognosis-related genes were used for combined analysis. APOL2, APOL3, and APOL6 were combined for OS and APOL3 and APOL6 were combined for RFS in the GSE14520 cohort (Table 3, Figure S1A-E). APOL3 and APOL4 were combined for RFS in TCGA cohort (Table S1, Figure S1F). Groups containing two poor prognosis indicators had the worst survival times whereas groups with two good prognosis indicators had the best survival times.
Prospective molecular mechanism exploration by GSEA
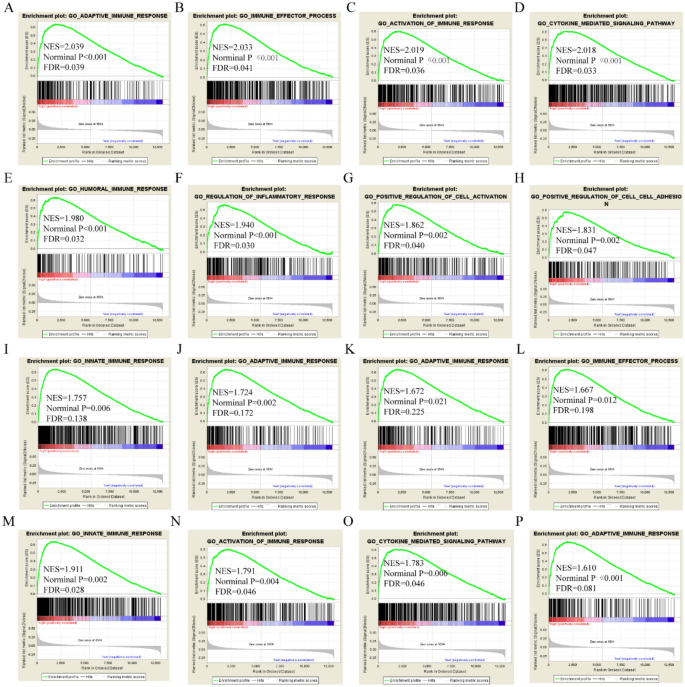
GSEA was performed to explore prospective molecular genome-wide mechanisms of APOL isoform involvement in HCC. APOL3 was involved in the adaptive immune response, immune effector processes, humoral immune response, positive cell activation, regulation of inflammatory responses, and cytokine-mediated signaling pathways by GO terms in the GSE14520 cohort (Fig. 5A–L). APOL3 was found to be involved in cell adhesion molecular cams, chemokine-signaling pathways, type 1 diabetes mellitus, and fatty acid metabolism by KEGG pathway in the GSE14520 cohort (Fig. 5M–P). APOL6 was found to be involved in the humoral immune response, fatty acid metabolism, immune effector processes, cytokine-mediated signaling pathways, and drug metabolism cytochrome P450 in the GSE14520 cohort (Figure S2).
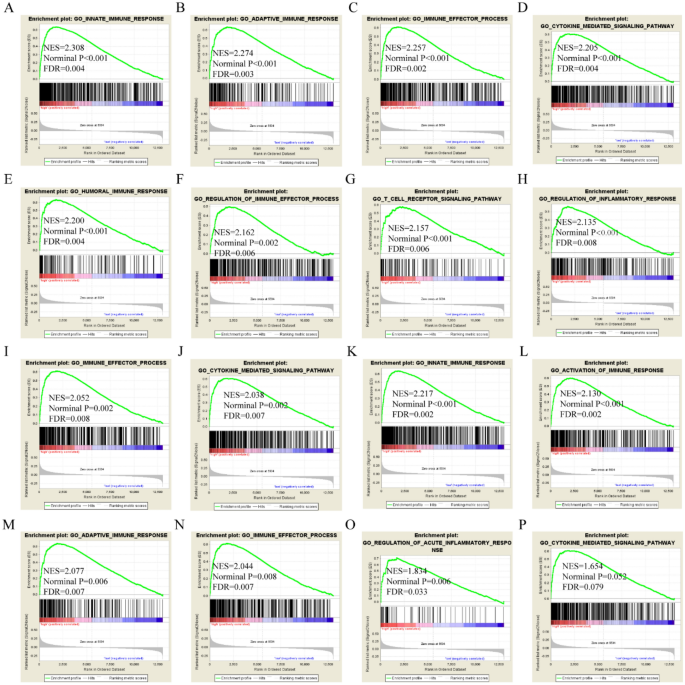
APOL3 was found to be involved in B-cell mediated immunity, activation of the immune response, the adaptive immune response, the humoral immune response, cytokine-mediated signaling pathways, chromosome centromeric region, histone binding, and chromatin binding by GO terms in the TCGA cohort (Fig. 6A–L). APOL3 was found to be involved in cell adhesion molecular cams, type 1 diabetes mellitus, chemokine signaling pathways, and fatty acid metabolism by KEGG pathways in the TCGA cohort (Fig. 6M–P). APOL6 was found to be involved in the immune effector response, B-cell mediated immunity, regulation of inflammatory responses, cytokine-mediated signaling pathways, JAK-STAT signaling pathways, and cell adhesion molecular cams in the TCGA cohort (Figure S3).
Risk score model and nomogram construction
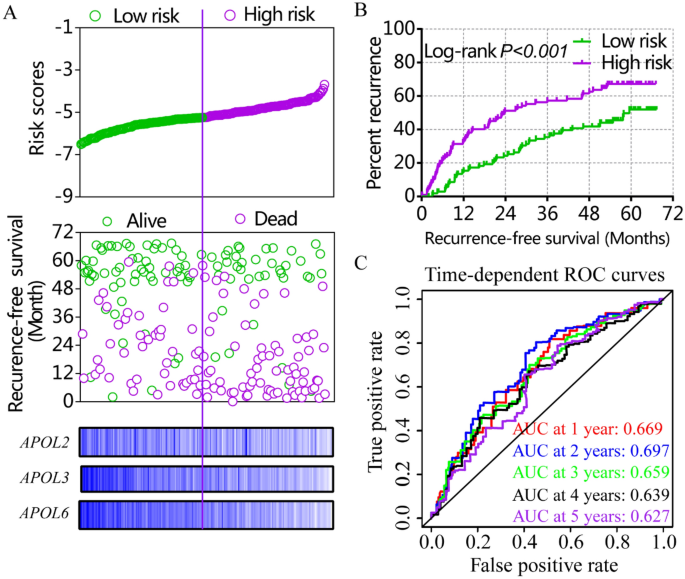
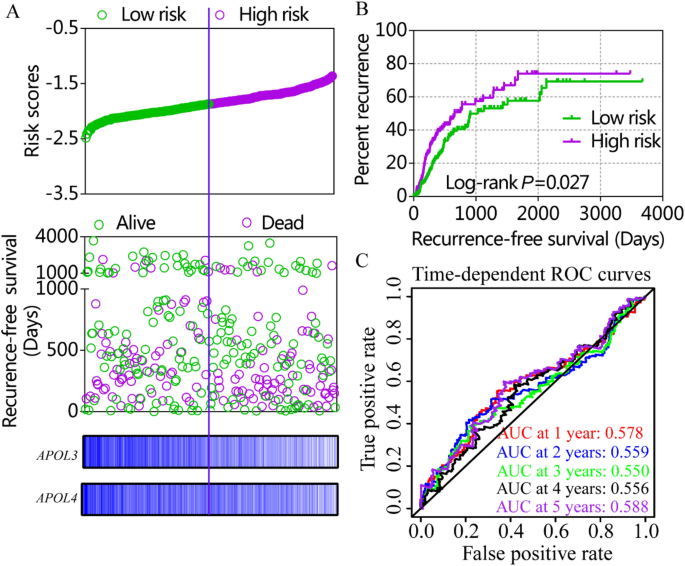
Risk score models were constructed using APOL3 and APOL6 for OS (Figure S4, Table 4) and APOL2, APOL3 and APOL6 for RFS in the GSE14520 cohort (Fig. 7, Table 4). Risk score models were constructed using APOL3 and APOL4 for RFS in the TCGA cohort (Fig. 8, Table 4). Risk score ranking, patient survival status, heat maps of APOL expression isoforms, Kaplan–Meier plots and time-dependent ROC curves for 1-, 2-, 3-, 4-, and 5-year survival were included in the models. ROC curves for the GSE14520 cohort, including OS and RFS models, showed better prognoses than for the TCGA cohort. Detailed prognostic analysis results of low- and high- risk groups were shown in Table S2.
Risk score model, Kaplan–Meier plots and time-dependent receiver operative characteristic curves for recurrence-free survival in the GSE14520 cohort. (A): Risk score model with risk score, survival status, heatmap of APOL2, 3 and 6. (B): Kaplan–Meier plots by low and high recurrence-risk groups; (C): Time-dependent receiver operative characteristic curves for recurrence-free survival at 1, 2, 3, 4, and 5 years.
Risk score model, Kaplan–Meier plots and time-dependent receiver operative characteristic curves for recurrence-free survival in the TCGA cohort. (A): Risk score model with risk score, survival status, and heatmap for APOL3 and 4. (B): Kaplan–Meier plots by low and high recurrence-risk groups. (C): Time-dependent receiver operative characteristic curves for recurrence-free survival at 1, 2, 3, 4, and 5 years.
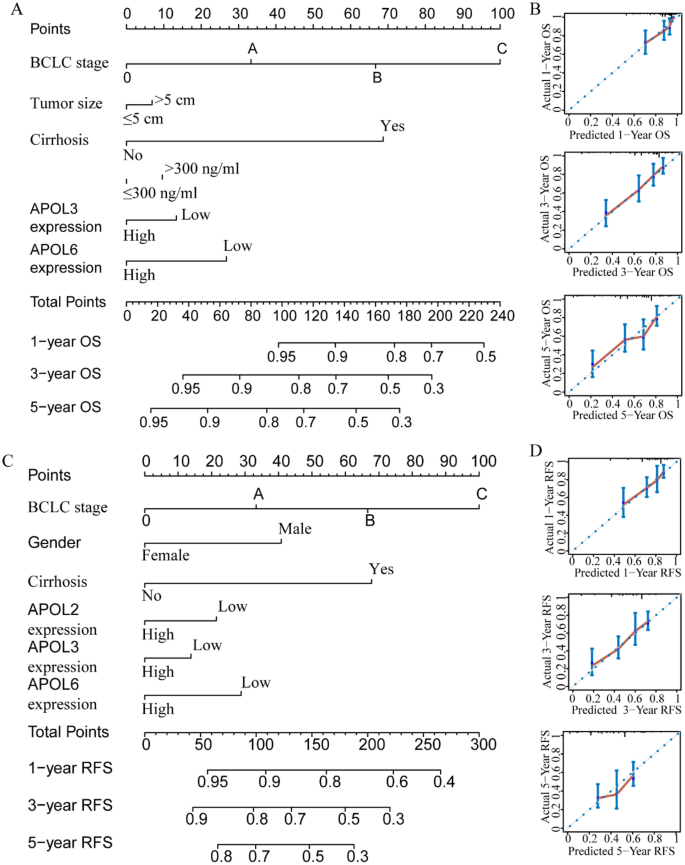
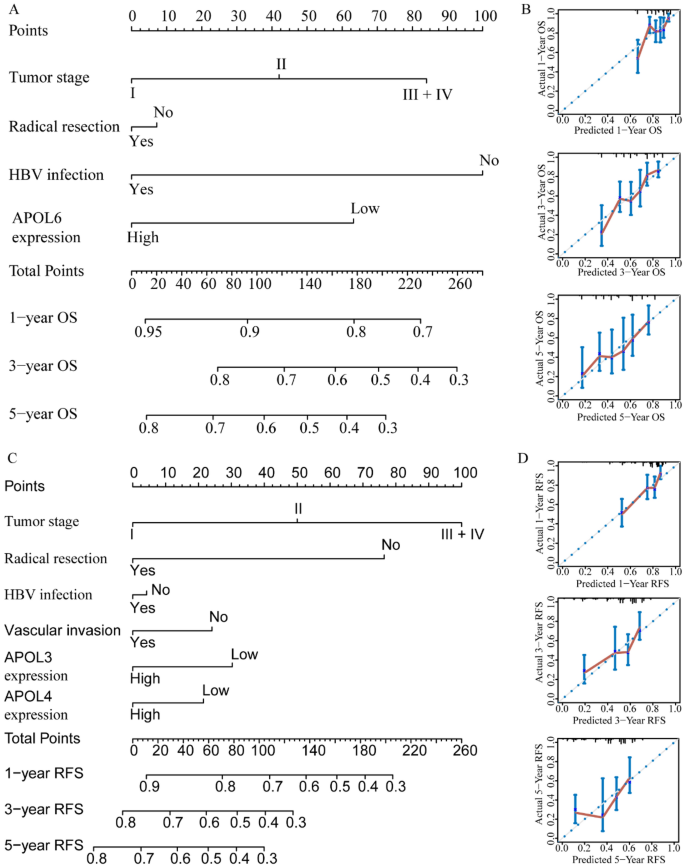
For the GSE14520 cohort, nomograms were constructed using tumor size, cirrhosis, α-fetoprotein (AFP), Barcelona Clinic Liver Cancer (BCLC) stage, APOL3 and APOL6 expressions for OS (Fig. 9A,B). Nomograms were constructed using sex, cirrhosis, BCLC stage, APOL2, APOL3 and APOL6 expression for RFS (Fig. 9C,D). For the TCGA cohort, nomograms were constructed using tumor stage, radical resection, HBV infection, and APOL6 expression for OS (Fig. 10A,B). Nomograms were constructed for tumor stage, radical resection, HBV infection, vascular invasion, APOL3 and APOL4 expression for RFS (Fig. 10C,D). Small tumor size; female sex; lack of cirrhosis; BCLC stage 0; high expression of APOL2, APOL3 and APOL6; and low AFP levels indicated higher survival rate in the GSE14520 cohort. Early tumor stage, radical resection, low APOL3 expression, high APOL4 and APOL6 expressions, vascular invasion and HBV infection indicated higher survival rates in the TCGA cohort.
Prognosis-predicted nomograms and inner validation for 1, 3, and 5 years in the GSE14520 cohort. (A-–B): Overall survival predicting nomogram using tumor size, cirrhosis, BCLC stage, AFP levels, APOL3 and APOL6 expression for 1, 3, and 5 years and inner validation for 1, 3, and 5 years. (C–D): Recurrence-free survival predicting nomogram using sex, cirrhosis, BCLC stage APOL2, APOL3 and APOL6 expression and inner validation for 1, 3, and 5 years for 1, 3, and 5 years.
Prognosis-predicting nomograms and inner validation for 1, 3, and 5 years in the TCGA cohort. (A–B): Overall survival-predicting nomogram using tumor stage, radical resection, hepatitis B virus infection status, and APOL6 expression and inner validation for 1, 3, and 5 years. (C–D): Recurrence-free survival-predicting nomogram using tumor stage, radical resection, hepatitis B virus infection status, vascular invasion, and APOL3 and APOL4 expression and inner validation for 1, 3, and 5 years.
Co-expression, protein-chemical interaction networks and matrix
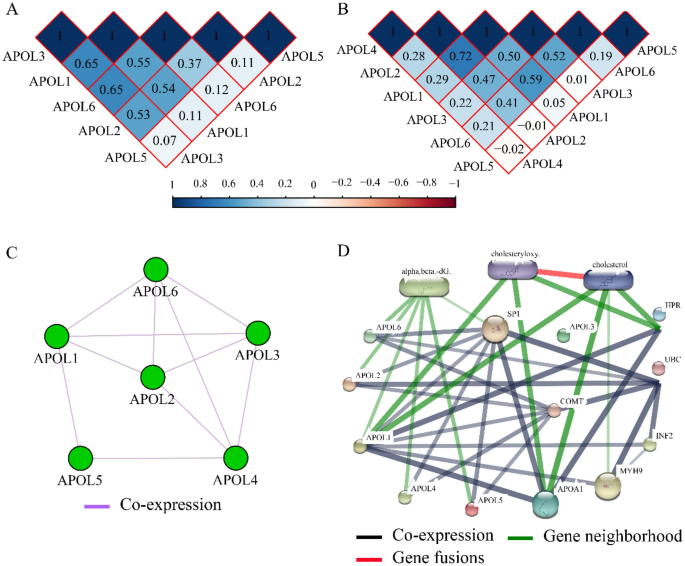
Co-expression matrixes of APOL isoforms indicated that all five APOL isoforms were positively correlated in the GSE14520 cohort. All other isoforms were positively correlated except for a negative correlation among APOL5, APOL2, and APOL4 in TCGA cohort (Fig. 11A,B). All these genes were co-expressed at the gene level (Fig. 11C). Protein-chemical interaction networks revealed that these proteins were also co-expressed at the protein level and were associated with α, β-dG, cholesteryloxy, and cholesterol in gene neighborhoods (Fig. 11D). In addition, visualized GO terms enriched by APOLs were indicated and involved in lipoprotein binding, extracellular region, lipoprotein metabolic process, et al. (Figure S5).
Co-expression matrix and protein-chemical compound interaction networks for APOL1-6. (A): Co-expression matrix for APOL1, 2, 3, 5, and 6 in the GSE14520 cohort. (B): Co-expression matrix for APOL1-6 in the TCGA cohort/ (C): Co-expression network for APOL1-6 genes. (D): Protein-chemical compound interaction networks for APOL1-6 and compounds.
Validation of diagnostic analysis and prognosis significance by Oncomine and HCCDB databases
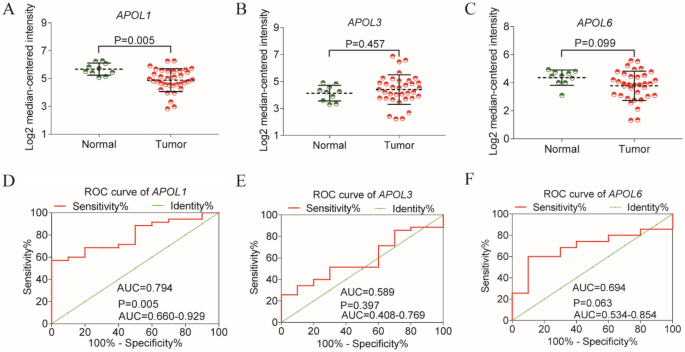
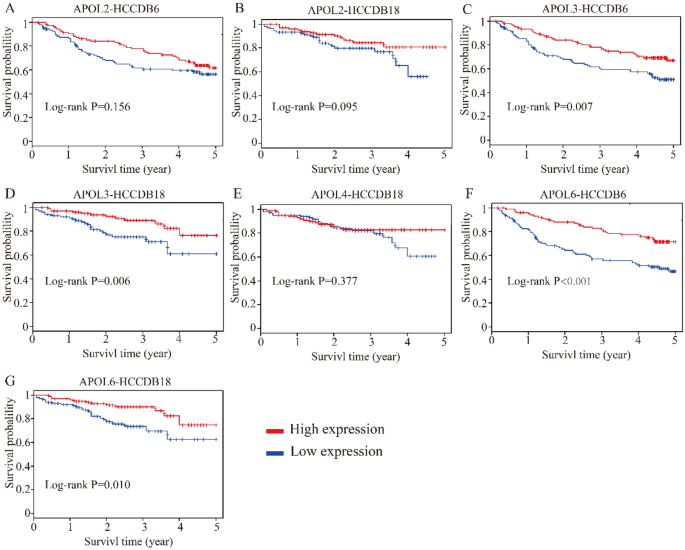
Differential expressions and diagnostic values of APOL1 and APOL3 were consistently validated in Oncomine database (AUC = 0.794, 0.589, Fig. 12A,C,D,F). Oncomine is a classic sample database in the field of cancer and can perform expression data, expression characteristics, gene set modules, etc. We applied it for validation of gene expression data. Strangely, APOL6 was showed weak diagnostic values in Oncomine database (AUC = 0.694, Fig. 12B,E). Furthermore, prognosis-related APOL isoforms were further validated in HCCDB database. HCCDB to serve as a one-stop online resource for exploring HCC gene expression with user-friendly interfaces, with integrating data from TCGA and GTEx. We applied it for validation of prognostic significance. APOL3 and APOL6, consistent in both TCGA and GSE14520 dataset, showed prognostic significance in two datasets of HCCDB as well (Log-rank P = 0.007, 0.006, Fig. 13C,D; Log-rank P < 0.001, = 0.010, Fig. 13F,G). However, APOL2 and APOL4, prognosis-related significance in TCGA or GSE14520 dataset, did not show prognostic significance (all P > 0.05, Fig. 13A,B,E).
Validation of survival analysis of APOL2, 3, 4 and 6 in HCCDB database. (A–B): Survival analysis of APOL2 in HCCDB6 and HCCDB18 datasets. (C–D): Survival analysis of APOL3 in HCCDB6 and HCCDB18 datasets. (E): Survival analysis of APOL4 in HCCDB18 dataset. (F-G): Survival analysis of APOL6 in HCCDB6 and HCCDB18 datasets.
Discussion
We explored prospective diagnostic capacity and prognostic significance and the mechanisms of APOL isoform involvement in HCC using GSE14520 and TCGA cohorts. We found that APOL1, 3, and 6 were differentially expressed in tumor and non-tumor tissues in both cohorts. In addition, both APOL1 and APOL6 had diagnostic abilities for HCC in the cohorts. In addition, diagnostic values of APOL1 and weak APOL6 were validated in Oncomine database. APOL3 and APOL6 showed prognostic significance for OS whereas APOL2, APOL3 and APOL6 showed prognostic significance for RFS in the GSE14520 cohort. However, APOL6 showed prognostic significance for OS whereas APOL3 and APOL4 showed prognostic significance for RFS in the TCGA cohort. These results indicated that APOL6 might be associated with OS and APOL3 might be associated with RFS of HCC patients. Moreover, APOL3 and APOL6, prognosis-related significance in both TCGA and GSE14520 datasets, were consistently validated their prognostic significance in HCCDB database; whereas APOL2 and APOL4, prognosis-related significance in TCGA or GSE14520 dataset, were not validated in HCCDB database. Prospective molecular mechanism exploration suggested that APOL3 and APOL6 might participate in HCC initiation and progression via the humoral immune response, regulation of the inflammatory response, cytokine-mediated signaling pathways, chemokine signaling pathways, fatty acid metabolism, and cell adhesion molecular cams. We constructed and used risk score models and nomograms to predict the survival of HCC patients using prognosis-related genes and clinical factors. We constructed co-expression interaction networks of APOL isoforms and visualized prospective GO-term networks. RT-PCR was performed on differentially expressed genes for APOL1, 3 and 6 and to validate the diagnostic ability of APOL1 and 6.
The six APOL isoforms are a cluster spanning a region of 619 kb on chromosome 2210. Lowry et al. found that APOL proteins are expressed in human placenta in a study identifying novel diagnostic biomarkers for pregnancy pathologies10. The liver is the main source of APOL1 proteins and has highly efficient secretory activity13. APOL1 has been widely studied for its expression and variants with many diseases, especially with kidney diseases. Coding variants within the APOL1 gene have a high frequency in recent populations of African ancestry and the highest odds ratio association with complicated renal diseases34, 35. APOL1 is a trypanolytic factor that confers resistance to Trypanosoma brucei brucei, containing Trypanosoma brucei rhodensience and Trypanosoma brucei gambsience parasites36, 37. Trypanosoma brucei rhodensience is found mainly in Eastern and Southern Africa while Trypanosoma brucei gambsience is found mainly in Western Africa38. APOL1 is suggested to function in natural selection due to the unique traits of these parasites in sub-Saharan Africa34. APOL1 variations increase the risk of kidney diseases in African Americans. Nonsynonymous variants coded by G1 and the coding region deletion G2 in APOL1 are sequence variants that have strong relationships to focal segmental glomerulosclerosis and hypertension-attributed end-stage kidney disease. After controlling for risk variants in APOL1, the association between kidney diseases and MYH9 sequence variants34.
Studies conducted for the Jackson Heart Study and Women’s Health Initiative indicated that a person with two risk variants may have a twofold risk for cardiovascular disease, even though these studies had no data on mortality and contained only 12 patients with myocardial infarction39. A prospective investigation conducted over two decades in older adults suggested that the APOL1 genotype was associated with albuminuria, peripheral atherosclerosis, risk of myocardial infarction and death40. Evidence indicates that APOL1 mRNA and protein are expressed in podocytes, renal tubule cells, and glomerular endothelial cells41, 42. APOL1 protein is also expressed in the blood, which may be the reason for its significance in pathology12. However, little is known about the association between APOL1 expression or genetic variants with malignancies. Our study demonstrated that APOL1 mRNA was not associated with HCC prognosis but showed a strong diagnostic ability for HCC.
APOL2 protein expression is markedly stimulated by interferon-γ in normal human bronchial epithelial cells while APOL2 mRNA is increased in normal human lung fibroblasts and smooth muscle cells16. Lowering expression of APOL2 with siRNA facilitates cytotoxicity induced by interferon-γ, with a significant drop in cell viability via MTT and CyQUANT NF cell proliferation assays and an increase in hypodiploid sub-G1 cell distribution in cell cycle assays16. Furthermore, depletion of APOL2 promotes membrane damage, DNA fragmentation and chromatin condensation induced by interferon-γ by Hoechst and propidium iodide-double staining, DNA laddering assays and transmission electron microscopy 16. These findings indicate a new function for APOL2: anti-apoptotic ability in human bronchial epithelial cells from cytotoxic effects by interferon-γ and maintaining airway epithelial layer integrity16. Tsuang et al. found that APOL1, 2 and 4 genes are located on chromosome 22q12.3–13.1 and upregulated in brains of schizophrenic patients18. They conducted a family-based association study using 130 single nucleotide polymorphisms (SNPs) in APOL1-6 family members in 112 African-American, 114 European-American, 109 Chinese and 42 Japanese families with schizophrenia. They concluded that seven SNPs in APOL1, 2 and 4 are associated with schizophrenia in these families18. Similar to APOL1, few reports are available about APOL2 and malignancies. Our study found APOL2 mRNA was associated with HCC recurrence in an HBV-related HCC cohort. In addition, APOL4 mRNA was associated with HCC recurrence in a TCGA cohort but not in a GSE14520 cohort. Due to the inconsistence of APOL2 and APOL4 in two cohorts, we did not perform GSEA for mechanical exploration. As the above literatures mentioned, literature reports of several SNP of them, anti-apoptotic and cell proliferation ability of APOL2, and our findings would be a direction of future study concerning APOL2 and APOL4 in HCC prognosis and mechanical pathways.
APOL3 was a risk locus in a family-based association analysis of 42 hereditary prostate cancer families17. In addition, APOL3 is differentially expressed in tumors and controls in oral squamous cell carcinoma. This information might be helpful for selecting possible biomarkers for oral squamous cell carcinoma43. However, APOL3 expression in HCC has not been reported. Our study found that APOL3 was associated with HCC prognosis and recurrence. In addition, mechanism exploration suggested that APOL3 involvement in HCC might be via the humoral immune response, regulation of the inflammatory response, cytokine-mediated signaling pathways, chemokine signaling pathways, fatty acid metabolism, and cell adhesion molecular cams. The above literature indicated rs2097465 and rs132656 located within the APOL3 were associated with prostate cancer initiation as well as differential APOL3 was involved in immune response in oral squamous cell carcinoma. This suggests further concentration on rs2097465 and rs132656 loci as well as immune response of APOL3 in HCC initiation and progression.
A SNP of APOL5, rs2076672, was identified by parallel independent component analysis linked to structural components in a European-American study with 18 schizophrenia patients and 33 healthy control individuals44. A new locus, rs2016586 of APOL5, has a suggestive association with childhood body mass index45. APOLs have documented associations with HDL, rather low or very low-density lipoproteins. Wu et al. found that APOL SNPs are not associated with low-density lipoprotein cholesterol levels in a physiogenomic analysis of statin-treated46. To date, APOL5 has not been associated with tumors. We found that APLO5 mRNA was not associated with HCC diagnosis and prognosis.
APOL6 was identified as a novel Bcl-2 homology 3-only protein in a mining-approach using public databases47. Overexpression of wild-type APOL6 induces mitochondria-mediated apoptosis in p53-null colorectal cancer cells, characterized by the release of cytochrome c and Smac/DIABLO from mitochondria and activation of caspase-947. Hu et al. showed that APOL6 is a downstream target of interferon-γ and upregulated by interferon-γ, which sensitizes atherosclerotic lesion-derived cells to Fas-induced apoptosis48. APOL6 expression partly co-localizes with activated caspase 3 in activated smooth muscle cells in atherosclerotic lesions and promotes reactive oxygen species generation, caspase activation, and apoptosis48. Furthermore, APOL6-induced cell apoptosis might be a potential therapeutic target for treating atherosclerosis and cardiovascular disease48. Apart from these studies, APLO6 has not been reported it with other tumors. Our study demonstrated that APOL6 was associated with HCC prognosis and might be a potential diagnostic biomarker for HCC. Mechanism exploration indicated involvement in HCC might be via the immune effector response, B-cell mediated immunity, cytokine-mediated signaling pathways, JAK-STAT signaling pathways, and cell adhesion molecular cams. The above studies indicated APOL6 mainly play its role via Fas-induced and mitochondria-mediated apoptosis, reactive oxygen species generation, and caspase activation. Taking present study and previous reports, further studies concerning APOL6 should be mainly focused on immune response and cell apoptosis aspects.
Apart from present study of APOLs in HCC diagnosis and prognosis, Xiaofeng Wang et al. reported HSP90α49, exosomal hnRNPH150, circulating tumor cells51 and Glypican-352, etc. in HCC diagnosis. Wang et al. reported cirRNA cirRHOT153, cirPRKCI54, circulating tumor cells51, GALAD model55. Even though many attempts on HCC early diagnosis and prognostic surveillance, mostly used for early diagnosis still AFP, and (or) AFP-L3, PIVKA-II56 and surveillance using abdominal ultrasound every 6 months57.
Although we report associations between APOL isoform expression and HCC patients, our study had some limitations. First, other cohorts are needed to validate the significance of APOL isoforms with HCC patients, especially the diagnosis- and prognosis-related genes. Second, prognosis-related genes need further in vivo and in vitro functional trials, especially focusing on immune response, cell apoptosis-related pathways, caspase cascades, cytokine-medicated pathways, to explore their concrete mechanisms of involvement in HCC. Third, potential target drugs need to be explored for APOL targets for future HCC treatment of medical community. Then, both diagnostic and prognostic biomarkers should be further validated in more medical centers. Then, a combination of biomarkers with AFP (or) AFP-L3, PIVKA-II and abdominal ultrasound for early diagnosis and surveillance is novel clue for future direction.
Conclusion
This study explored prospective diagnostic capacity and prognostic significance as well as mechanisms of APOL isoforms involvement in HCC. We found that APOL1, 3, and 6 were differentially expressed in tumor and non-tumor tissues. Both APOL1 and APOL6 had diagnostic ability for HCC in TCGA and GSE14520 cohorts. These findings were validated by Oncomine database. Prognostic significance analysis indicated that APOL6 was associated with OS and APOL3 was associated with RFS of HCC patients in both TCGA and GSE14520 datasets. And their prognostic significance was further consistently validated in HCCDB database as well. Prospective molecular mechanism exploration suggested that APOL3 and APOL6 were associated with HCC prognosis via the immune response, inflammatory response, cytokine-mediated signaling pathways, and fatty acid metabolism. We constructed and used risk score models and nomograms to predict the survival of HCC patients using prognosis-related genes and clinical factors.
Data availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- HCC:
-
Hepatocellular carcinoma
- APOL:
-
Apolipoprotein
- HBV:
-
Hepatitis B virus
- AUC:
-
Area under curve
- TCGA:
-
The cancer genome atlas
- BCLC:
-
Barcelona clinic liver cancer
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- GSEA:
-
Gene set enrichment analysis
- BP:
-
Biological process
- CC:
-
Cellular component
- MF:
-
Molecular function
- GO:
-
Gene ontology
- OS:
-
Overall survival
- MST:
-
Median survival time
- CI:
-
Confidence interval
- HR:
-
Hazard ratio
- ROC:
-
Receiver operating characteristic
- GGI:
-
Gene-gene interaction
- DAVID:
-
Database for annotation, visualization and integrated discovery
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68(6), 394–424 (2018).
Beasley, R. P. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer 61(10), 1942–1956 (1988).
Yu, M. W. & Chen, C. J. Hepatitis B and C viruses in the development of hepatocellular carcinoma. Critic. Rev. Oncol. Hematol. 17(2), 71–91 (1994).
Lai, Y. L. et al. The contribution of matrix metalloproteinase-1 genotypes to hepatocellular carcinoma susceptibility in Taiwan. Cancer Genom. Proteomics 14(2), 119–125 (2017).
Forner, A., Llovet, J. M. & Bruix, J. Hepatocellular carcinoma. Lancet (London, England). 379(9822), 1245–1255 (2012).
Poon, R. T. P. Prevention of recurrence after resection of hepatocellular carcinoma: A daunting challenge. Hepatology. 54(3), 757–759 (2011).
Tralhão, J. G., Dagher, I., Lino, T., Roudié, J. & Franco, D. Treatment of tumour recurrence after resection of hepatocellular carcinoma Analysis of 97 consecutive patients. Eur. J. Surg. Oncol. 33(6), 746–751 (2007).
Liu, X., Gao, H., Zhang, J. & Xue, D. FAM83D is associated with gender, AJCC stage, overall survival and disease-free survival in hepatocellular carcinoma. Biosci. Rep. 39(5), BSR20181640 (2019).
Huang, W. et al. Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology 61(6), 1920–1933 (2015).
Page, N. M., Butlin, D. J., Lomthaisong, K. & Lowry, P. J. The human apolipoprotein L gene cluster: identification, classification, and sites of distribution. Genomics 74(1), 71–78 (2001).
Li, X. et al. Status of lipid and lipoprotein in female breast cancer patients at initial diagnosis and during chemotherapy. Lipids Health Dis. 17(1), 91 (2018).
Duchateau, P. N. et al. Apolipoprotein L, a new human high density lipoprotein apolipoprotein expressed by the pancreas. Identification, cloning, characterization, and plasma distribution of apolipoprotein L. J. Biol. Chem. 272(41), 25576 (1997).
Cheng, D. et al. Biogenesis and cytotoxicity of APOL1 renal risk variant proteins in hepatocytes and hepatoma cells. J. Lipid Res. 56(8), 1583–1593 (2015).
Timmins, J. M. et al. Targeted inactivation of hepatic Abca1 causes profound hypoalphalipoproteinemia and kidney hypercatabolism of apoA-I. J. Clin. Investig. 115(5), 1333–1342 (2005).
Wan, G. et al. Apolipoprotein l1, a novel BH3-only lipid binding protein, induces autophagic cell death. J. Biol. Chem. 4(8), 1079–1082 (2008).
Liao, W. et al. A novel anti-apoptotic role for apolipoprotein L2 in IFN-γ-induced cytotoxicity in human bronchial epithelial cells. J. Cell Physiol. 226(2), 397–406 (2011).
Johanneson, B. et al. Family-based association analysis of 42 hereditary prostate cancer families identifies the apolipoprotein L3 region on chromosome 22q12 as a risk locus. Hum. Mol. Genet. 19(19), 3852–3862 (2010).
Takahashi, S. et al. Association of SNPs and haplotypes in APOL1, 2 and 4 with schizophrenia. Schizophr. Res. 104(1–3), 153–164 (2008).
Liu, Z., Lu, H., Jiang, Z., Pastuszyn, A. & Hu, C. A. Apolipoprotein L6, a novel proapoptotic Bcl-2 homology 3–only protein, induces mitochondria-mediated apoptosis in cancer cells11howard hughes medical institute research aids to university of New Mexico cancer research and treatment center. Am. Cancer Mol. Cancer Res. 3(1), 21–31 (2005).
Page, N. M., Butlin, D. J., Lomthaisong, K. & Lowry, P. J. The human apolipoprotein l gene cluster: identification, classification, and sites of distribution ☆. Genomics 74(1), 71–78 (2001).
Thomas, E. A. & Sutcliffe, J. G. The neurobiology of apolipoproteins in psychiatric disorders. Mol. Neurobiol. 26(2–3), 369–388 (2002).
Roessler, S. et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 70(24), 10202–10212 (2010).
Roessler, S. et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology 142(4), 957–66.e12 (2012).
Lossos, I. S. et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. New Engl. J. Med. 350(18), 1828–1837 (2004).
Alizadeh, A. A. et al. Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Blood 118(5), 1350–1358 (2011).
Wang, X. et al. Diagnostic and prognostic value of mRNA expression of phospholipase C β family genes in hepatitis B virus-associated hepatocellular carcinoma. Oncol. Reps. 41(5), 2855–2875 (2019).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11), 2498 (2003).
Montojo, J. et al. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics 26(22), 2927 (2010).
Szklarczyk, D. et al. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucl. Acids Res. 44(D1), D380–D384 (2016).
Maere, S., Heymans, K. & Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21(16), 3448–3449 (2005).
Wurmbach, E. et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 45(4), 938–947 (2007).
Lian, Q. et al. HCCDB: A database of hepatocellular carcinoma expression atlas. Genomics Proteom. Bioinform. 16(4), 269–275 (2018).
Liao, X. et al. Distinct diagnostic and prognostic values of minichromosome maintenance gene expression in patients with hepatocellular carcinoma. J. Cancer 9(13), 2357–2373 (2018).
Genovese, G. et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science (New York, NY). 329(5993), 841–845 (2010).
Freedman, B. I. et al. The apolipoprotein L1 (APOL1) gene and nondiabetic nephropathy in African Americans. J. Am. Soc. Nephrol. 21(9), 1422–1426 (2010).
Perez-Morga, D. et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science (New York, NY). 309(5733), 469–472 (2005).
Vanhamme, L. et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature 422(6927), 83–87 (2003).
Simarro, P. P., Jannin, J. & Cattand, P. Eliminating human African trypanosomiasis: Where do we stand and what comes next?. PLoS Med. 5(2), e55 (2008).
Ito, K. et al. Increased burden of cardiovascular disease in carriers of APOL1 genetic variants. Circ. Res. 114(5), 845–850 (2014).
Mukamal, K. J. et al. APOL1 genotype, kidney and cardiovascular disease, and death in older adults. Arterioscler. Thromb. Vasc. Biol. 36(2), 398–403 (2016).
Ma, L. et al. Localization of APOL1 protein and mRNA in the human kidney: Nondiseased tissue, primary cells, and immortalized cell lines. J. Am. Soc. Nephrol. 26(2), 339–348 (2015).
Madhavan, S. M. et al. APOL1 localization in normal kidney and nondiabetic kidney disease. J. Am. Soc. Nephrol. 22(11), 2119–2128 (2011).
Suhr, M. L. et al. Gene expression profile of oral squamous cell carcinomas from Sri Lankan betel quid users. Oncol. Rep. 18(5), 1061–1075 (2007).
Jagannathan, K. et al. Genetic associations of brain structural networks in schizophrenia: a preliminary study. Biol. Psychiatry 68(7), 657–666 (2010).
Namjou, B. et al. EMR-linked GWAS study: Investigation of variation landscape of loci for body mass index in children. Front. Genet. 4, 268 (2013).
Ruaño, G. et al. Physiogenomic analysis of statin-treated patients: Domain-specific counter effects within the ACACB gene on low-density lipoprotein cholesterol?. Pharmacogenomics 11(7), 959–971 (2010).
Liu, Z., Lu, H., Jiang, Z., Pastuszyn, A. & Hu, C. A. Apolipoprotein l6, a novel proapoptotic Bcl-2 homology 3-only protein, induces mitochondria-mediated apoptosis in cancer cells. Mol. Cancer Res. 3(1), 21–31 (2005).
Zhaorigetu, S., Yang, Z., Toma, I., McCaffrey, T. A. & Hu, C. A. Apolipoprotein L6, induced in atherosclerotic lesions, promotes apoptosis and blocks Beclin 1-dependent autophagy in atherosclerotic cells. J. Biol. Chem. 286(31), 27389–27398 (2011).
Wang, X. et al. The regulatory mechanism of Hsp90alpha secretion and its function in tumor malignancy. Proc. Natl. Acad. Sci. USA 106(50), 21288–21293 (2009).
Xu, H., Dong, X., Chen, Y. & Wang, X. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin. Chem. Lab. Med. 56(3), 479–484 (2018).
Ahn, J. C. et al. Detection of circulating tumor cells and their implications as a biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatology. 73(1), 422–436 (2021).
Zhou, F., Shang, W., Yu, X. & Tian, J. Glypican-3: A promising biomarker for hepatocellular carcinoma diagnosis and treatment. Med. Res. Rev. 38(2), 741–767 (2018).
Wang, L. et al. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol. Cancer 18(1), 119 (2019).
Qi, S. X. et al. Role and mechanism of circ-PRKCI in hepatocellular carcinoma. World J. Gastroenterol. 25(16), 1964–1974 (2019).
Yang, J. D. et al. GALAD score for hepatocellular carcinoma detection in comparison with liver ultrasound and proposal of GALADUS score. Cancer Epidemiol. Biomark. Prev. 28(3), 531–538 (2019).
Luo, P. et al. Current status and perspective biomarkers in AFP negative HCC: Towards screening for and diagnosing hepatocellular carcinoma at an earlier stage. Pathol. Oncol. Res. 26(2), 599–603 (2020).
Singal, A. G., Lampertico, P. & Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J. Hepatol. 72(2), 250–261 (2020).
Acknowledgements
This manuscript was supported by 2022 Henan Province Medical Science and Technology Joint Construction Project (LHGJ20220317), 2023 Provincial Natural Science Foundation Project of Henan Province (232300420038), 2023 Provincial Natural Science Foundation Youth Fund Project of Henan Province (232300420249) and The present study was supported by The Key Scientific Research Project Plan of Henan University (Grant No.20A320037) and 2022 Henan Province Key R&D and Promotion Special Support Project (The role of OSBP2 in the regulation of malignant phenotype of pancreatic cancer).
Author information
Authors and Affiliations
Contributions
Conceptulization: X.-K.W., Y.-X.G., M.W., R.-F.L., Data curation: X.-K.W., Y.-X.G., M.W., X.-D.Z., Formal analysis: X.-K.W., Y.-X.G., M.W., R.-F.L., Funding acquisition: X.-K.W., R.-F.L., Investigation: X.-K.W., K.L., M.W., S.H., Methodology: X.-K.W., X.-D.Z., K.L., Z.-Y.L., M.-S.W., Project administration: X.-K.W., R.-F.L., S.H., Resources: X.-K.W., Y.-X.G., Z.-Y.L., X.-D.Z., S.H., Software: X.-K.W., Y.-X.G., M.W., M.-S.W., X.-D.Z.ang, Supervision: R.-F.L., S.H., Validation: X.-K.W., Y.-X.G., M.W., K.L.s, Visualization: X.-K.W., X.-D.Z., M.-S.W., Original draft: X.-K.W., Y.-X.G., M.W., Reviw & editing: X.-K.W., S.H., R.-F.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, XK., Guo, YX., Wang, M. et al. Identification and validation of candidate clinical signatures of apolipoprotein L isoforms in hepatocellular carcinoma. Sci Rep 13, 20969 (2023). https://doi.org/10.1038/s41598-023-48366-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-48366-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.