Abstract
Taste perception is crucial and impairments, which can be linked to pathologies, can lead to eating disorders. It is triggered by taste compounds stimulating receptors located on the tongue. However, the tongue is covered by a film containing saliva and microorganisms suspected to modulate the taste receptor environment. The present study aimed to elucidate the links between taste sensitivity (sweetness, sourness, bitterness, saltiness, umami) and the salivary as well as the tongue microbiota using shotgun metagenomics. 109 bacterial species were correlated with at least one taste. Interestingly, when a species was correlated with at least two tastes, the correlations were unidirectional, indicating a putative global implication. Some Streptococcus, SR1 and Rickenellaceae species correlated with five tastes. When comparing both ecosystems, saliva appears to be a better taste predictor than tongue. This work shows the implication of the oral microbiota in taste and exhibits specificities depending on the ecosystem considered.
Similar content being viewed by others
Introduction
As taste plays a key role in the perception of food and consequently influences food choice, it is associated with the avoidance of potentially poisonous molecules, which usually taste bitter, or the attraction towards energy-rich foods, for example, those tasting sweet. Therefore, impairments in this sensation, which can be linked to pathologies, can lead to eating disorders and malnutrition1. Taste perception is the sensation elicited by taste molecules when they stimulate the tongue's taste receptors, and it is admitted that five basic tastes exist, sweet, salty, sour, bitter and umami. Taste perception varies strongly between individuals2, 3, and the originating factors of this variability are incompletely understood. If polymorphisms in genes involved in taste perception contribute to this variability4, other factors linked to the oral environment, such as saliva, could also contribute to the modulation of this perception5, 6.
Anatomically, the tongue is a muscle covered by a specialized mucosa with irregular topography. This structure contains numerous depressions and constitutes an ecological site favourable to the accumulation of saliva, food and cellular debris and microorganisms7. Therefore, the lingual mucosal surface is covered by a biological film comprising desquamating cells, resting saliva and microorganisms; some studies have established relationships between the composition of this film and taste perception8, 9. The main hypotheses explaining these relationships are that this film could act as a physical barrier that limits the access of high molecular weight taste molecules, such as bitter compounds, to taste receptors10 or that bacterial metabolism could modulate taste perception. This last phenomenon includes the use of sugars and amino acids as substrates and the production of organic acids affecting sweet, umami and sour sensations7, 11.
Recently, the implication of the tongue microbiota has gained interest, and several groups have tried to establish links between it and taste perception, aided by the increasing accessibility of sequencing techniques, such as 16S rRNA gene metabarcoding. It is difficult to determine the specific implication of the tongue microbiota in basic taste perception; some authors focused on the specific fat taste in obese subjects12, 13, and others on the global taste sensitivity elicited by the bitter PROP (6-n-propylthiouracil)14. Recently, two studies revealed contradictory findings regarding the relationships between the basic tastes and the oral microbiota15, 16; Cattaneo et al.15 found association between tongue dorsum microbiota and gustatory functions whereas Fluitman et al.16 did not. It can be concluded from these previous works that the relationships between the tongue microbiota and the different basic tastes are complicated to resolve. One possible explanation is the lack of precision of the 16S rRNA gene sequencing that provides information at the genus level when specific species or subspecies are involved. Another hypothesis is that the tongue dorsum is not the best site to assess. Indeed, whole saliva appears a more interesting fluid for analysis than particular oral sites, especially as it is easy to sample and has potential as a biomarker source17. In addition, the bacterial composition of saliva is well-characterized in comparison with other oral niches18. Some researchers have focused on salivary microbiota and taste perception19, 20 and observed differences in the ability to correctly identify different taste qualities depending on the abundance of Bacteroidota19 or with taste-preference gene polymorphisms20. Others observed association between taste compound intake21 or food allergy22 and salivary microbiota composition. However, only one study23 investigated both lingual and salivary microbiota and highlighted differences in the associations between taste and lingual and salivary microbiota according to the site. A specific positive association between bitterness and Bacteroidota was observed in the lingual film, whereas a negative association between Actinobacteriota and saltiness was observed when focusing on saliva. However, the targeted method used, qPCR of the phyla, did not provide information on the specific genera or species.
Thus, the objective of the present work was to elucidate whether sensitivity to the five primary tastes is linked to oral microbiota in human with a special focus on the microbiota of the tongue dorsum and whole resting saliva microbiota. Shotgun metagenomics, which allows identification of species was used to investigate these relationships in greater depth than previous studies performed using 16S rRNA gene sequencing.
Methods
Subject selection
One hundred subjects, 50 females and 50 males aged 20 to 64 (mean: 41 ± 14) years were recruited. The exclusion criteria were as follows: pregnant women; smokers, those with food allergies; those who had been taking medication longer than one month; those who underwent previous radiotherapy of the head or neck; those using antibiotics, undergoing dental treatment or using antiseptic mouthwash in the preceding month; and individuals with Sjögren syndrome, lupus, dermatomyositis, lichen planus, Behçet, oral mycosis, gingivitis, gingivostomatitis, recurrent aphthae, and inflammatory bowel diseases.
Taste sensitivity determination and biological sampling
Participants were required to visit the laboratory twice. They were instructed not to brush their teeth, eat or drink anything other than water at least 2 h before sample collection, which occurred between 9 and 11 a.m. During the first visit, a sample of unstimulated saliva was collected in a container on ice by passive drooling for 5 min. For participants who did not secrete a minimum of 2 ml, extra time was allowed until they reached this volume. After a short rest, tongue film was collected as follows: participants swallowed saliva, immediately stuck their tongue out as far as possible, and maintained this pose while the film was collected by scraping the tongue with a plastic stick from the root to the apex in a single scraping motion. The same experimenter performed all the sampling to ensure repeatability. The saliva and tongue film samples were frozen at 80 °C immediately and stored until the analyses were conducted. After a short rest, taste sensitivity for the five basic tastes, sweet (fructose), salty (sodium chloride), sour (citric acid), bitter (quinine hydrochloride), and umami (monosodium glutamate), was determined using the T@sty test™ (Patent WO2015/165880) following the procedure described in Martin and Neyraud24. Briefly, the T@sty test is presented in the form of several edible test sheets, each allowing for one given taste sensitivity measurement. Each test sheet consists of six triplets of detachable precut discs (wafer paper) that need to be placed on the first third of the tongue. The triplets all consist of one disc containing the taste substance and two neutral discs. The taste compounds, dissolved in deionized water, were printed on the surface of the discs using a food-grade inkjet printer. The position of the tasty disc varies randomly depending on the triplets and the test sheets. The surface concentration of the taste compound and the intensity of taste gradually increases from the first to the sixth triplets. Distilled water is printed in the same manner on the neutral discs to make them look the same as the tasty discs. For each test sheet, and thus for each taste, the subjects had to successively evaluate the six triplets, starting with the triplet containing the lowest concentration located at the top of the sheet and then moving downwards.
The measured scores vary from 1 to 6 depending on the last triplet where the subject detect the taste concentration. A high score signifies a high taste sensitivity. At the end of the visit, a T@sty test™ kit was given to the participants, who conducted the test at home before the second visit. The second visit occurred approximately 2 weeks after the first and the same procedures were conducted. Overall, 2 unstimulated saliva and tongue film samples and 3 repeat T@sty test™ data were obtained for all participants.
DNA extraction and shotgun sequencing
Saliva samples were thawed and centrifuged at 17,000 ×g for 10 min. DNA from both saliva pellets and lingual film samples was extracted following the publicly available MGP SOP 002 V1 protocol (https://mgps.eu/standard-operating-procedure/) inspired by the International Human Microbiome Standards (IHMS) guidelines and adapted for low-biomass samples. DNA was quantitated using Qubit Fluorometric Quantitation (ThermoFisher Scientific, Waltham, US) and qualified using DNA size profiling on a Fragment Analyser (Agilent Technologies, Santa Clara, US). A total of 422 ± 300 ng of high molecular weight DNA (> 10 kbp) was used to build the library. Shearing of DNA into fragments of approximately 150 bp was performed using an ultrasonicator (Covaris, Woburn, US), and DNA fragment library construction was performed using the Ion Plus Fragment Library and Ion Xpress Barcode Adapters Kits (ThermoFisher Scientific, Waltham, US). Purified and amplified DNA fragment libraries were sequenced using the Ion Proton Sequencer (ThermoFisher Scientific, Waltham, US), generating 22.1 million reads ± 4.3 of 150 bp (on average) per sample.
Microbial gene count table
Reads were cleaned using ALIENTRIMMER v0.4 to (i) remove resilient sequencing adapters and (ii) trim low-quality nucleotides at the 3’ side using a quality and length cutoff of 20 and 45 bp, respectively. Cleaned reads were subsequently filtered using a human reference sequence (human genome version: Homo_sapiens_chm13_t2t_v1_1) with an identity score threshold of 90%. Gene abundance profiling was performed using the 8.4 million gene catalogue of the human oral microbiome [https://doi.org/10.15454/WQ4UTV]. Filtered high-quality reads were mapped with an identity threshold of 95% to the 8.4 million-gene oral catalogue using Bowtie2 (v2.3.5) included in METEOR v3.2 software (https://forgemia.inra.fr/metagenopolis/meteor). A gene abundance profiling table was generated through a two-step procedure using METEOR v3.2. First, reads mapped to a unique gene in the catalogue were attributed to their corresponding genes. Second, reads that mapped with the same alignment score to multiple genes in the catalogue were attributed according to the ratio of their unique mapping counts. The gene abundance table was processed for rarefaction, normalization, and further analysis using the MetaOMineR (momr, v1.31) R package. Read counts were rarefied to decrease technical bias due to different sequencing depths and avoid any artefacts of sample size on low abundance genes. The gene abundance table was rarefied to 2 million reads per sample by random sampling of 2 million mapped reads without replacement. The resulting rarefied gene abundance table was normalized according to the fragments per kilobase per million mapped reads (FPKM) approach to give the gene abundance profile table.
Metagenomic species pangenome (MSP) profiles
Metagenomic species pangenomes (MSPs) are coabundant gene groups corresponding to microbial species. A total of 853 MSPs were clustered using MSPminer25. MSP abundances were estimated as the mean abundance of the 100 genes defining a robust centroid of the cluster (if more than 10% of these genes demonstrated positive signals). MSP richness (MSP count) was calculated directly from the rarefied MSP abundance matrix. Bacterial gene richness (gene count) was calculated by counting the number of genes detected at least once in a given sample, using the average number of genes counted in 10 independent rarefaction experiments. Finally, an occurrence filter was used, leaving 666 species present in at least one sample.
Data analysis
A principal component analysis (PCA; biplot representation) of the covariance matrix provided a multidimensional representation of the links between the scores obtained for the different tastes. The participants were represented using different symbols depending on their age and sex. A second PCA was performed on saliva and lingual film species to depict the dataset variability. By correlating principal components to descriptive variables, such as age, sex and salivary flow, we aimed to evaluate whether species-major information is linked to these participants characteristics. Spearman correlation tests were used to evaluate the significance of correlations.
Correlations between phylum, genus and species expression quantified in saliva and lingual film were tested for each of the 5 tastes using the Spearman correlation test. p values < 0.05 were considered statistically significant. Features significantly correlated to at least one taste were selected and depicted through heatmaps showing significant Spearman correlation values. A positive correlation means that a higher abundance of the microbial taxa corresponds to a higher sensitivity to the taste. We chose to perform selection based on raw p values rather than FDR-adjusted p values to allow a wider selection of entities of interest.
Classifier models were built with each taste score using saliva species abundancies or lingual species abundancies as the input to predict whether each subject was “high” or “low” in this specific taste. “High” and “low” statuses were defined using the median value of the taste score. Participants with taste scores higher than the median were defined as “high” for this specific taste, and those with lower scores were defined as “low”. The classifiers were PLS DA (discriminant analysis) models built on a training set comprising 80% of the participants and tested on a test set of participants (the remaining 20%). This data split (training and test sets) was reproduced a hundred times to remove any potential bias due to the randomized splitting between two sets. With each data splitting, the AUC (Area Under the Receiver Operating Characteristics) on the test set was used as predictive score to determine classifier performances. An excellent model has AUC near to 1 whereas a poor model has an AUC near 0. When AUC is 0.5, the model has no class separation capacity. The CARET R package was used to compute these classifier models (https://CRAN.R-project.org/package=caret).
A flow chart of the different steps of the study is presented Fig. 1.
Ethics approval and consent to participate
Written informed consent was obtained from each participant in the study. The protocol was approved by the Ethics Committee: Comité de Protection des Personnes Ouest 5 (CPP no. 2016-A01954-47).
Results
Taste sensitivity and subject characteristics
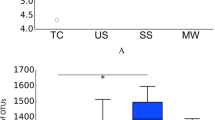
Subjects displayed heterogeneous sensitivity to the different tastes, as shown in Fig. 2A. Taste sensitivity for sourness appears linked with bitterness and umami with saltiness. The sensitivity scores associated with the different taste modalities decrease significantly with age (Fig. 2B), and a significant sex effect was observed for all taste modalities except saltiness, with females being more sensitive than males (Fig. 2C).
Taste sensitivity of the panel (A) Biplot representation of the distribution of the participants (N = 100). Each dot represents the average of 3 tests. Females are represented by circles, and males by squares. The intensity of colour decreases with age. (B) Average taste sensitivity scores for 4 age classes (N = 25). Different letters indicate significant differences (p ≤ 0.05) between the age groups for a given taste. (C) Average sensitivity scores for males (N = 50) and females (N = 50). A star indicates a significant difference (p ≤ 0.05) between groups. A high score signifies a high taste sensitivity.
Saliva and lingual film microbiota
A total of 666 microbial species were identified in the saliva and lingual film samples (Fig. 3A). Most of these species (86%) were common to both oral habitats. The number of nonshared species was slightly higher in saliva than in lingual film (12% and 3%, respectively), and the same observations were obtained at the genus level (8% and 3%, respectively), for a total of 115 genera. The 12 phyla identified are common to saliva and lingual film. Interestingly, when restricting the analysis to the species present in at least 10% of individuals (application of a 10% occurrence filter on each body site, giving 405 species in the saliva and 293 species in the lingual film), the proportion of shared species between the two ecosystems is lower, with almost all the nonshared species identified in the saliva (114 species specific to saliva and 2 specific to lingual film). The same observations were observed to a lesser extent for genera, but only one phylum was specific to saliva. This finding shows that the most prevalent species are not always found in the lingual film, while the less prevalent species are most often found in these samples. In addition to species richness, the relative abundance of some phyla varied between the two different ecosystems. This is illustrated at the individual and group levels, where a higher abundance of Bacteroidota and a lower abundance of Proteobacteria can be observed in saliva compared to that in lingual film (Fig. 3B). Moreover, approximately 90% of the species whose relative abundance significantly differed between the saliva and tongue dorsum were more abundant in the saliva (270 species vs. 29 species, Supplementary Table S1). Notably, there was a significantly higher correlation between lingual and saliva samples from the same participants than among samples from different participants within the same ecosystem (R = 0.75 between 2 samples of the same subjects; R = 0.46 between samples from the salivary ecosystem and R = 0.53 between samples from the lingual ecosystem). This finding points towards a stronger heterogeneity of the microbiota between the same ecosystems in different participants than for both ecosystems within the same subject (Fig. 3C). Regarding the characteristics of the subjects, neither salivary nor lingual microbiota was affected by sex; however, the lingual microbiota was significantly affected by age, and the salivary microbiota was affected by the resting salivary flow rate (Table 1).
Bacterial composition of saliva and lingual film. (A) Venn diagrams showing the number of features (resp. phylum, genus and species) unique to the saliva (in blue) or lingual film (in red) and common to both (in purple), with (above) or without (below) occurrence filtering. (B) Relative abundance of the different phyla in the saliva and lingual film at the group level on the left (relative abundance average) and at the individual level on the right. (C) Box plot representation of the Spearman correlation coefficients. The green boxplot displays correlations between saliva samples; the blue box displays correlations between lingual samples, and the red box displays correlations between saliva and lingual samples from the same subject. A star indicates a significant difference (p ≤ 2.22–16) between samples.
Relationships between taste sensitivity and oral microbiota
No significant correlation between the sensitivity to the five tastes and microbial richness (MSP count) was observed for either the saliva or the lingual film (Table 2). Correlations were made between individual taste sensitivity scores and microbes at the phylum, genus and species taxonomic levels, as shown in Fig. 4. Saliva and lingual microbiota are both informative, with more significant correlations in the salivary ecosystem; 86 in saliva species vs. 69 in lingual film, consistent with the higher number of species identified at this site (see above and Fig. 3A). Generally, approximately twice as many positive correlations as negative correlations between species and taste are found in the saliva, whereas the opposite is true in the lingual film. In the saliva, umami and sour tastes display more correlations with microbial taxa than other tastes, i.e., 37 and 44 correlations at the species level, similar to umami in the lingual film (34 correlations). In the saliva, most of the correlations were positive with sweetness (18 positive vs. 6 negative correlations at the species level), saltiness (17 vs. 5) and umami (30 vs. 7), whereas in the lingual film, most of the correlations were positive with sourness (18 vs. 4) and negative with sweetness (3 vs. 14) and bitterness (3 vs. 16). Other combinations are rather evenly distributed. It is worth mentioning that the Spearman coefficient distribution is similar between the different tastes at the genus and species levels but varies at the phylum level, with the highest positive and negative values observed with umami taste in saliva and lingual film data and the highest negative values observed with sour and umami taste in saliva and lingual film, respectively (Supplementary Fig. S1). Note that only the difference between sour and umami is significant (Wilcoxon p value = 0.02).
Heatmap showing the Spearman correlation among differential phylum, genus, species expression and taste sensitivity scores in saliva and lingual film. Phyla, genera and species highlighted in yellow are common to the saliva and lingual film. Positive correlations are represented in red, and negative ones in blue. A positive correlation means that a higher abundance of the microbial taxa corresponds to a higher sensitivity to the taste. (p value < 0.05).
Four phyla were significantly associated with umami taste: Fusobacteriota and Patescibacteria were positively correlated, and Actinobacteria and Firmicutes were negatively correlated. Actinobacteria is the only genus appearing in both saliva and film ecosystems and is also negatively correlated with sour taste in the saliva. At the genus and species levels, a large portion of bacterial taxa that correlated with at least one taste sensitivity in the lingual film also correlated with one taste in the saliva (8 genera out of 36 and 46 species out of 109, highlighted in yellow in Fig. 4). Generally, correlations occur in the same direction but not always for the same taste. For instance, Rothia and Streptococcus genera are negatively correlated with the sour taste in the saliva and with the umami taste in the lingual film. Among the most abundant species in the oral area (> 10–6 in saliva or film), Veillonella atipyca, Porphyromonas gingivalis, Neisseria sp. oral taxon 014, Streptococcus salivarius, Atopobium parvulum 2, Prevotella salivae and Rothia mucilaginosa 1 display negative correlations with one or several taste sensitivities in both oral sites, and Prevotella shahii and unclassified Saccharimonadaceae display positive correlations. While over half of the species correlated with a single taste, some species correlated with four or five tastes, all in the same direction: Streptococcus parasanguinis, Streptococcus gordonii, P. gingivalis and Prevotella jejuni had negative correlations, and unclassified Rikenellaceae 3, SR1 bacterium human oral taxon HOT-345 (SR1 HOT-345), and Prevotellaceae bacterium Marseille-P2826 had positive correlations. Hence, P. gingivalis, S. parasanguinis, S. gordonii and P. jejuni were negatively correlated with three or four similar tastes in both saliva and lingual film. In contrast, Prevotellaceae bacterium Marseille positively correlated with all tastes except saltiness at both sites. To go beyond correlations, we selected the species that displayed correlations for all tastes in saliva (i.e., S. parasanguinis, S. gordonii for negative correlations; unclassified Rikenellaceae 3 and SR1 HOT-345 for positive correlations); for each species, the participants were divided into two groups (n = 50) according to the proportion of each species in their saliva microbiota, and the taste scores were compared between the two groups. For the two Streptococcus species, taste scores were significantly lower for the group with the highest proportion of the studied species for all the tastes except salty for S. parasanguinis (Fig. 5). For SR1 HOT-354 and Rikenellaceae 3, sweet, salty, and umami scores were significantly higher for the groups with the highest proportion of the species. Differences between taste scores were also found for the species that correlated with four tastes and the lingual film, but to a lesser extent (Supp. Figure 2). These results validate the tendency highlighted by the correlations.
Boxplots showing the distribution of sensitivity scores of the participants according to their high (N = 50) or low (N = 50) abundance in the four species correlated significantly to the sensitivity to the five tastes in saliva. The number of stars (*p < 0.05; **p < 0.01; ***p < 0.001; ns: nonsignificant) indicates the significance level of the Wilcoxon test depicting the difference between the two groups.
For most taxa, the genus level recapitulated the direction of the correlation. For example, all selected Streptococcus and Rothia species showed negative correlations, while all selected Saccharimonadaceae species showed positive correlations. Interestingly, all selected Veillonella species displayed negative correlations except Veillonella tobetsuensis. For Prevotella, the direction of correlations is dependent on the species. For example, Prevotella salivae is negatively correlated with sweet and sour perceptions, whereas Prevotella nigrescens is positively correlated with these tastes.
The impact of a bacterial species does not seem to be linked to its abundance or prevalence since most of the species that correlate with taste sensitivity are not particularly abundant (< 10–6) or prevalent (< 70) (Supplementary Table 1). To identify if the microbiota from both oral sites studied can predict taste sensitivity, prediction models performances were determined through AUC computation (see Methods) (Fig. 6). It appears that the saliva presents better taste-predictive capacities than the lingual film. Nevertheless, the predictive scores remain of mediocre quality, with the highest qualities observed when predicting salty or sour sensitivity using saliva microbiome species (mean AUC predictive score = 0.65 and 0.63, respectively).
Discussion
Comparison between saliva and lingual film microbiota
The objective of the present work was to elucidate whether oral microbiota composition is related to taste perception. We focused on two ecosystems, the salivary and lingual film microbiota, which is present on the tongue dorsum and cover the taste papillae. The latter is suspected to modulate the concentration of taste compounds in the vicinity of taste receptors7, whereas the resting saliva microbiota may reflect the various niches of the oral cavity and can be relevant as a future source of markers. We first identified 666 different species, which is comparable with the current number of 774 oral species reported in the expanded Human Oral Microbiome Database26. This represents an important diversity, especially considering that we studied participants from the same living area. The second interesting observation is that most species are significantly more abundant in the saliva than in the lingual film, but the lingual film appears to be characterized by a lower presence of most of the shared species between individuals than that observed in the saliva.
On average, the saliva and lingual film microbiota profiles are characterized by an overrepresentation of the Proteobacteria, Firmicutes and Bacteroidota phyla comparable to the profiles found recently by other authors using metagenomics27 and 16S rRNA data from the eHOMD28,29,30. Our study shows that saliva contains more Bacteroidota and less Actinobacteria than lingual film, consistent with our previous results obtained using qPCR on a smaller number of participants23. However, the eHOMD data reported this difference in Bacteroidota but not in Proteobacteria, and no differences were observed by Liu et al.27. In addition to the different origins of the three panels, US young adults (eHOMD), Chinese27 and French (present study) individuals and the different analytic methods, it is worth noting that the sampling protocols differed. For instance, the sampling in the present study consisted of scrapping the tongue dorsum by an experimenter, whereas in the study of Liu et al. the sampling was performed by the participants themselves31. Self-sampling by the participants is generally softer than when performed by an experimenter, resulting in sampling of the superficial layer of the lingual microbiota, which may be more heterogeneous. We collected all the microbial material in the tongue dorsum, including microorganisms installed in the deep layers of the lingual film close to the taste receptors.
In addition to the observed specificities in the profiles, saliva and lingual film microbiota vary importantly between individuals, as already reported18, 27; however, the causative factors of this variability have not been fully identified. We reported an effect of saliva flow on salivary microbiota. This is not surprising since saliva has antibacterial properties, especially due to its protein composition26, 27. This fluid contains various compounds, including glycoproteins and sugars, that can be used as substrates for microorganisms11, modulating the microbiota composition differently. Saliva flow also plays a role in cleaning the oral cavity, favoring the unhooking of microorganisms from the various niches of the oral cavity. It is, however, less surprising that the lingual film is not correlated with the saliva flow rate; its microbiota is firmly anchored to the lingual mucosa. Age was also significantly correlated with the lingual film microbiota but not with that of the saliva. This lack of correlation was unexpected since it is known that the microbiota of the different niches of the oral cavity varies with age32.
Oral microbiota and taste
From a global view, microbial richness is not correlated with any specific taste sensitivity in the lingual film or saliva. No correlation with microbial richness was observed with 6-n-propylthiouracyl (bitter) perception by Cattaneo et al.14 or with linoleic acid (fatty acid) perception by Besnard et al.13. Altogether, these observations suggest that the link between taste sensitivity and single taxa must be considered. Moreover, regarding the hypothesis that bacterial metabolites could modulate the concentration of the taste molecules near the taste receptors, we could expect opposite correlations for the same taxon. For example, some saccharolytic and acidogenic bacteria could impact sweet and sour taste perceptions in opposite ways. In the context of this hypothesis, it seemed important to properly test the perception of the 5 tastes simultaneously in all individuals and not a single molecule as a marker of general perception. Surprisingly, no bacterial taxon (regardless of the taxonomic level) correlated positively with one taste and negatively with another. Without rejecting the hypothesis, it could be assumed that active metabolic pathways imply intricate interspecies interactions that cannot be analysed in this study. In contrast, this study highlights some bacterial taxa that correlate in the same direction for several tastes, especially some Streptococcus and Prevotella species. These observations cannot be made at the genus level, probably because it represents the mean of species with and without correlation with taste perception. This must be why Cattaneo et al.15 did not observe correlations using 16S RNA gene sequencing. Our correlations at the genus level do not match exactly with theirs. Lachnoanaerobaculum, Fusobacterium and Rothia are in the same direction but not always for the same taste, and both studies highlight original genera. Comparing with a study from the same group, we only observed one common genus, Catonella, correlated to taste14. These differences between studies could arise from the various approaches employed, especially the sequencing techniques and the selected populations.
Streptococcus, which is the most abundant genus in the saliva and on the tongue, is negatively correlated only with sourness in the saliva and only with umami in the lingual film. This finding was confirmed at a lower taxonomic rank since none of the species of this genus positively correlated with taste. S. gordonii and S. parasanguinis are remarkable since they are negatively correlated with all taste modalities in the saliva. This example illustrates the importance of considering the species level in such studies, with the genus level being an average of the contribution of species that could have opposite effects. This finding highlights the added value of shotgun metagenomics, which allows quantification at the species level.
Such an implication of Streptococcus in taste had already been observed in elderly patients with a link between a global lower taste perception and the abundance of culturable salivary Streptococci8 and in children with a link between a 6-n-propylthiouracil lower taste perception and the abundance of culturable salivary S. mutans33. Streptococci are important primary colonizers of the mouth because of their ability to adhere to tooth surfaces. The nature and number of adhesins vary among Streptococcus species, and S. gordonii is particularly adept at adhering to various surfaces and coaggregating with various bacterial species due to its numerous adhesins34, 35. Wilbert et al.36 observed that oral bacterial biofilms on the lingual papillae are organized in patches and that the special organization of the two Streptococcus species they studied was different, with S. salivarius forming patches and S. mitis forming layers at the biofilm surface. We assume that S. gordonii and S. parasanguinis would impact the structure of the lingual biofilm in a way that increases constraints for the diffusion of the taste molecules to the taste receptors. Moreover, these Streptococcus species can compete with other species by producing H2O2 and bacteriocins, which can also modify the biofilm structure37.
The Prevotella genus was the second most abundant genus in terms of abundance at both oral sites, consistent with previous studies38. The species belonging to this genus are particularly heterogeneous regarding correlations, especially Prevotellaceae bacterium Marseille-P2826, for which positive correlations were found for 4 tastes, and P. jejuni, which is negatively correlated with 4 tastes. These correlations occur in both oral sites. The physiology and functionality of this genus have been poorly studied to date, mainly because of culturing difficulties. Recent advances in metagenomics have uncovered the large diversity of the Prevotella genus, and the oral cavity hosts the largest diversity38. The different Prevotella species correlating in the same direction are not the closest phylogenetically.
Conclusion
One of the aims of the present work was to identify key microorganisms from the oral microbiota involved in taste perception by exploring two potentially involved oral ecosystems. We found several correlations between the different taste modalities and various bacterial species, some of which were common to both ecosystems. Our study reveals the importance of 2 main oral bacterial genera for taste perception. The abundance of Streptococcus species, especially S. gordonii and S. parasanguinis, is correlated with a reduced taste sensitivity, whereas the type of correlation is species-dependent for the Prevotella genus. When trying to answer the question of which ecosystem is the most linked to taste sensitivity, it appears from our modelling approach that the saliva microbiota is a better taste predictor than that of the lingual film. This result is somewhat surprising since we expected that the lingual microbiota, at the interface between the oral cavity and the taste receptors, would be implied directly in this perception. In this context, saliva appears to be the sum of the microbiota of the various niches of the oral cavity. Therefore, one possible explanation is that the implication of the salivary microbiota is to find at the oral level by studying the global metabolic activity involving species consortia. In the overall context of taste perception, a next step would be to understand the contribution of the oral microbiota alongside other factors involved in taste perception such as the genetics of taste receptors, the number of taste buds or the biochemistry of saliva.
Data availability
Raw shotgun sequencing data that support the findings of this study have been deposited in the European Nucleotide Archive with accession codes PRJEB60621 (https://www.ebi.ac.uk/ena/browser/view/PRJEB60621). Correspondence and material request can be addressed to eric.neyraud@inrae.fr.
References
Brondel, L., Jacquin, A., Meillon, S. & Penicaud, L. Taste: Physiology, roles and dysfunction. Nutr. Clin. Metab. 27(3), 123–133 (2013).
Puputti, S., Hoppu, U. & Sandell, M. Taste sensitivity is associated with food consumption behavior but not with recalled pleasantness. Foods 8(10), 444 (2019).
Landis, B. N. et al. “Taste strips”—A rapid, lateralized, gustatory bedside identification test based on impregnated filter papers. J. Neurol. 256(2), 242–248 (2009).
Garcia-Bailo, B., Toguri, C., Eny, K. M. & El-Sohemy, A. Genetic variation in taste and its influence on food selection. OMICS 13(1), 69–80 (2009).
Mounayar, R. et al. Nutri-metabolomics applied to taste perception phenotype: Human subjects with high and low sensitivity to taste of fat differ in salivary response to oleic acid. OMICS 18(11), 666–672 (2014).
Neyraud, E. Role of saliva in oral food perception. Monogr. Oral. Sci. 24, 61–70 (2014).
Neyraud, E. & Morzel, M. Biological films adhering to the oral soft tissues: Structure, composition, and potential impact on taste perception. J. Texture Stud. 50(1), 19–26 (2019).
Solemdal, K. et al. Association between oral health and body cell mass in hospitalised elderly. Gerodontology 29(2), e1038-1044 (2012).
Ohno, T., Uematsu, H., Nozaki, S. & Sugimoto, K. Improvement of taste sensitivity of the nursed elderly by oral care. J. Med. Dent. Sci. 50(1), 101–107 (2003).
Dsamou, M. et al. Salivary protein profiles and sensitivity to the bitter taste of caffeine. Chem. Senses 37(1), 87–95 (2012).
Takahashi, N. Oral microbiome metabolism: From “Who are they?” to “what are they doing?”. J. Dent. Res. 94(12), 1628–1637 (2015).
Besnard, P. et al. Fatty taste variability in obese subjects: The oral microbiota hypothesis. OCL 27, 38 (2020).
Besnard, P. et al. Obese subjects with specific gustatory papillae microbiota and salivary cues display an impairment to sense lipids. Sci. Rep. 8(1), 6742 (2018).
Cattaneo, C. et al. New insights into the relationship between taste perception and oral microbiota composition. Sci. Rep. 9(1), 3549 (2019).
Cattaneo, C., Riso, P., Laureati, M., Gargari, G. & Pagliarini, E. Exploring associations between interindividual differences in taste perception, oral microbiota composition, and reported food intake. Nutrients 11(5), 1167 (2019).
Fluitman, K. S. et al. Associations of the oral microbiota and Candida with taste, smell, appetite and undernutrition in older adults. Sci. Rep. 11(1), 23254 (2021).
Wong, D. T. W. Salivaomics. J. Am. Dent. Assoc. (1939) 143(10 Suppl), 19S-24S (2012).
Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature 486(7402), 207–214 (2012).
Mameli, C. et al. Taste perception and oral microbiota are associated with obesity in children and adolescents. PLoS One 14(9), e0221656 (2019).
Esberg, A., Haworth, S., Hasslof, P., Lif Holgerson, P. & Johansson, I. Oral microbiota profile associates with sugar intake and taste preference genes. Nutrients 12(3), 681 (2020).
Esberg, A. et al. Using oral microbiota data to design a short sucrose intake index. Nutrients 13(5), 1400 (2021).
D’Auria, E. et al. Alteration of taste perception, food neophobia and oral microbiota composition in children with food allergy. Sci. Rep. 13(1), 7010 (2023).
Feng, Y. et al. The associations between biochemical and microbiological variables and taste differ in whole saliva and in the film lining the tongue. Biomed. Res. Int. 2018, 2838052 (2018).
Martin, C. & Neyraud, E. Impact of very hot drink consumption habits, age, and sex, on taste sensitivity. Foods 10(5), 1139 (2021).
Onate, F. P. et al. MSPminer: Abundance-based reconstitution of microbial pan-genomes from shotgun metagenomic data. Bioinformatics 35(9), 1544–1552 (2019).
Escapa, I. F. et al. New Insights into human nostril microbiome from the expanded human oral microbiome database (eHOMD): A Resource for the Microbiome of the human aerodigestive tract. Msystems 3(6), 10–1128 (2018).
Liu, X. et al. Metagenome-genome-wide association studies reveal human genetic impact on the oral microbiome. Cell Discov. 7(1), 117 (2021).
Chen, T. et al. The human oral microbiome database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010, baq013 (2010).
Eren, A. M., Borisy, G. G., Huse, S. M. & Mark Welch, J. L. Oligotyping analysis of the human oral microbiome. Proc. Natl. Acad. Sci. 111(28), E2875-2884 (2014).
Segata, N. et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13(6), R42 (2012).
Zhu, J. et al. Over 50,000 metagenomically assembled draft genomes for the human oral microbiome reveal new taxa. Genom. Proteom. Bioinform. 20(2), 246–259 (2022).
Xu, X. et al. Oral cavity contains distinct niches with dynamic microbial communities. Environ. Microbiol. 17(3), 699–710 (2015).
Shetty, V., BL, P. & Hegde, A. M. PROP test: Prediction of caries risk by genetic taste perception among the visually impaired children. Spec. Care Dentist. 34(1), 34–40 (2014).
Abranches, J. et al. Biology of oral streptococci. Microbiol. Spectr. 6(5), 10–1128 (2018).
Kolenbrander, P. E. et al. Bacterial interactions and successions during plaque development. Periodontology 2006(42), 47–79 (2000).
Wilbert, S. A., Mark Welch, J. L. & Borisy, G. G. Spatial ecology of the human tongue dorsum microbiome. Cell Rep. 30(12), 4003-4015 e4003 (2020).
Lamont, R. J., Koo, H. & Hajishengallis, G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 16(12), 745–759 (2018).
Tett, A., Pasolli, E., Masetti, G., Ercolini, D. & Segata, N. Prevotella diversity, niches and interactions with the human host. Nat. Rev. Microbiol. 19(9), 585–599 (2021).
Acknowledgements
The authors thank A. David, A. Bougaud and H. Brignot for their valuable technical assistance and Dr. M. Bournez for her valuable contribution to the ethical procedures.
Funding
This work was supported by grants from the French “Investissements d’Avenir” program, project ISITE-BFC (contract ANR-15-IDEX-0003) TOM, the French Institut Carnot Qualiment (project MORGoM), the Conseil Régional Bourgogne, Franche-Comte (PARI grant) and FEDER (European Funding for Regional Economical Development).
Author information
Authors and Affiliations
Contributions
H.L. and H.M.B. and E.N. conceived and designed the research. C.Ma. and C.Mo. and B.Q. and N.P. and E.N. developed the experimental procedures and conducted the experiments. C.T. and S.F. and N.P. analysed the data. H.L. and C.T. and S.F. and N.P. and H.M.B. and EN wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Licandro, H., Truntzer, C., Fromentin, S. et al. The bacterial species profiles of the lingual and salivary microbiota differ with basic tastes sensitivity in human. Sci Rep 13, 20339 (2023). https://doi.org/10.1038/s41598-023-47636-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-47636-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.