Abstract
Tephritid fruit fly pests pose an increasing threat to the agricultural industry due to their global dispersion and a highly invasive nature. Here we showcase the feasibility of an early-detection SEPARATOR sex sorting approach through using the non-model Tephritid pest, Ceratitis capitata. This system relies on female-only fluorescent marker expression, accomplished through the use of a sex-specific intron of the highly-conserved transformer gene from C. capitata and Anastrepha ludens. The herein characterized strains have 100% desired phenotype outcomes, allowing accurate male–female separation during early development. Overall, we describe an antibiotic and temperature-independent sex-sorting system in C. capitata, which, moving forward, may be implemented in other non-model Tephritid pest species. This strategy can facilitate the establishment of genetic sexing systems with endogenous elements exclusively, which, on a wider scale, can improve pest population control strategies like sterile insect technique.
Similar content being viewed by others
Introduction
The Mediterranean fruit fly (medfly), Ceratitis capitata (Tephritidae), is among the most notorious and vigorous agricultural pests. It has a vast host range of over 260 fruit and vegetable species, and a worldwide distribution characterized by ongoing rapid expansions1,2,3,4. Its invasive nature is nourished by factors like increased food trading fueled by the growing demand for produce1 and climate change3,5,6,7. The medfly often acts as a model for other Tephritid pests such as the Mexican fruit fly, Anastrepha ludens, due to its easier laboratory-based rearing and a shorter life cycle.
Tephritid pests are targeted for population control through sterile insect technique (SIT) as part of area-wide integrated pest management (AW-IPM) programs implemented internationally8. SIT relies on mass rearing, irradiation-based sterilization of flies and their sequential release into the wild9,10. The aim of this strategy is to promote mating of released sterile males with wild females, suppressing population growth10. In general, both males and females have been released simultaneously in SIT11. Male-only releases, however, were shown to be more cost-effective than co-releases with females12. Additionally, male-only release schemes reduce crop damage invoked by egg laying and encourage wider sterile male dispersal allowing greater mating frequency with wild females13,14.
To eliminate the necessity for sex-sorting at adulthood in SIT and allow male-only releases, multiple cost-effective genetic sexing strains (GSSs) were developed in Tephritids15,16,17,18. Traditionally, GSS requires a selective marker, for instance conditional female development arrest or puparium coloration which is linked to the Y chromosome through translocation15. Such strains have been implemented in C. capitata SIT rearing19; and developed in multiple other Tephritid species16,17,18. The largest problem with Y-linked GSSs is the associated genetic instability caused by recombination and thus desired phenotype loss15. Another complication with the currently used GSSs is the partial sterility of the males which creates rearing difficulties20. To tackle this, novel scalable sexing strategies are required, boasting superior genetic stability and lower fitness costs11,21.
To address genetic instability of Y-linked GSSs, multiple Y-independent GSSs have been developed in fruit flies22,23,24,25,26. The majority of Y-independent GSS approaches rely on the sex-specific alternative splicing properties of a sex determination gene, transformer (tra)22,24,26. Among Tephritids, the role of tra is best characterized in the medfly, although its functions are well-conserved across the family27. The default mechanism of medfly embryonic development involves maternal and zygotic C. capitata tra (Cctra) and C. capitata tra2 (Cctra2) which enable a positive feedback loop of female-specific splicing of Cctra28,29,30. This female form of Cctra is needed for female-specific splicing of downstream doublesex and fruitless genes, conserved across the Tephritidae family27,31,32,33,34,35. In XY male embryos, the female-specific splicing of tra is blocked by Maleness on Y (MoY) factor, transcribed from the Y chromosome36. In the presence of MoY, tra is spliced with stop codon-containing male-specific exons resulting in premature translation arrest and thus the lack of functional Cctra production37. For GSS establishment, the C. capitata tra intron was used to induce female-specific lethality in two tetracycline-dependent systems in the medfly22,25 and similarly in A. ludens26. Although highly efficient, these sexing strains require tetracycline supply for line maintenance which affects mitochondrial function and fitness of released males38,39,40,41, as well as becoming costly for production on a mass scale22,25,26. Hence alternative approaches would be beneficial to alleviate the reliance of Y-independent GSSs on tetracycline.
In this study, we showcase a sex-sorting method in C. capitata by exploiting Sexing Element Produced by Alternative RNA-splicing of a Transgenic Observable Reporter (SEPARATOR). This strategy was previously used in Drosophila melanogaster as a proof of principle, and further demonstrated in Aedes aegypti mosquitoes and enabled sex-sorting of early L1 is through large particle flow cytometry using a complex object parametric analysis and sorting (COPAS) instrument42,43. The transgenic SEPARATOR lines generated here harbor a cassette with one constituent, and one tra intron-containing fluorescence markers. Thus, phenotypically, the males and females can be easily distinguished through fluorescence screening from early development. We demonstrate the feasibility of such a system using both the endogenous C. capitata and A. ludens tra introns. Our work provides a basis for accessible cross-species transfer across Tephritid family members with a shorter research history, including A. ludens. Moreover, the strategy described here can serve as a cornerstone for seamless endogenous GSS generation through insertion of the tra intron into genes such as white pupae and ebony for their sex-specific expression15,18. Such an approach is feasible due to the recent successful implementation of homology-directed repair in the medfly44,45.
Results
Establishment and characterization of fluorescent SEPARATOR strains
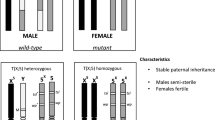
To engineer the SEPARATOR genetic cassettes for Ceratitis capitata, we utilized the transformer (tra) intron of C. capitata or A. ludens to generate 795H1 or 795K1 construct, respectively. Importantly, both constructs included the DsRed coding sequence separated by the tra intron and expressed under the Opie2 promoter, and a dominant Hr5-IE1-eGFP marker (Fig. 1a). We used germline transformation to induce piggyBac-reliant integrations for 795H1 and 795K1 constructs (Supplementary Table 1). Two separate strains for each construct were successfully characterized through inverse PCR (H-001 and H-002 for 795H1; K-001 and K-002 for 795K1) (Supplementary Fig. 1 and Supplementary Table 2). Homozygosity for all strains was achieved via sibling crosses at G2 thereafter. We noted no DsRed+ /GFP+ males or DsRed-/GFP+ females for any of the 4 homozygous strains during routine screenings for 8 consecutive generations (from G3 to G10). This was indicative of sex-specific splicing of DsRed, and thus the success of tra intron selection for both C. capitata and A. ludens. For verification, we expanded the strains and characterized the entire populations at G9 and G10 (Fig. 2a). As expected, no DsRed+ /GFP+ males or DsRed-/GFP+ females were detected. Transgenic individuals with only two phenotypes: DsRed + /GFP + females (48.8%) and DsRed-/GFP+ males (51.2%) were noted across the four strains (Supplementary Table 3). Chi-squared tests for observed and expected sex ratios showed no statistical significance for any of the four strains. Overall, 100% transgenic flies expressed the desired fluorescence phenotypes across both C. capitata (795H1) and A. ludens (795K1) tra intron-harboring medflies across G9 and G10 with a total of 3787 flies counted.
SEPARATOR system for sex-sorting of Ceratitis capitata. (A) A schematic representation of the Sexing Element Produced by Alternative RNA-splicing of Transgenic Observable reporter (SEPARATOR) cassettes, 795H1 and 795K1. Both cassettes contain two functional elements: constituent Hr5IE1-eGFP-SV40 and Opie2-DsRed-p10, in which the DsRed coding sequence is separated by a transformer (tra) intron. The tra intron in 795H1 and 795K1 originates from Ceratitis capitata and Anastrepha ludens, respectively. (B) In the SEPARATOR strains the males produce eGFP only, whilst females express both eGFP and DsRed. The representative images of larval, pupal and adult stages of wild-type (WT), and homozygous male and female individuals are compared against each other under bright light, and green fluorescence protein (GFP), and red fluorescence protein (RFP) filters. All images were taken for the H-002 strain harboring the 795H1 construct. Images for all life stages were taken under bright light, and green fluorescence protein (GFP), and red fluorescence protein (RFP) filters.
Characterization of transgenic SEPARATOR strains. (A) A stack graph showing the fluorescence phenotype distributions by sex of the four homozygous transgenic strains at the 9th and 10th generations with over 3500 adult flies screened. Only the desired DsRed + /GFP + females and DsRed-/GFP + males were observed, whilst no DsRed + /GFP + males or DsRed-/GFP + females were observed. (B) Egg laying rates within strain crosses for all four strains was compared to wild-type through the egg laying rates within a 5-h period. (C) Egg hatching rate within strain crosses for all four strains was compared to wild-type through the hatching rates of eggs laid within a 5-h period. (A–C) H-001 and H-002 strains have the 795H1 cassette harboring the endogenous Ceratitis capitata transformer (tra) intron, while K-001 and K-002 carry the 795K1 cassette with the Anastrepha ludens tra intron. (A) Chi-squared tests showed no statistical significance in sex ratio distortion for any of the four strains. (B, C) Dunn test statistical significance was established at follows: p < 0.05 = * and the wild-type—transgenic strain significance is displayed on the graphs. (B, C) The bar levels represent the mean value whilst the dots represent raw values of the replicates. SEPARATOR stands for Sexing Element Produced by Alternative RNA-splicing of a Transgenic Observable reporter. (A–C) were constructed in RStudio.
Fitness of fluorescent SEPARATOR strains
To assess whether the SEPARATOR cassettes are associated with notable fitness costs, we assessed the rates of egg laying and egg hatching rate for the four homozygous strains (Supplementary Table 4). Sibling crosses of homozygous individuals were conducted for each strain alongside wild-type controls. The egg laying rates were variable across wild-type and transgenic strains, although no statistically significant differences between wild-type and transgenic lines were determined through Kruskal–Wallis test and Dunn’s multiple comparison test (Fig. 2b). It is of note, however, that there was a notable reduction in H-002 strain egg production. Meanwhile, the egg hatching rate of the 795H1-harboring strains with the endogenous tra intron was similar to wild-type (Fig. 2c). The exogenous intron-containing 795K1 strains had reduced egg hatching rate compared to non-transgenic flies. Specifically, the egg hatching rate of K-001 was significantly reduced (p = 0.019), while the egg hatching rate of K-002 was reduced insignificantly (p = 0.072).
Fluorescence patterns of SEPARATOR strains
We observed that the GFP fluorescence signal across the 4 homozygous strains was visibly similar. DsRed signal, however, was visually more intense in the females harboring the endogenous C. capitata tra intron (H-001 and H-002), compared to the females with the A. ludens tra intron (K-001 and K-002), irrespective of GFP fluorescence intensity in homozygous individuals (Supplementary Fig. 2). We further investigated the patterns of fluorescence marker expression throughout the C. capitata life cycle (Fig. 1b). We observed a clear distinction between males and females immediately upon hatching as larvae and increasingly throughout all later stages of the life cycle. Specifically, the signal was easily recognizable throughout the first, second and third instar larvae; pupae and adult development stages. At the late egg stage, GFP fluorescence is very clear in all individuals, whilst DsRed fluorescence was more difficult to differentiate (Supplementary Fig. 3). This is because all eggs had a DsRed signal of variable intensity, which was stronger than in their wild-type counterparts.
We additionally carried out reverse-transcription PCR (RT-PCR) to amplify Opie2-DsRed for molecular verification of the sex-specific nature of DsRed splicing. cDNA, synthesized from total RNA, and gDNA were used to validate this phenomenon from adult males and females harboring either 795H1 or 795K1 (Supplementary Fig. 4). We observed gDNA-derived fragments of similar sizes in both sexes, and cDNA-derived fragments of different sizes in males and females. The female-specific cDNA-derived fragments for both C. capitata (795H1) and A. ludens (795K1) tra introns expectedly amplified as single shortest fragments. One of the two male-specific transcripts was detected for 795H1 males, while multiple bands were observed for 795K1 males. These were more challenging to distinguish for the 795K1-harboring males as there are 5 alternative male-specific transcripts of tra in Anastrepha due to the presence of 3 male-specific exons32. We deduced that DsRed in both cassettes is spliced in a similar fashion to the tra gene whereby DsRed translation is interrupted with male-specific stop codons. We also concluded that the A. ludens tra intron, despite incomplete sequence similarity, is functionally suitable to replace its endogenous counterpart in C. capitata.
Discussion
In this study, we demonstrate a novel SEPARATOR approach to distinguish males from females in the global agricultural pest C. capitata with observed accuracy of 100%. This Y chromosome-independent and genetically stable strategy utilizes sex-specific splicing of tra, the master gene of sex-determination in Tephritids29. We tested sex-specifically spliced tra introns from two species belonging to different genera within the Tephritidae family, the endogenous C. capitata and the exogenous A. ludens. PiggyBac-mediated transformations were used for line establishment in this study due to their high success rates across model and non-model insect species alike46. To alleviate the insertion site-dependent effects on construct performance, we assessed 2 strains for each construct. In the system described above, females translate both a constitutive eGFP and the sex-specifically spliced DsRed fluorescence markers. The males, however, only produce eGFP as DsRed is not translated. The sex-specific splicing hypothesis is supported by the observed differential splicing patterns in males and females, altogether showing similar patterns of splicing to tra itself (Supplementary Fig. 4). Both fluorescence signals are easily detectable at the early first instar larval stage and throughout all later stages of the C. capitata life cycle. Hence, there is an inherent suitability of this system to be implemented in larger-scale automated high-throughput sex separation. Furthermore, the system demonstrated in this study does not rely on antibiotics supply either for line maintenance22,25,26 or for sex selection24. Its cost efficiency, therefore, reinforces its scalability for SIT improvement purposes. Although the strains described here can be separated by sex at early development, the COPAS optical sorter system may be unsuitable for their separation as it requires water in which survival of C. capitata is immediately limited. Waterless optical sorter systems are therefore required to allow the most-efficient use of the SEPARATOR strains in SIT.
Importantly, here we observed that the A. ludens tra intron is capable of seamlessly mediating sex-specific splicing of DsRed in C. capitata. This highlights the conserved functional nature of tra and Tephritid sex determination altogether27. When evaluating the fitness of transgenic lines, we noted high variation in egg laying among them, which may be explained by external stressors, integration site-specific effects and behavioral variation within the short experimental window. Additionally we noted a statistically significant reduction in one of the A. ludens tra intron-containing strains, whilst the endogenous intron-containing strains had similar egg hatching to wild-type controls (Fig. 2c). This, in part, can be attributed to integration site-specific effects of the construct as we observed notable differences between the two strains harboring the A. ludens intron cassette. Nonetheless, this strategy is particularly useful in A. ludens as transgenesis in this species has a limited research history compared to C. capitata26,47,48. The described system is also suitable for other cross-species applications due to the conserved function of tra throughout the wider Tephritidae family32,49,50.
Remarkably, we observed no instances of DsRed-/GFP + females, although this could be expected due to mutations in DsRed. On a mass rearing scale, however, this may become more frequent, hence a positive-selection system can be explored next whereby double-positive males are selected upon screening. This could employ pre-stop codon male-specific exons of tra or, alternatively, other genes spliced in a sex-specific manner such as dsx51. Such positive selection system will mitigate risks of accidental female releases due to marker mutations. Additionally, the SEPARATOR system described here can serve as a backbone for novel strategies such as sex-specific expression of traditionally used selective markers. Specifically, this system can employ pupal coloration-involved genes such as white pupae and ebony can be targeted, which are both implemented in currently used Y-linked GSSs15,18. This can be achieved through site-specific integration of the tra intron into the coding region of the gene via homology-directed repair. This is particularly compelling for the use in the medfly as a protocol for such integration has just recently been used for gene drive establishment in the species45. Similarly, this strategy can be highly beneficial for mass rearing purposes if used in the Y-linked GSS-engaged temperature sex lethal (tsl) locus. To achieve this, however, tsl gene characterization in the medfly needs to be achieved52.
Methods
Plasmid design and construction
Both 795H1 and 795K1 were constructed via Gibson assembly as described previously43 into an existing piggyBac plasmid containing Opie2 expressing DsRed and Hr5Ie1 expressing eGFP. This plasmid was linearized using AvrII and BamHI restriction endonucleases. The tra intron of either C. capitata or A. ludens was PCR amplified from the corresponding genomic DNA preps and then inserted into the DsRed coding sequence immediately downstream of an ATG start codon (Supplementary Table 5). Complete sequence maps and plasmids are deposited at Addgene.org (#205482 for 795H1 and #205485 for 795K1).
C. capitata rearing
The wild-type Benakeion strain of C. capitata used in this study was obtained from the Saccone Lab (University of Naples “Federico II”). Adult food consisted of a mixture of yeast and glucose in equal proportions and larvae were maintained using a carrot-based diet53. Flies were consistently kept in 12:12 h light: dark daily cycle at 26 °C and relative humidity of 65%.
C. capitata germline transformation
Plasmid microinjections were carried out into wild-type Benakeion strain embryos as previously described54. The donor 795H1 and 795K1 piggyBac plasmids (500 ng/ml) were microinjected in combination with a helper plasmid encoding the ihyPBase transposase (300 ng/ml)55. The hatched G0 larvae were manually transferred onto larval diet and the surviving adults were crossed to virgin wild-type flies in a reciprocal manner. Fluorescence microscopy was used to identify marker-positive G1 progeny at the pupal stage of development, and selected adults were individually crossed with wild-type flies. Once established, homozygous transgenic lines were maintained through sibling crosses of 10 males and 20 females.
Inverse PCR
Unique piggyBac construct integrations were analyzed through inverse PCR in selected marker positive G1 individuals. In brief, genomic DNA (gDNA) was first extracted via a modified protocol developed previously56. The inverse PCR protocol was adapted from an established protocol57 utilizing Sau3AI (New England Biolabs®) and HinP1I (New England Biolabs®) restriction endonucleases for initial gDNA digestion. piggyBac-specific primers from58 were used for sequential PCR amplification. Sanger sequencing was carried out using Genewiz Inc. services and the resulting sequences were analyzed by blasting and thus mapping them to the latest C. capitata genome reassembly (GenBank GCA_905071925.1).
DsRed splicing confirmation
Adult males and females from transgenic lines were separately collected in TRIzol reagent (Ambion). RNA was extracted using an adjusted protocol from59 and Maxima H Minus First Strand cDNA Synthesis Kit with dsDNase (ThermoFisher) was used for cDNA synthesis. In parallel, gDNA was extracted as described above. PCR was performed using Phusion High-Fidelity PCR Master Mix with HF Buffer (New England Biolabs®) or RedTaq DNA Polymerase 2X Master Mix (VWR Life Science) using primers designed in Geneious Prime 2023.1.2 (Supplementary Table 5). The amplicons were visualized using a 1% agarose gel.
Sex sorting assay
For all homozygous strains two consecutive generations, G9 and G10, were screened to confirm system efficiency. Parental crosses between 10 male and 20 female homozygous individuals were established and eggs were collected twice 3 days apart for each cross. The offspring were reared until adulthood under regular conditions. At adulthood, all flies were assessed by 3 phenotypic parameters. These included phenotypic sex characteristics (male or female), as well as fluorescence marker phenotypes determined via separate screening with the GFP (GFP + or GFP-) and RFP (DsRed + or DsRed-) filters. MVX-ZB10 Macro Zoom Fluorescence Microscope System (Olympus) was used for all imaging and fluorescence screening.
Fitness assays
To assess fitness costs of two copies of sex-sorting genetic cassette, the number of eggs laid and rate of egg hatching of homozygous cassette-carrying and wild-type flies were evaluated. Genetic crosses between 15 males and 25 females from each strain (H-001, H-002, K-001, K-002) were set up in biological triplicates. Simultaneously, crosses of 15 male and 25 female wild-type Benakeion strain adults were also established in triplicates. After 5 days, eggs oviposited within a 5-h window were placed onto black filter paper on top of the larval diet and counted using ImageJ. The hatching rate was established 4 days after initial egg collection similarly through counting the remaining unhatched eggs.
Figure generation and statistical analysis
All graphs were generated in RStudio using the ggplot() function. Statistical analyses including chi-squared, Kruskal–Wallis and Dunn tests were similarly performed in RStudio. Diagrams, gel and image-centered figures were constructed in Microsoft PowerPoint.
Data availability
Complete plasmid sequences are available at addgene.org (#205482 and #205485). All data used to generate figures is provided in the Supplementary materials. Medfly transgenic lines are available upon request to A.M.
References
Papadopoulos, N. T. Fruit fly invasion: historical, biological, economic aspects and management. In: Anonymous Trapping and the Detection, Control, and Regulation of Tephritid Fruit Flies. Springer, pp. 219–252 (2014)
De Meyer, M., Robertson, M. P., Peterson, A. T. & Mansell, M. W. Ecological niches and potential geographical distributions of Mediterranean fruit fly (Ceratitis capitata) and Natal fruit fly (Ceratitis rosa). J. Biogeogr. 35(2), 270–281 (2008).
Gutierrez, A. P., Ponti, L., Neteler, M., Suckling, D. M. & Cure, J. R. Invasive potential of tropical fruit flies in temperate regions under climate change. Commun. Biol. 4(1), 1–14 (2021).
Liquido, N. J., Shinoda, L. A. & Cunningham, R. T. Host plants of the Mediterranean fruit fly (Diptera: Tephritidae): An annotated world review. (Entomological Society of America, 1991). https://books.google.co.uk/books?id=dSSzmQEACAAJ
Gilioli, G. et al. Non-linear physiological responses to climate change: The case of Ceratitis capitata distribution and abundance in Europe. Biol. Invasions 24(1), 261–279 (2022).
Lehmann, P. et al. Complex responses of global insect pests to climate warming. Front. Ecol. Environ. 18(3), 141–150 (2020).
Vera, M. T., Rodriguez, R., Segura, D. F., Cladera, J. L. & Sutherst, R. W. Potential geographical distribution of the Mediterranean fruit fly, Ceratitis capitata (Diptera: Tephritidae), with emphasis on Argentina and Australia. Environ. Entomol. 31(6), 1009–1022 (2002).
Klassen, W. & Vreysen, M.J.B. (2021) Area-wide integrated pest management and the sterile insect technique. In Sterile insect technique. 75–112
Knipling, E. F. Possibilities of insect control or eradication through the use of sexually sterile males. J. Econ. Entomol. 48(4), 459–462 (1955).
Krafsur, E. S. Sterile insect technique for suppressing and eradicating insect population: 55 years and counting. J. Agric. Entomol. 15(4), 303–317 (1998).
Lutrat, C. et al. Sex sorting for pest control: It’s raining men!. Trends Parasitol. 35(8), 649–662 (2019).
Caceres, C. Mass rearing of temperature sensitive genetic sexing strains in the Mediterranean fruit fly (Ceratitis capitata). Genetica 116(1), 107–116 (2002).
Hendrichs, J., Franz, G. & Rendon, P. Increased effectiveness and applicability of the sterile insect technique through male-only releases for control of Mediterranean fruit flies during fruiting seasons. J. Appl. Entomol. 119(1–5), 371–377 (1995).
Rendón, P., McInnis, D., Lance, D. & Stewart, J. Medfly (Diptera: Tephritidae) genetic sexing: Large-scale field comparison of males-only and bisexual sterile fly releases in Guatemala. J. Econ. Entomol. 97(5), 1547–1553 (2004).
Franz, G., Bourtzis, K. & Caceres, C. Practical and operational genetic sexing systems based on classical genetic approaches in fruit flies, an example for other species amenable to large-scale rearing for the sterile insect technique. In Sterile Insect Technique. 575–604 (2021)
McCombs, S. D. & Saul, S. H. Translocation-based genetic sexing system for the oriental fruit fly (Diptera: Tephritidae) based on pupal color dimorphism. Ann. Entomol. Soc. Am. 88(5), 695–698 (1995).
McInnis, D. O. et al. Development of a pupal color-based genetic sexing strain of the melon fly, Bactrocera cucurbitae (Coquillett) (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 97(5), 1026–1033 (2004).
Ramírez-Santos, E. et al. A novel genetic sexing strain of Anastrepha ludens for cost-effective sterile insect technique applications: Improved genetic stability and rearing efficiency. Insects 12(6), 499 (2021).
Augustinos, A. A. et al. Ceratitis capitata genetic sexing strains: Laboratory evaluation of strains from mass-rearing facilities worldwide. Entomol. Exp. Appl. 164(3), 305–317 (2017).
Franz, G. Recombination between homologous autosomes in medfly (Ceratitis capitata) males: Type-1 recombination and the implications for the stability of genetic sexing strains. Genetica 116(1), 73–84 (2002).
Papathanos, P. A. et al. Sex separation strategies: Past experience and new approaches. Malar. J. 8(2), 1–8 (2009).
Fu, G. et al. Female-specific insect lethality engineered using alternative splicing. Nat. Biotechnol. 25(3), 353–357 (2007).
Heinrichs, J. C. & Scott, M. J. A repressible female-specific lethal genetic system for making transgenic insect strains suitable for a sterile-release program. Proceed. Natl. Acad. Sci. 97(15), 8229–8232 (2000).
Kandul, N. P., Liu, J., Hsu, A. D., Hay, B. A. & Akbari, O. S. A drug-inducible sex-separation technique for insects. Nat. Commun. 11(1), 1–10 (2020).
Ogaugwu, C. E., Schetelig, M. F. & Wimmer, E. A. Transgenic sexing system for Ceratitis capitata (Diptera: Tephritidae) based on female-specific embryonic lethality. Insect Biochem. Mol. Biol. 43(1), 1–8 (2013).
Schetelig, M. F., Targovska, A., Meza, J. S., Bourtzis, K. & Handler, A. M. Tetracycline-suppressible female lethality and sterility in the Mexican fruit fly, Anastrepha ludens. Insect Mol. Biol. 25(4), 500–508 (2016).
Bopp, D., Saccone, G. & Beye, M. Sex determination in insects: Variations on a common theme. Sex. Dev. 8(1–3), 20–28 (2014).
Gomulski, L. M. et al. Gene discovery in an invasive tephritid model pest species, the Mediterranean fruit fly, Ceratitis capitata. BMC Genomics 9, 1–15 (2008).
Pane, A., Salvemini, M., Bovi, P. D., Polito, C. & Saccone, G. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 129(15), 3715–3725. https://doi.org/10.1242/dev.129.15.3715 (2002).
Salvemini, M. et al. Ceratitis capitata transformer-2 gene is required to establish and maintain the autoregulation of Cctra, the master gene for female sex determination. Int. J. Dev. Biol. 53(1), 109–120 (2003).
Laohakieat, K., Isasawin, S. & Thanaphum, S. The transformer-2 and fruitless characterisation with developmental expression profiles of sex-determining genes in Bactrocera dorsalis and B. correcta. Sci. Rep. 10(1), 1–13 (2020).
Ruiz, M. F. et al. The gene doublesex of Anastrepha fruit flies (Diptera, Tephritidae) and its evolution in insects. Dev. Genes Evol. 217, 725–731 (2007).
Saccone, G., Pane, A. & Polito, L. C. Sex determination in flies, fruitflies and butterflies. Genetica 116, 15–23 (2002).
Schetelig, M. F., Milano, A., Saccone, G. & Handler, A. M. Male only progeny in Anastrepha suspensa by RNAi-induced sex reversion of chromosomal females. Insect Biochem. Mol. Biol. 42(1), 51–57 (2012).
Shearman, D. & Frommer, M. The Bactrocera tryoni homologue of the Drosophila melanogaster sex-determination gene doublesex. Insect Mol. Biol. 7(4), 355–366 (1998).
Meccariello, A. et al. Maleness-on-the-Y (MoY) orchestrates male sex determination in major agricultural fruit fly pests. Science 365(6460), 1457–1460 (2019).
Primo, P. et al. Targeting the autosomal Ceratitis capitata transformer gene using Cas9 or dCas9 to masculinize XX individuals without inducing mutations. BMC Genet. 21(2), 1–11 (2020).
Chatzispyrou, I. A., Held, N. M., Mouchiroud, L., Auwerx, J. & Houtkooper, R. H. Tetracycline antibiotics impair mitochondrial function and its experimental use confounds research. Cancer Res. 75(21), 4446–4449 (2015).
Elzeinová, F. et al. Adverse effect of tetracycline and doxycycline on testicular tissue and sperm parameters in CD1 outbred mice. Exp. Toxicol. Pathol. 65(6), 911–917 (2013).
Jiao, Y. X. & Yang, Y. Combating mitochondrial toxicity: Looking at the past and where to go. Toxicol. Adv. 5(2), 5 (2023).
O’Shea, K. L. & Singh, N. D. Tetracycline-exposed Drosophila melanogaster males produce fewer offspring but a relative excess of sons. Ecol. Evol. 5(15), 3130–3139 (2015).
Weng, S. C., Antoshechkin, I., Marois, E. & Akbari, O. S. Efficient sex separation by exploiting differential alternative splicing of a dominant marker in Aedes aegypti. BioRxiv. 2023-06 (2023).
Liu, J., Rayes, D., & Akbari, O. S. (2023). A fluorescent sex-sorting technique for insects with the demonstration in Drosophila melanogaster. BioRxiv. 2023.08.11.553026.
Aumann, R. A., Häcker, I. & Schetelig, M. F. Female-to-male sex conversion in Ceratitis capitata by CRISPR/Cas9 HDR-induced point mutations in the sex determination gene transformer-2. Sci. Rep. 10(1), 1–12 (2020).
Meccariello, A., Hou, S., Davydova, S., Fawcett, J., Siddall, A., Leftwich, P. T., Krsticevic, F., Papathanos, P. A., & Windbichler, N. Gene drive and genetic sex conversion in the global agricultural pest Ceratitis capitata. BioRxiv. 2023.08.16.553191 (2023).
Chomczynski, P. & Mackey, K. Substitution of chloroform by bromo-chloropropane in the single-step method of RNA isolation. Anal. Biochem. 225(1), 163–164 (1995).
Handler, A. M. Use of the piggyBac transposon for germ-line transformation of insects. Insect Biochem. Mol. Biol. 32(10), 1211–1220 (2002).
Meza, J. S., Nirmala, X., Zimowska, G. J., Zepeda-Cisneros, C. S. & Handler, A. M. Development of transgenic strains for the biological control of the Mexican fruit fly, Anastrepha ludens. Genetica 139, 53–62 (2011).
Meza, J. S., Schetelig, M. F., Zepeda-Cisneros, C. S. & Handler, A. M. Male-specific Y-linked transgene markers to enhance biologically-based control of the Mexican fruit fly, Anastrepha ludens (Diptera: Tephritidae). BMC Genet. 15, 1–7 (2014).
Lagos, D., Koukidou, M., Savakis, C. & Komitopoulou, K. The transformer gene in Bactrocera oleae: the genetic switch that determines its sex fate. Insect Mol. Biol. 16(2), 221–230 (2007).
Peng, W., Zheng, W., Handler, A. M. & Zhang, H. The role of the transformer gene in sex determination and reproduction in the tephritid fruit fly, Bactrocera dorsalis (Hendel). Genetica 143, 717–727 (2015).
Sollazzo, G. et al. Genomic and cytogenetic analysis of the Ceratitis capitata temperature-sensitive lethal region. G3 Genes Genomes Genet. https://doi.org/10.1093/g3journal/jkad074 (2023).
Pane, A., De Simone, A., Saccone, G. & Polito, C. Evolutionary conservation of Ceratitis capitata transformer gene function. Genetics 171(2), 615–624 (2005).
Sollazzo, G. et al. Temperature sensitivity of wild-type, mutant and genetic sexing strains of Ceratitis capitata. Insects 13(10), 943 (2022).
Meccariello, A. et al. Engineered sex ratio distortion by X-shredding in the global agricultural pest Ceratitis capitata. BMC Biol. 19(1), 1–14 (2021).
Eckermann, K. N. et al. Hyperactive piggyBac transposase improves transformation efficiency in diverse insect species. Insect Biochem. Mol. Biol. 98, 16–24 (2018).
Holmes, D. S. & Bonner, J. Preparation, molecular weight, base composition, and secondary structure of giant nuclear ribonucleic acid. Biochemistry 12(12), 2330–2338 (1973).
Scolari, F. et al. Fluorescent sperm marking to improve the fight against the pest insect Ceratitis capitata (Wiedemann; Diptera: Tephritidae). New Biotechnol. 25(1), 76–84 (2008).
Galizi, R. et al. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat. Commun. 5(1), 3977 (2014).
Funding
This work was supported by cooperative agreements (22-8130-1007-CA and AP22PPQs&t00C188) between the United States Department of Agriculture (USDA)—Animal and Plant Health Inspection Service (APHIS)—Plant Protection and Quarantine (PPQ) and Imperial College London and University of California—San Diego, respectively. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture, an equal opportunity employer.
Author information
Authors and Affiliations
Contributions
O.S.A, N.P.K, A.M and E.B conceived the project and directed the research. J.L and N.P.K designed constructs and J.L performed the cloning. A.M and S.D designed the experiments and S.D performed the medfly and molecular validation experiments. A.M performed the germline transformation. A.M and S.D analyzed the data. All authors contributed to writing and editing of the manuscript and approved the final article.
Corresponding author
Ethics declarations
Competing interests
O.S.A is a founder of Agragene, Inc. and Synvect, Inc. with equity interest. N.P.K is a founder of Synvect, Inc. with equity interest. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. All other authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Davydova, S., Liu, J., Kandul, N.P. et al. Next-generation genetic sexing strain establishment in the agricultural pest Ceratitis capitata. Sci Rep 13, 19866 (2023). https://doi.org/10.1038/s41598-023-47276-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-47276-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.