Abstract
Studies indicate EGFL7 as an important gene in controlling angiogenesis and cancer growth, including in colorectal cancer (CRC). Anti-EGFL7 agents are being explored, yet without promising results. Therefore, the role of EGFL7 in CRC carcinogenesis should be investigated. This study aimed to evaluate the prognostic value of EGFL7 expression in CRC and the signaling pathways influenced by this gene. EGFL7 expression was evaluated through immunohistochemistry in 463 patients diagnosed with CRC and further associated with clinicopathological data, angiogenesis markers and survival. In silico analyzes were performed with colon adenocarcinoma data from The Cancer Genome Atlas. Analysis of enriched gene ontology and pathways were performed using the differentially expressed genes. 77.7% of patients presented low EGFL7 expression, which was associated with higher lymph node spread and invasion of lymphatic vessels, with no impact on survival. Additionally, low EGFL7 expression was associated with high VEGFR2 expression. Finally, we found in silico that EGFL7 expression was associated with cell growth, angiogenesis, and important pathways such as VEGF, Rap-1, MAPK and PI3K/Akt. Expression of EGFL7 in tumor cells may be associated with important pathways that can alter functions related to tumor invasive processes, preventing recurrence and metastatic process.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the fourth most lethal and incident neoplasia around the world, being responsible for about 10% of the diagnostics and deaths associated with cancer1. The standard treatment relies in surgery, radiotherapy and/or chemotherapy, but the choice of treatment depends on disease stage, pathological characteristics, microsatellite instability status, genomic alterations, among others2.
About 35% of the patients present with metastatic disease, and up to 50% of those who do not present metastasis at the diagnosis will evolve with metastasis at some point of the disease3. In fact, angiogenesis and lymphangiogenesis are important factors in tumor maintenance and metastasization4. The lymphatic vessels can be influenced by tumor-derived growth factors, which can lead to lymphatic remodeling, immune function modulation, facilitating metastasization to lymph nodes and distant organs5. The survival, proliferation and migration of endothelial cell during the process of lymphangiogenesis depends on activation of VEGFR2/VEGFR3 receptors by VEGFC/VEGFD, stimulating the activation of protein kinase C of ERK1, ERK2 or PI3K-AKT pathways5.
Besides the canonical activators, studies have pointed to alternative ligands capable of modulating vessels growth. Among them, EGFL7 has been studied6, since it has a specific role in vascular tubulogenesis and angiogenesis regulation7. It has been described as an alternative Notch ligand, integrin and MAPK pathways8. Studies have shown that high EGFL7 expression is associated with poor prognosis in different tumor types, such as gastric cancer and CRC9,10.
The use of bevacizumab, an anti-VEGF monoclonal antibody11, combined with chemotherapy in mCRC patients showed promising results such as reduction of tumor size, increased overall survival / progression-free survival in patients with liver metastases and reduction of circulating EGFL7, associated with VEGFA reduction11,12,13. In 2013, Johnson and colleagues proposed the use of parsatuzumab, an anti-EGFL7 monoclonal antibody, for solid tumors treatment14. However, the phase-II studies associating chemotherapy + bevacizumab with parsatuzumab in CRC and non-small cell lung cancer did not show favorable or significant results15,16. Given the evidences that high EGFL7 expression led to poor prognostic events in CRC, and parsatuzumab studies did not show promisor results, the contribution of EGFL7 in CRC development, maintenance and metastization should be better investigated in order to propose more efficient treatment protocols.
Therefore, the aim of the present study is to evaluate the prognostic role of EGFL7 expression in a series of a well characterized CRC cohort and evaluate in silico the biological functions and pathways associated with differential expression of EGFL7.
Material and methods
Patients and tissue samples
Colorectal cancer samples were obtained from a well-characterized series of 463 patients who had undergone surgery at University of Minho17. The samples were collected from patients who underwent surgical excision of the primary tumor at Hospital of Braga (Portugal) between January of 2005 and January of 2010 and were classified by an experienced pathologist. Tumor localization was recorded and classified as colon and rectum (between anal verge and 15 cm at rigid rectoscopy). Hematoxylin and eosin staining was performed, and representative areas of the tumor were selected for tissue microarray construction. Each case was represented in the tissue microarray (TMA) by at least two cores of 0.6 mm.
Imunohistochemistry
The 5-μm-thick sections were deparaffinized and rehydrated, and immunostaining was performed according to Brunhara et al.18. There was performed antigen retrieval for 20 min at 98 °C in tris–EDTA buffer and endogenous peroxidase and protein blocking were performed using Novolink Polymer Detection System (Leica Biosystems, UK). The slides were subsequently incubated with rabbit polyclonal anti-EGFL7 antibody (catalog number ab115786, Abcam, Cambridge, MA) 1:100 for 90 min at room temperature (RT). Post-primary antibody and polymer from Novolink Polymer Detection System were then placed on the slides (30 min each at RT) and chromogen color development was accomplished with 3,3’-diaminobenzidine (DAB), with a Gill-2 hematoxylin counterstain. Endothelial cells were used as an internal positive control since this antibody also labels endothelium.
The slides were blindly scored by an expert pathologist (G.A.L.) using 0 to 3 + scores19. The expression of EGFL7 was considered low if the score was 0–2 + ; otherwise, the expression was considered high.
The immunostaining data was associated with clinicopathological (age, gender, clinical and personal history of CRC, clinical presentation, duration of symptoms, location, CEA, presence of metastasis, tumor size, histological type, differentiation, lymph node invasion, vascular and lymphatic invasion, clinical staging and recurrence, follow-up and status) and immunohistochemical data of angiogenesis/lymphoangiogenesis markers—VEGFA, VEGFC, VEGFR2 and VEGFR3, retrieved from Martins et al.17. Finally, survival analysis was performed using Kaplan–Meier curves. Overall survival was defined as the period of analysis until death from any cause; relapse-free survival was defined as the period of analysis until any relapse detected.
The population of study was characterized by descriptive analysis, and the association between EGFL7 immunostaining and clinicopathological or angiogenesis/lymphoangiogenesis markers was performed using chi-square or Fisher’s exact test. Log rank test was performed to compare overall and relapse-free survival of patients presenting low and high EGFL7 expression. In order to analyze the impact of EGFL7 on prognosis for each tumor stage, the patients were separated by stage (1 to 4) and log rank test was performed to compare overall and relapse-free survival depending on EGFL7 expression. The results were considered statistically significant when P ≤ 0.05.
In silico analysis
In order to analyze the molecular impact associated with EGFL7 expression in CRC, RNA sequencing data of colon adenocarcinoma (COAD) from TCGA was analyzed. In silico analysis was performed using RTCGAToolbox20 and TCGAbiolinks21. Normalized Illumina HiSeq RSEM data from COAD was obtained, and Z-score of EGFL7 reads was calculated for each patient. The patients were stratified in high EGFL7 expression (above 3rd quartile, n = 109), and low expression (below 1st quartile, n = 109).
The analysis of differentially expressed genes was performed using eBayes test implemented in LIMMA22, using as contrast the patients with high vs. low EGFL7 expression. The genes presenting False Discovery Rate (FDR) ≤ 0.05 and Fold Change ≥|2.0| were considered differentially expressed.
The list of differentially expressed genes was submitted to Enriched Gene Ontology (GO) gene set enrichment analyses and Kyoto Encyclopedia of Genes and Genomes (KEGG)23,24 enrichment analyses using clusterProfiler25. Similarity of the terms was determined using enrichplot package implemented in R. The GO and KEGG terms were considered statistically significant when FDR ≤ 0.05.
Ethical approval
This study protocol was reviewed and approved by the Ethics Committee of University of Minho (number 32/2013) and Barretos Cancer Hospital (number 1955/2020). This study was performed in line with the principles of the Declaration of Helsinki.
Consent participate
Written informed consent was obtained from participants to participate in the study.
Results
Imunohistochemistry
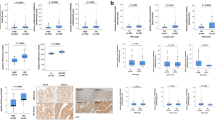
We found high expression of EGFL7 protein by immunohistochemistry in 103 out of 463 patients (22.3%). The staining was essentially cytoplasmatic in all cases (shown in Fig. 1). There was found no association of EGFL7 expression and clinopatiological features (Table 1). The analysis of EGFL7 expression and pathological data showed that low EGFL7 expression was associated with presence of spread to lymph nodes and lymphatic vessel invasion (P < 0.05, Table 1). Moreover, there was found association between low EGFL7 expression and the expression of VEGFR2 (P < 0.05, Table 2).
Representative immunohistochemical expression of EGFL7 in normal colorectal tissue (A and B) and colon adenocarcinoma (C and D). A. enterocytes weakly cytoplasmatic positive (+/3+); B. enterocytes strongly positive (+++/3+) with minor background on stromal cells; C. adenocarcinoma with negative expression; D. adenocarcinoma mildly citoplasmatic positive (++/3+). Scale bars of 20 micrometers.
Overall and recurrence-free survival was not different between low and high expression of EGFL7 in colorectal cancer (shown in Fig. 2). The overall survival (OS) of 50% of the patients was close to 110 months. Regarding tumor localization (ascending colon, descending colon, or rectum, shown in Supplementary Fig. 1), there was no difference in OS and RFS in the comparison of patients with low and high expression of EGFL7. Finally, although obvious differences in survival can be noted among tumor stages (stages 1, 2, 3 or 4, shown in Supplementary Fig. 2), the comparison of the curves (low EGFL7 expression vs. high EGFL7 expression) within each stage (stages 1, 2, 3 or 4) was not statistically different (P > 0.05).
In silico analysis
In order to characterize the pathways and biological processes associated with EGFL7 expression, we performed in silico analysis using RNA sequencing data from The Cancer Genome Atlas (TCGA). Differential expression of patients presenting high EGFL7 expression vs. low EGFL7 expression presented 1,718 differentially expressed genes, being 103 downregulated and 1,615 upregulated (Supplementary Table 1).
Enriched gene ontology (GO) gene set enrichment analysis revealed 24 enriched terms. Of note, 8 terms (shown in Fig. 3) were aggregated under a cluster associated with growth/development. The genes present in the GO terms are found in Supplementary Table 2.
Enriched KEGG analysis revealed 75 enriched pathways. Of note, we found a cluster which include several pathways associated with cancer development (Focal adhesion, ECM-receptor interaction, PI3K-Akt signaling pathway, MAPK signaling pathway, Ras signaling pathway, RAP1 signaling pathway and Proteoglycans in cancer) (shown in Fig. 4). The genes present in the KEGG terms are found in Supplementary Table 2. Of note, Wnt signaling pathway was also enriched in our analysis (shown in Fig. 4, Supplementary Table 2).
Discussion
The present study aimed to evaluate the prognostic potential of EGFL7 expression in patients diagnosed with CRC and to propose biological processes and pathways altered by the differential expression of this gene. We found that the low expression of EGFL7 in neoplastic cells was associated with greater lymph node involvement and lymphatic vessel invasion, possibly due to dysregulation of carcinogenesis-related processes (cell growth, cell adhesion, angiogenesis) through important pathways such as proteoglycans in cancer, Ras, Rap1, MAPK and PI3K/Akt.
There are several studies regarding EGFL7 expression in different tumor types. The high expression of this gene was found as a marker of worse prognosis in several tumor types9,10,26. In addition, the interaction of EGFL7 and EGFR leads to the activation of important downstream pathways related to the development and growth of several types of tumors. High expression of EGFL7 in gliomas26 and in metastatic gastric cancer27 promotes the activation of the AKT and ERK pathways through the interaction in EGFR; in hepatocellular cancer28 EGFL7 promotes metastasis through activating FAK phosphorylation by binding EGFR. Additionally, in renal cancer29, the activation of the EGFL7/EGFR/FAK pathway induces the migration of endothelial cells, inducing the formation of vascular tubes contributing to tumor progression. Overall, the EGFL7/EGFR signaling pathway may play an important role in intratumoral angiogenesis, metastasis and invasion8. We previously observed that high EGFL7 expression was associated with worse clinical outcome in patients diagnosed with pilocytic astrocytoma18, and worse survival and lower Karnofsky Performance Score in glioblastoma30.
Hansen et al. analyzed the expression of EGFL7 to estimate vessel area in mCRC, and found positive association with KRAS mutation31. Although the mechanism is still poorly characterized, possibly the increase in MAPK pathway can upregulate the expression of EGFL7, leading to an increase in (lymph)angiogenesis and tumor aggressiveness32. Subsequently, they described that the intratumoral endothelial expression of this protein is higher in primary tumors of patients diagnosed with stage II or III CRC who had recurrence33. Finally, this group described reduction of circulating EGFL7 of mCRC patients after chemotherapy13. In addition, a worse prognosis was found in those who had high amounts of baseline cir-EGFL7 before treatment13. In patients with liver metastases who underwent bevacizumab-based chemotherapy followed by surgical resection, low intratumoral expression of EGFL7 mRNA in metastases was associated with higher disease-free survival34. In addition to these encouraging data, the study by Hansen et al.33 suggests an important predictive value of EGFL7-positive vascular area in relation to first-line chemotherapy and bevacizumab for CRC and suggests the use of a dual VEGFA-EGFL7 blocking mechanism. In contrast, our data suggest that EGFL7 expression in tumor parenchyma is not associated with differences in overall survival and, additionally, low expression is associated with increased lymph node spread and invasion of lymphatic vessels of metastatic colorectal cancer. We suggest that the literature data found analyzing only EGFL7 expression in vessels13,31,33 may be more associated with angiogenesis per se than to the effects of EGFL7 expression.
This evidence can be indirectly supported by ongoing studies, such as Garcia-Carbonero and colleagues15, who showed that treatment with parsatuzumab (anti-EGFL7 antibody that selectively blocks the interaction of EGFL7 and endothelial cells) failed to improve the efficacy of FOLFOX + bevacizumab combination in patients with mCRC. Similarly, another phase II randomized clinical trial16 showed that administration of parsatuzumab to non-small cell lung cancer patients did not improve treatment (bevacizumab + carboplatin/placlitaxel) efficacy; still, patients who received parsatuzumab had lower progression-free survival than placebo arm. Therefore, the effect of blocking EGFL7 expression by parsatuzumab led to exiguous results compared to blocking VEGF, suggesting that the main effect of this combination was associated with angiogenesis and not to EGFL7 expression.
Our in silico data show enriched biological processes and pathways related to growth, mesenchyme development, regulation of PI3K/Akt, MAPK, Wnt signaling and Rap1 pathway, thus strengthening the potential molecular mechanism of EGFL7 in mediating CRC35. Activation of the PI3K/Akt pathway together with mTOR can regulate several biological processes important for growth, metabolism, autophagy, and angiogenesis36. This pathway regulates angiogenesis by increasing VEGF secretion, modulating the expression of NO and angiopoietins36,37. The binding of VEGF to receptors on endothelial cells stimulates the activation of this pathway, which is essential for endothelial cell migration, being fundamental for the development of blood vessels37. VEGFR2 activation increases signaling of several pathways, such as MAPK and PI3K/Akt/mTOR38, found altered in our in silico analysis. Altogether, the positive expression of VEGFR2 may have increased lymphangiogenesis in patients with low EGFL7 expression. This, therefore, led to greater lymph node spread, since the lymphatic pathway is the main route of spreading of neoplastic cells in CRC6. Importantly, in CRC, the involvement of lymph node and lymphatic invasion are important factors to be considered when determining treatment6.
In addition to the dysregulation of the widely studied pathways in carcinogenesis cited above, our in silico analysis also found other less studied pathways and biological processes, such as the Rap1. RAP1 has potential to regulate and mediate Ras functions, as well as being related to many of the characteristics of cancer39, acting as a central regulator of adhesion, motility cellularity, cell polarity, and migration39. Furthermore, RAP1 promotes vascular endothelial growth factor receptor 2 (VEGFR2) activation and angiogenesis through integrins. Thus, RAP1 plays an important role in invasion and metastasis due to its regulation of cell adhesion and cytoskeletal remodeling through ERK/MAPK signaling and integrin activation40. There is evidence that RAP1 activation promotes tumorigenesis in several systems39. In CRC, activation of RAP1 resulted in impaired cell adhesion and increased cell–matrix adhesion, inducing the spread of neoplastic cells. Therefore, activation of RAP1 is associated with several biological processes such as cellular metabolism, cytoskeletal remodeling, cell proliferation, migration and metastasis through the regulation of downstream pathways such as ERK, AKT, FAK and Wnt36,40.
We conclude, therefore, that the low expression of EGFL7 in the tumor cells of patients diagnosed with CRC may be associated with high expression of VEGF2, thus leading to an increase in lymphatic invasion and greater lymphangiogenesis. Our in silico analysis indicates that EGFL7 expression is associated with important pathways related to carcinogenesis and lymphangiogenesis. Further studies are needed to validate the findings identified in silico, and to lighten the association of these results with clinicopathological findings to elucidate the mechanism of EGFL7 in the genesis of CRC, in order to propose adequate treatment approaches for colorectal cancer using EGFL7 as possible biomarker.
Data availability
The datasets analyzed during the current study are available in the TCGA repository, [https://www.cancer.gov/tcga].
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries CA Cancer. J. Clin. https://doi.org/10.3322/caac.21660 (2021).
Boland, C. R., Sinicrope, F. A., Brenner, D. E. & Carethers, J. M. Colorectal cancer prevention and treatment. Gastroenterology 118, S115-128 (2000).
Piawah, S. & Venook, A. P. Targeted therapy for colorectal cancer metastases: A review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer 125, 4139–4147. https://doi.org/10.1002/cncr.32163 (2019).
Adams, R. H. & Alitalo, K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat. Rev. Mol. Cell Biol. 8, 464–478. https://doi.org/10.1038/nrm2183 (2007).
Stacker, S. A. et al. Lymphangiogenesis and lymphatic vessel remodelling in cancer Nature reviews. Cancer 14, 159–172. https://doi.org/10.1038/nrc3677 (2014).
Sundlisaeter, E. et al. Lymphangiogenesis in colorectal cancer–prognostic and therapeutic aspects. Int. J. Cancer 121, 1401–1409. https://doi.org/10.1002/ijc.22996 (2007).
Parker, L. H. et al. The endothelial-cell-derived secreted factor Egfl7 regulates vascular tube formation. Nature 428, 754–758. https://doi.org/10.1038/nature02416 (2004).
Hong, G. et al. EGFL7: Master regulator of cancer pathogenesis, angiogenesis and an emerging mediator of bone homeostasis. J. Cell Physiol. https://doi.org/10.1002/jcp.26792 (2018).
Deng, Q. J., Xie, L. Q. & Li, H. Overexpressed MALAT1 promotes invasion and metastasis of gastric cancer cells via increasing EGFL7 expression. Life Sci. 157, 38–44. https://doi.org/10.1016/j.lfs.2016.05.041 (2016).
Fan, C. et al. The expression of Egfl7 in human normal tissues and epithelial tumors. Int. J. Biol. Markers 28, 71–83. https://doi.org/10.5301/JBM.2013.10568 (2013).
Rosen, L. S., Jacobs, I. A. & Burkes, R. L. Bevacizumab in colorectal cancer: Current role in treatment and the potential of biosimilars target. Oncol 12, 599–610. https://doi.org/10.1007/s11523-017-0518-1 (2017).
Cremolini, C. et al. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: Updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 16, 1306–1315. https://doi.org/10.1016/S1470-2045(15)00122-9 (2015).
Hansen, T. F., Andersen, R. F., Olsen, D. A., Sorensen, F. B. & Jakobsen, A. Prognostic importance of circulating epidermal growth factor-like domain 7 in patients with metastatic colorectal cancer treated with chemotherapy and bevacizumab. Sci. Rep. 7, 2388. https://doi.org/10.1038/s41598-017-02538-x (2017).
Johnson, L. et al. Anti-EGFL7 antibodies enhance stress-induced endothelial cell death and anti-VEGF efficacy. J. Clin. Investig. 123, 3997–4009. https://doi.org/10.1172/JCI67892 (2013).
Garcia-Carbonero, R. et al. Randomized phase II trial of parsatuzumab (Anti-EGFL7) or placebo in combination with FOLFOX and bevacizumab for first-line metastatic colorectal cancer. Oncologist 22, 375-e330. https://doi.org/10.1634/theoncologist.2016-0133 (2017).
von Pawel, J. et al. Randomized phase II Trial of Parsatuzumab (Anti-EGFL7) or placebo in combination with carboplatin paclitaxel, and bevacizumab for first-line nonsquamous non-small cell lung cancer. Oncologist 23, 654-e658. https://doi.org/10.1634/theoncologist.2017-0690 (2018).
Martins, S. F. et al. Clinicopathological correlation and prognostic significance of VEGF-A, VEGF-C, VEGFR-2 and VEGFR-3 expression in colorectal cancer. Cancer Genom. Proteom. 10, 55–67 (2013).
Brunhara, B. B. et al. Evaluation of the prognostic potential of EGFL7 in pilocytic astrocytomas. Neuropathology https://doi.org/10.1111/neup.12698 (2020).
Nikolic, I. et al. EGFL7 ligates alphavbeta3 integrin to enhance vessel formation. Blood 121, 3041–3050. https://doi.org/10.1182/blood-2011-11-394882 (2013).
Samur, M. K. RTCGAToolbox: A new tool for exporting TCGA Firehose data. PLoS One 9, e106397. https://doi.org/10.1371/journal.pone.0106397 (2014).
Colaprico, A. et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44, e71. https://doi.org/10.1093/nar/gkv1507 (2016).
Smyth, K. G. Limma: Linear models for microarray data. Bioinformatics and Computational Biology Solutions using R and Bioconductor Springer, New York, NY. doi: https://doi.org/10.1007/0-387-29362-0_23 (2005).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592. https://doi.org/10.1093/nar/gkac963 (2023).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
Wang, F. Y. F. et al. EGFL7 is an intercellular EGFR signal messenger that plays an oncogenic role in glioma. Cancer Lett. 384, 9–18. https://doi.org/10.1016/j.canlet.2016.10.009 (2017).
Luo, B. H. et al. Epidermal growth factor-like domain-containing protein 7 (EGFL7) enhances EGF receptor-AKT signaling, epithelial-mesenchymal transition, and metastasis of gastric cancer cells. PLoS One 9, e99922. https://doi.org/10.1371/journal.pone.0099922 (2014).
Wu, F. et al. Novel role for epidermal growth factor-like domain 7 in metastasis of human hepatocellular carcinoma. Hepatology 50, 1839–1850. https://doi.org/10.1002/hep.23197 (2009).
Xu, H. F. et al. Targeting EGFL7 expression through RNA interference suppresses renal cell carcinoma growth by inhibiting angiogenesis. Asian Pacific J. Cancer Prev. 15, 3045–3050. https://doi.org/10.7314/apjcp.2014.15.7.3045 (2014).
da Costa, B. H. B. et al. EGFL7 expression profile in IDH-wildtype glioblastomas is associated with poor patient outcome. J. Pathol. Transl. Med. 56, 205–211. https://doi.org/10.4132/jptm.2022.04.22 (2022).
Hansen, T. F., Nielsen, B. S., Jakobsen, A. & Sorensen, F. B. Intra-tumoural vessel area estimated by expression of epidermal growth factor-like domain 7 and microRNA-126 in primary tumours and metastases of patients with colorectal cancer: A descriptive study. J. Transl. Med. 13, 10. https://doi.org/10.1186/s12967-014-0359-y (2015).
Massimiani, M. et al. Epidermal growth factor-like domain 7 promotes migration and invasion of human trophoblast cells through activation of MAPK, PI3K and NOTCH signaling pathways. Mol. Hum. Reprod. 21, 435–451. https://doi.org/10.1093/molehr/gav006 (2015).
Hansen, T. F., Nielsen, B. S., Sorensen, F. B., Johnsson, A. & Jakobsen, A. Epidermal growth factor-like domain 7 predicts response to first-line chemotherapy and bevacizumab in patients with metastatic colorectal cancer. Mol. Cancer Ther. 13, 2238–2245. https://doi.org/10.1158/1535-7163.MCT-14-0131 (2014).
Stremitzer, S. et al. Expression of genes involved in vascular morphogenesis and maturation predicts efficacy of bevacizumab-based chemotherapy in patients undergoing liver resection. Mol. Cancer Ther. 15, 2814–2821. https://doi.org/10.1158/1535-7163.MCT-16-0275 (2016).
Yeung, S. et al. Abstract 3295: Inhibiting vascular morphogenesis in tumors: EGFL7 as a novel therapeutic target. Cancer Res. 71, 3295–3295. https://doi.org/10.1158/1538-7445.Am2011-3295 (2011).
Shahcheraghi, S. H. et al. Wnt/beta-catenin and PI3K/Akt/mTOR signaling pathways in glioblastoma: Two main targets for drug design: A review. Curr. Pharm. Des. 26, 1729–1741. https://doi.org/10.2174/1381612826666200131100630 (2020).
Karar, J. & Maity, A. PI3K/AKT/mTOR pathway in angiogenesis. Front Mol. Neurosci. 4, 51. https://doi.org/10.3389/fnmol.2011.00051 (2011).
Simons, M., Gordon, E. & Claesson-Welsh, L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 17, 611–625. https://doi.org/10.1038/nrm.2016.87 (2016).
Shah, S., Brock, E. J., Ji, K. & Mattingly, R. R. Ras and Rap1: A tale of two GTPases. Semin. Cancer Biol. 54, 29–39. https://doi.org/10.1016/j.semcancer.2018.03.005 (2019).
Bos, J. L., de Rooij, J. & Reedquist, K. A. Rap1 signalling: Adhering to new models. Nat. Rev. Mol. Cell Biol. 2, 369–377. https://doi.org/10.1038/35073073 (2001).
Funding
This work was supported by The São Paulo Research Foundation (grant number 2021/14253–4 to Bidinotto LT); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) fellowships to Oliveira C and Golçalves PG; National Council for Scientific and Technological Development (CNPq) (productivity fellowship to Reis RM).
Author information
Authors and Affiliations
Contributions
C.O., S.F.F.M., P.G.G., A.L.F. performed development of methodology, acquisition and analysis of the data; GAL and CO performed the histopathological analysis of the patients; S.F.F.M. and A.L.F. performed the acquisition of the material; S.F.F.M. and L.T.B. performed the statistical analysis; C.O. and L.T.B. performed the writing of the paper; R.M.R. and A.L.F. performed the conception of the study, design and review of the paper; L.T.B. performed the conception of the study, design, writing of the paper, interpretation of the data, and review of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
de Oliveira, C., Martins, S.F.F., Gonçalves, P.G. et al. Low EGFL7 expression is associated with high lymph node spread and invasion of lymphatic vessels in colorectal cancer. Sci Rep 13, 19783 (2023). https://doi.org/10.1038/s41598-023-47132-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-47132-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.