Abstract
Calcific aortic valve stenosis (CAVS) is associated with an increased risk of atrial fibrillation (AF) in observational studies, but whether these associations are causal has not been determined. This study aimed to explore the potential causal relationship between CAVS and AF via Mendelian randomization (MR). Genetic variants from the genome-wide association study (GWAS) summary data of the European population for CAVS were used to investigate the association with AF. The inverse variance weighted (IVW) approach was used to obtain the primary causal inference, and several sensitivity analysis approaches, such as the MR‒Egger and weighted median (WM), were performed to assess the robustness of the results. A total of nineteen valid and independent genetic SNPs associated with CAVS were obtained from the GWAS database. Genetically predicted CAVS (OR: 1.105; 95% CI: 1.072–1.139; p = 8.60E−11) was associated with an increased risk of AF. Similar results were discovered in the sensitivity analyses by using MR Egger and weighted median approaches. An MR design was used to reduce confounding variables and the potential for reverse causality bias. The results provide genetic evidence that CAVS considerably increased the risk of AF.
Similar content being viewed by others
Considering the worldwide aging trend and the development of wearable and smartphone-based devices for the monitoring of heart rate and rhythm, the prevalence of atrial fibrillation (AF) is growing globally1. According to the findings from the Framingham Heart Study (FHS), the prevalence of atrial fibrillation has tripled in the last 50 years2. The Global Burden of Disease project estimated that the number of patients with AF was very large, with approximately 46.3 million individuals in 20163. This trend is predicted to continuously deteriorate in the future. For instance, the prevalence of AF in the European region in 2010 was about 9 million among individuals aged over 55 years old and is expected to reach 14 million by 20604,5, and at least 72 million individuals in Asian areas will be diagnosed with AF by 20506. Under these circumstances, targeted preventive measures need to be taken to prevent AF according to the risk factors for AF.
Several population-based studies have recently identified risk factors for AF, including left atrial (LA) enlargement, heart failure, valvular heart disease, hypertension, and aging7,8,9,10,11. Additionally, observational studies have revealed a potential relationship between AF and risk in patients at high risk for aortic stenosis (AS) 12,13,14,15,16. However, observational studies still have limitations due to confounders and reverse causality, making it challenging to assess the causal relationship between AS and AF.
Mendelian randomization (MR) is a method of using naturally occurring random allocation of genetic variation as a genetic instrument, with results similar to randomized controlled trials (RCTs) and less affected by confounding factors and reverse causality17. This study aimed to evaluate the causal relationship between CAVS and the risk of AF using MR methods.
Methods
Study design
MR analysis is based on three key assumptions: (1) exposure variables and genetic instrumental variation are consistently associated. (2) the outcome variables should not be associated with genetic instrumental variation. (3) only via the exposure variable can genetic instrumental variation significantly affect the outcome, but not in any other way.
An overview of the study design is shown in Fig. 1. We performed MR analysis using summary statistics from genome-wide association studies (GWAS). Genetic variants associated with CAVS were identified as instrumental variation, CAVS was used as the exposure and AF as the outcome, and the exposure and outcome datasets were harmonized to perform MR analyses, examining MR analysis assumptions and sensitivity analyses.
Selection of instrumental SNPs
To select SNPs associated with CAVS as genetic instrumental variables for MR analysis, appropriate SNPs were selected based on a threshold of genome-wide significance (p < 5×10−8) and Linkage disequilibrium (r2 < 0.001). When harmonizing exposure and outcome data, palindromic SNPs that could lead to potential strand ambiguity were excluded. The F statistic was calculated to evaluate the strength of the instrument. F ≥ 10 indicates the association was not driven by weak instrument bias. This study included validated and independent genetic variants associated with CAVS as instrumental variables (see Supplementary Table S1 online).
Data sources of calcific aortic valve stenosis and atrial fibrillation
The CAVS GWAS data from the FinnGen Project Database18, totaled 9153 cases and 368124 controls of European ancestry19. The FinnGen Project Database is a collaborative project led by the Institute for Molecular Medicine Finland (FIMM), the national biomedical infrastructure in Finland, which integrates clinical data, genetic data, and biological samples from all regions of Finland to create a large research resource.
Pooled data related to AF were obtained from a large GWAS meta-analysis study that included six contributing studies, namely, The Nord‐Trøndelag Health Study (HUNT), deCODE, Michigan Genomics Initiative (MGI), DiscovEHR, UK Biobank, and the Arial Fibrillation Genomics (AFGen) consortium20. A total of 60,620 cases and 970,216 controls of European ancestry. AF was diagnosed using ICD‐9 or ICD‐10 codes. Participants in the control group had diagnoses without any ICD-9 or ICD-10 codes. This study tested the relationship between AF and 34,740,186 genetic variations, identified 142 independent risk variants at 111 loci, and prioritized 151 functional candidate genes that potentially have an association with AF. The study design was described in detail in other publications20. The exposure and outcome information of GWAS summary data can be found in Supplementary Table S2 online.
Statistical analysis
Two-sample Mendelian randomization (MR) was employed to assess the association between CAVS and AF. Three distinct two-sample MR approaches, including inverse variance weighted (IVW), weighted median (WM), and MR-egger, were utilized to evaluate the relationship between exposures (CAVS) and outcomes (AF). The IVW method, assuming no average pleiotropic effect, was considered the primary MR method for assessing potential causal effects21. Cochran’s Q test was initially performed to assess heterogeneity in individual causal effects, with p < 0.05 indicating the presence of heterogeneity. Subsequently, random-effect IVW was performed if heterogeneity was detected. Otherwise, the fixed-effect IVW method was adopted. The MR Egger approach was used to identify and assess directional pleiotropy effects in MR analyses, providing a sensitivity analysis for result robustness22. The WM approach required at least 50% of the weights to come from valid instrumental variables, enabling consistent estimates of causal effects and offering sensitivity analysis for the robustness of the results 23.
Sensitivity analyses were conducted to validate the causal estimates and account for potential confounding factors. Horizontal pleiotropy was assessed using the MR-Egger intercept term, where deviations from zero indicate the presence of directional pleiotropy. In the presence of horizontal pleiotropy, the slope of the MR-Egger regression provides unbiased MR estimations22. Furthermore, the stability of the causal estimates was evaluated through a leave-one-out analysis, where each SNP was sequentially excluded to assess the potential impact of a single SNP on the overall results.
Statistical analyses were conducted in R software (version 4.1.0, R Foundation for Statistical Computing) utilizing the TwoSampleMR (version 0.5.6) and Mendelian Randomization (version 0.5.1) packages. Multiple comparisons were performed using the Bonferroni correction (corrected p = 0.05/X/Y, where X denotes the number of exposures and Y denotes the number of outcomes). The observation of two-sided p < 0.025 was considered significant evidence for a causal effect, and p < 0.05 indicated significance.
Results
Calcific aortic valve stenosis and atrial fibrillation
Under the prior presumed standards, nineteen genetic variants associated with CAVS were obtained to explore the effect of CAVS genetic instrumental variables on AF. The characteristics of the genetic variants associated with CAVS and AF can be found in Supplementary Table S1 online. All of the F-statistics, which serve as an indicator of the instruments’ strength, exceed the conventional threshold of 10, indicating that the association was not driven by weak instrument bias.
As shown in Table 1, Cochran’s Q test for CAVS with AF revealed no evidence of heterogeneity (p > 0.05), and the fixed-effect IVW model demonstrated that genetically predicted CAVS (1.105; 95% CI: 1.072–1.139; p = 8.60E−11) is associated with an increased risk of AF. Similar results were discovered for both exposures in the sensitivity analyses by the MR Egger and WM approach. Moreover, the MR‒Egger regression intercept term showed no evidence of a directed pleiotropy effect between genetic variants (intercept, − 0.008; p = 0.142).
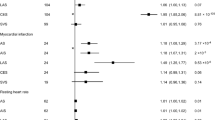
Figure 2 shows the forest plots that represent the estimated single SNP effects of CAVS on AF. Eight SNPs (rs1706003, rs78012551, rs17550940, rs118039278, rs3901734, rs1116262, rs11166276, rs12929673, and rs1800797) among the selected nineteen SNPs in CAVS were significant for the estimated single effect on AF risk, whereas the other SNPs, e.g., rs143466522, rs4129225, rs76665052, etc., were not significant for the risk of AF.
The results of the leave-one-out analysis are illustrated in Fig. 3. In the leave-one-out analysis, no substantial change was observed in the genetically predicted risk estimates of CAVS with the risk of AF after excluding one SNP each time, indicating that the association of CAVS with AF was not driven by a single SNP, and the findings were stable.
Sensitivity analysis
In addition, we conducted sensitivity analyses to assess the robustness of our findings by using two separate CAVS GWAS datasets. The first dataset included 13,765 cases and 640,102 controls of European ancestry24, while the second dataset involved 14,451 cases and 398,544 controls from the Million Veteran Program25(see Supplementary Table S2 online). The characterization of genetic variants associated with CAVS and AF in both groups is shown in Supplementary Table S1 online.
In the sensitivity analysis of CAVS-related SNPs and AF in both groups, we obtained consistent results with before (OR: 1.143; 95% CI: 1.107–1.181; p = 5.89E−16; OR: 1.194; 95% CI: 1.126–1.266; p = 3.49E-09), and Cochran's Q-test did not find heterogeneity evidence (p > 0. 05). Moreover, the MR-Egger regression intercept term showed no evidence of pleiotropic effect (intercept, − 0.908; p = 0.139; intercept, − 0.0128; p = 0.213) (see Supplementary Table S3 online).
The forest plots of the single SNP effect estimates of CAVS on AF for the two groups are shown in Supplementary Fig. S1 online. The leave-one-out analysis of genetic variants of CAVS on AF for the two groups is shown in Supplementary Fig. S2 online.
Discussion
Using two-sample Mendelian randomization, our data provided genetic evidence of a potential causal effect of CAVS on AF.
Our study shows that CAVS was associated with an increased risk of AF, which is consistent with previous observational studies. For instance, a population-based cohort study demonstrated a significant association between severe aortic valve stenosis and the risk of AF26. Similarly, a prospective study found that about one-third of patients without a history of AF who underwent Transcatheter Aortic Valve Implantation (TAVI) developed new-onset AF12. Furthermore, several other studies have reported that valvular heart disease increases the risk of AF by 1.8-fold and 3.4-fold in males and females, respectively27. The prevalence of AF was 9.1% in patients with mild to moderate AS and 33.7% in patients with severe stenosis28,29.
Mechanistically, there are several potential explanations. The accumulation of inflammatory cells in aortic valves is associated with remodeling and fibrosis 24,30, Alleles that increase the risk of AS are linked with elevated expression of IL6 and IL6-AS1 in fibroblasts31. On the one hand, fibroblasts, as the main cellular effectors of atrial fibrosis, contribute to AF by promoting fibrosis through the recruitment of inflammatory cells and excessive secretion of extracellular matrix (ECM) proteins32. Meanwhile, macrophages regulate fibrosis by producing proinflammatory cytokines (IL-6, ROS, and TNF-α) which can also lead to AF33. On the other hand, left atrial dilatation is believed to result from the stimulatory effect of IL-6 on matrix-metalloproteinase-2 (MMP2), a protease associated with atrial remodeling. Ultimately, IL-6 induces AF by inducing atrial remodeling 34. In addition, obesity, dyslipidemia, and calcification may contribute to aortic stenosis and be involved in the development of atrial fibrillation11,30,35,36. Pathophysiological studies have shown that AS can cause left ventricular outflow tract obstruction, resulting in left ventricular (LV) hypertrophy, and increases the left end-diastolic filling pressure resulting in left atrial (LA) dilatation, a key trigger of atrial fibrillation, which has been associated with the development of atrial fibrillation37,38,39,40,41.
This study provides genetic evidence that CAVS is associated with an increased risk of atrial fibrillation by using an MR design that minimizes confounding and reverse causation bias. We confirmed that the results were sufficiently robust by Sensitivity and multiplicity MR methods such as IVW, MR-Egger, and WM. Furthermore, there was no evidence of a directed pleiotropy effect between the genetic variants examined. Heterogeneity analysis revealed no significant variations among the SNPs studied. Additionally, leave-one-out analysis demonstrated that the overall effect was not driven by a single SNP, indicating the stability of our results.
Several limitations should be considered when interpreting our findings. Firstly, the use of summary-level statistics from published data prevented us from conducting nonlinear causal analyses. Secondly, the estimates of exposure and outcome were derived from aortic stenosis and atrial fibrillation cases obtained from linked hospital electronic health records, and the presence of comorbidities, disease progression, or severity was not evaluated. Lastly, the current study relies on genetic data collected from a predominantly European population, which, despite greater genetic homogeneity, limits the applicability of the current findings to other population groups.
Conclusion
Our data provided genetic evidence supporting a possible causal relationship between CAVS and AF. Utilizing a two-sample MR design that minimizes confounding and reverse causality bias, we demonstrate an association between CAVS and an increased risk of AF.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request
References
Mizuno, A. et al. Wearable devices to monitor and reduce the risk of cardiovascular disease: Evidence and opportunities. Annu. Rev. Med. 72, 459–471 (2021).
Schnabel, R. B. et al. 50 year trends in atrial fibrillation prevalence, incidence, risk factors, and mortality in the Framingham Heart Study: A cohort study. Lancet 386(9989), 154–62 (2015).
Benjamin, E. J. et al. Heart disease and stroke statistics-2019 update: A report from the American Heart Association. Circulation 139(10), e56–e528 (2019).
Krijthe, B. P. et al. Projections on the number of individuals with atrial fibrillation in the European Union, from 2000 to 2060. Eur. Heart J. 34(35), 2746–51 (2013).
Di Carlo, A. et al. Prevalence of atrial fibrillation in the Italian elderly population and projections from 2020 to 2060 for Italy and the European Union: The FAI Project. Europace 21(10), 1468–1475 (2019).
Chiang, C. E. et al. Stroke prevention in atrial fibrillation: An Asian perspective. Thromb. Haemost. 111(5), 789–97 (2014).
Baman, J. R. et al. Atrial fibrillation. JAMA 325(21), 2218–2218 (2021).
Chung, M. K. et al. Lifestyle and risk factor modification for reduction of atrial fibrillation: A scientific statement from the American Heart Association. Circulation 141(16), e750–e772 (2020).
Kornej, J. et al. Epidemiology of atrial fibrillation in the 21st century: Novel methods and new insights. Circ. Res. 127(1), 4–20 (2020).
Andrade, J. et al. The clinical profile and pathophysiology of atrial fibrillation: Relationships among clinical features, epidemiology, and mechanisms. Circ. Res. 114(9), 1453–68 (2014).
Sagris, M. et al. Atrial fibrillation: Pathogenesis, predisposing factors, and genetics. Int. J. Mol. Sci. 23(1), 6 (2021).
Amat-Santos, I. J. et al. Incidence, predictive factors, and prognostic value of new-onset atrial fibrillation following transcatheter aortic valve implantation. J. Am. Coll. Cardiol. 59(2), 178–88 (2012).
Nombela-Franco, L. et al. Timing, predictive factors, and prognostic value of cerebrovascular events in a large cohort of patients undergoing transcatheter aortic valve implantation. Circulation 126(25), 3041–53 (2012).
Stortecky, S. et al. Atrial fibrillation and aortic stenosis: Impact on clinical outcomes among patients undergoing transcatheter aortic valve implantation. Circ. Cardiovasc. Interv. 6(1), 77–84 (2013).
Chopard, R. et al. Baseline characteristics and prognostic implications of pre-existing and new-onset atrial fibrillation after transcatheter aortic valve implantation: Results from the FRANCE-2 registry. JACC Cardiovasc. Interv. 8(10), 1346–1355 (2015).
Mentias, A. et al. Impact of pre-existing and new-onset atrial fibrillation on outcomes after transcatheter aortic valve replacement. JACC Cardiovasc. Interv. 12(21), 2119–2129 (2019).
Lawlor, D. A. et al. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat. Med. 27(8), 1133–63 (2008).
Finnish Genetics of Health and Disease https://r9.finngen.fi/pheno/I9_CAVS_OPERATED.
Kurki M., et al. FinnGen: Unique genetic insights from combining isolated population and national health register data. medRxiv [Preprint]. (2022). https://doi.org/10.1101/2022.03.03.22271360.
Nielsen, J. B. et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 50(9), 1234–1239 (2018).
Burgess, S. et al. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37(7), 658–65 (2013).
Bowden, J. et al. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44(2), 512–25 (2015).
Bowden, J. et al. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40(4), 304–14 (2016).
Yu Chen, H. et al. Dyslipidemia, inflammation, calcification, and adiposity in aortic stenosis: A genome-wide study. Eur. Heart J. 44(21), 1927–1939 (2023).
Small, A. M. et al. Multiancestry genome-wide association study of aortic stenosis identifies multiple novel loci in the million veteran program. Circulation 147(12), 942–955 (2023).
Benjamin, E. J. et al. independent risk factors for atrial fibrillation in a population-based cohort: The Framingham Heart Study. JAMA 271(11), 840–844 (1994).
Kannel, W. B. et al. Prevalence, incidence, prognosis, and predisposing conditions for atrial fibrillation: Population-based estimates. Am. J. Cardiol. 82(7), 2N-9N (1998).
Greve, A. M. et al. Prognostic importance of atrial fibrillation in asymptomatic aortic stenosis: The Simvastatin and Ezetimibe in Aortic Stenosis study. Int. J. Cardiol. 166(1), 72–76 (2013).
Stortecky, S. et al. Atrial fibrillation and aortic stenosis: Impact on clinical outcomes among patients undergoing transcatheter aortic valve implantation. Circul. Cardiovasc. Interv. 6(1), 77–84 (2013).
Coté, N. et al. Inflammation is associated with the remodeling of calcific aortic valve disease. Inflammation 36(3), 573–581 (2013).
GTEx Consortium et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348(6235), 648–660 (2015).
Pellman, J. et al. Myocyte-fibroblast communication in cardiac fibrosis and arrhythmias: Mechanisms and model systems. J. Mol. Cell. Cardiol. 94, 22–31 (2016).
Kim, P. et al. Mechanoregulation of myofibroblast fate and cardiac fibrosis. Adv. Biosyst. 2(1), 1700172 (2018).
Marcus, G. M. et al. Interleukin-6 and atrial fibrillation in patients with coronary artery disease: Data from the Heart and Soul Study. Am. Heart J. 155(2), 303–309 (2008).
Larsson, S. C. et al. Overall and abdominal obesity and incident aortic valve stenosis: Two prospective cohort studies. Eur. Heart J. 38(28), 2192–2197 (2017).
Nalliah, C. J. et al. The role of obesity in atrial fibrillation. Eur. Heart J. 37(20), 1565–1572 (2015).
Seko, Y. et al. Association between atrial fibrillation, atrial enlargement, and left ventricular geometric remodeling. Sci. Rep. 8(1), 6366 (2018).
Tsang, T. S. et al. Left atrial volume: Important risk marker of incident atrial fibrillation in 1655 older men and women. Mayo Clin. Proc. 76(5), 467–75 (2001).
Wijffels, M. C. et al. Atrial fibrillation begets atrial fibrillation. A study in awake chronically instrumented goats. Circulation 92(7), 1954–68 (1995).
Sridhar, A. et al. Chronic heart failure and the substrate for atrial fibrillation. Cardiovasc. Res. 84(2), 227–36 (2009).
Sanders, P. et al. Electrical remodeling of the atria in congestive heart failure: Electrophysiological and electroanatomic mapping in humans. Circulation 108(12), 1461–8 (2003).
Author information
Authors and Affiliations
Contributions
C.C., L.C. and X.B.S. wrote the main manuscript text and S.P.L. conducted the MR analyses and prepared the graphs and tables. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chai, C., Li, S., Chen, L. et al. Causal association of calcific aortic valve stenosis and atrial fibrillation: a Mendelian randomization study. Sci Rep 13, 20284 (2023). https://doi.org/10.1038/s41598-023-47770-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-47770-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.