Abstract
Myosin IC, a single-headed member of the myosin I family, specifically interacts with anionic phosphatidylinositol 4,5-bisphosphate (PI[4,5]P2) in the cell membrane via the pleckstrin homology domain located in the myosin IC tail. Myosin IC is widely expressed and physically links the cell membrane to the actin cytoskeleton; it plays various roles in membrane-associated physiological processes, including establishing cellular chirality, lipid transportation, and mechanosensing. In this study, we evaluated the motility of full-length myosin IC of Drosophila melanogaster via the three-dimensional tracking of quantum dots bound to actin filaments that glided over a membrane-bound myosin IC-coated surface. The results revealed that myosin IC drove a left-handed rotational motion in the gliding actin filament around its longitudinal axis, indicating that myosin IC generated a torque perpendicular to the gliding direction of the actin filament. The quantification of the rotational motion of actin filaments on fluid membranes containing different PI(4,5)P2 concentrations revealed that the rotational pitch was longer at lower PI(4,5)P2 concentrations. These results suggest that the torque generated by membrane-bound myosin IC molecules can be modulated based on the phospholipid composition of the cell membrane.
Similar content being viewed by others
Introduction
Myosin IC is a single-headed, membrane-bound, actin-based motor protein that is widely expressed in both invertebrates and vertebrates1. Myosin IC comprises an N-terminal motor domain, a neck region containing a light chain-binding domain (LCBD), and a relatively short tail domain2. The motor domain, which hydrolyzes adenosine triphosphate (ATP), interacts with the actin filament to generate mechanical force. The LCBD of myosin IC contains three isoleucine–glutamine motifs that bind to calmodulins and calmodulin-like proteins and function as a mechanical lever arm2. The tail domain contains the pleckstrin homology (PH) domain that specifically interacts with phosphatidylinositol 4,5-bisphosphate (PI[4,5]P2) in the cell membrane. PI(4,5)P2 and its derivatives are considered minor cell membrane components and function as signaling molecules in various pathways3. Myosin IC physically connects and generates forces between the cortical actin filaments and membrane lipids4,5. Furthermore, it plays different roles in various physiological processes, including cargo transportation4,6, lipid raft trafficking7, and mechanosensing in the stereocilia of the inner ear8. Myosin IC and myosin ID, another member of the myosin I family, play key roles in establishing the left–right axis in the bilateria. In Drosophila melanogaster, myosin IC overexpression modifies the cell shape chirality and alters the left–right axis of organs9,10. Recently, a detailed kinetics analysis of Drosophila myosin IC was performed11; however, the relationship between the motor function-associated mechanical properties of membrane-bound myosin IC and its physiological role remains unclear.
The cell membrane, which separates the cytoplasm from the extracellular environment, also serves as a two-dimensional reaction interface for membrane proteins and signaling molecules for several functions, including cell migration and cell–cell interactions12. Previous in vitro studies have investigated the motor properties of myosin family members associated with supported lipid bilayers (SLBs)13,14,15, a widely used cell membrane model system16,17. Although membrane fluidity may decrease the transfer of the force generated by motor proteins to cytoskeletal filaments, motor proteins anchored to the SLBs can move cytoskeletal filaments13,18,19,20,21. Regarding myosin IC, actin filament gliding has been observed in SLBs containing physiological concentrations of PI(4,5)P214,15,22. Furthermore, myosin IC-induced asymmetric motility of actin filaments, where gliding actin filaments turn in leftward circles, was increased on SLBs14. Further investigation of motility resulting from the interaction of membrane-bound myosin IC and actin filaments is needed to understand the biophysical properties of myosin IC and its physiological functions in the cell.
Some myosins, such as myosin II, V, VI, and X, exhibit unique mechanical properties and can generate a directional force along (on-axis) and perpendicular to (off-axis) the longitudinal axis of the actin filament. The off-axis force generated by myosin II (heavy meromyosin) has been observed via supercoiling formation caused by the rotational motion of a gliding actin filament around its longitudinal axis, with its leading part selectively fixed to the glass substrate23. This torque component of the cross-bridge force was also indicated by detecting the orientation of individual fluorophores sparsely labeled to the actin filament, which was free to glide over a surface coated with myosin II24,25 or V25. The helical motion of myosin V, VI, or X around the freely suspended actin filament was observed through defocused imaging26,27, polarized light imaging28, or Parallax, a technique for three-dimensional (3D) nanometer-scale tracking29,30. These in vitro experiments were only performed using myosins anchored to rigid surfaces, such as glass coverslips, microbeads, or quantum dots (QDs). However, the effects on torque generation of myosin motors anchored to a fluid lipid bilayer closer to the intracellular environment have not yet been thoroughly examined.
Herein, we investigated if unconventional, lipid membrane-bound single-headed myosin IC molecules could generate torque to rotate the gliding actin filaments along their longitudinal axes. We performed in vitro gliding assays on SLBs using full-length myosin IC molecules of Drosophila melanogaster (Drosophila myosin IC) and observed the displacement of QDs bound to actin filaments using a three-dimensional prismatic optical tracking (tPOT) microscope31,32. We report that a rotational motion is induced by myosin IC anchored to PI(4,5)P2-containing SLBs in gliding actin filaments around their longitudinal axes, a motion we termed corkscrewing. Furthermore, we assessed the gliding velocity and pitch of the corkscrewing motion based on varying PI(4,5)P2 concentrations in SLBs to determine the involvement of a lipid composition-dependent regulatory mechanism for torque generation by myosin IC. Additionally, we discussed the possible underlying mechanisms of torque generation by myosin IC molecules and their intracellular functions.
Result
SLB-bound myosin IC drove the left-handed corkscrewing motion of actin filaments
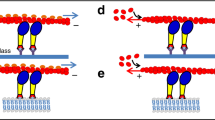
To gain insights into the motility of myosin IC anchored to a diffusive lipid environment, we examined the 3D motion of a QD bound to a gliding actin filament driven by SLB-bound full-length Drosophila myosin IC. SLBs are highly viscous fluids and can function as a substrate that transmits the myosin-generated force to drive the gliding of actin filaments13,14,15,18. SLBs are formed by fusing small unilamellar vesicles (SUVs) comprising the neutral phospholipid 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) with the anionic phospholipid PI(4,5)P2 on hydrophilic glass coverslips. We used SLBs containing 6% PI(4,5)P2 and 94% DOPC for the standard gliding assay. To assess the lipid fluidity of SLBs, fluorescence recovery after photobleaching (FRAP) experiments were performed (Supplementary Fig. 1a). The diffusion coefficient of the lipids in SLBs (DLIPID) with 0.05% (mol/mol) ATTO647N–1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) as the fluorescent tracer was 2.7 ± 0.4 µm2 s−1 (n = 5, 4 independent SLBs; Supplementary Fig. 1b and Supplementary Table 1) by fitting the fluorescence recovery curve14; this was consistent with the values reported in previous studies14,19,33,34. Furthermore, we prepared SLBs containing 1% (mol/mol) 1,2-distearoyl-sn-glycero-3-phosphoethanolamine–N-(biotinyl[polyethyleneglycol]-2000) (DSPE–PEG-2000-biotin) in a flow chamber and added fluorescent streptavidin (Avi-488) to the flow chamber so that Avi-488 was anchored to DSPE–PEG-2000-biotin. To measure the diffusivity of Avi-488 in the SLBs, individual Avi-488 molecules were tracked using a total internal reflection fluorescence (TIRF) microscope to obtain the single-molecule trajectories (Supplementary Fig. 1c). The ensemble average diffusion coefficient of Avi-488 (DAvi) was 1.1 ± 0.3 µm2 s−1 (n = 56 tracked molecules, 11 independent experiments; Supplementary Fig. 1d and Supplementary Table 1) by fitting the mean square displacement as a function of time; the result was consistent with that of a previous study (a diffusion coefficient of 1.5 ± 0.1 µm2 s−1 for Avi-488)19. The obtained values indicate that the fluidity of PI(4,5)P2-containing SLBs, which interact with the PH domain of the myosin IC tail, is sufficient to facilitate the lateral diffusion of myosin IC on the SLB surface in the corkscrewing assay (Fig. 1a).
3D observation of the corkscrewing motion of myosin IC-driven gliding actin filaments. (a) Schematic of the in vitro corkscrewing assay. Cover glass surface is coated with PI(4,5)P2-containing SLBs. Sparsely biotinylated and Alex647-labeled actin filaments bound to QDs (λ = 525 nm) are driven by the PI(4,5)P2-bound myosin IC via the PH domain. (b) Schematic of the tPOT microscope (not to scale). The x- and y-positions of actin filament-bound QDs were obtained from the image pairs split by the prism. The z-position of the QD was obtained from the relative y-position of the split image pair. (c) Split image pairs captured by the tPOT microscope. The top panel illustrates a split image pair of a biotinylated-Alex647-labeled actin filament (marked by red rectangles). The other panels illustrate the time series of paired images of a QD bound to the actin filament gliding on the myosin IC-coated surface (pairs of red and cyan arrowheads). Scale bar = 1 µm; interval between panels = 50 s. (d) The 3D plot of QD revealing the left-handed corkscrewing motion of the myosin IC-driven actin filament. The color indicates the observation time (see color bar). (e) The yz-trajectory of the QD shown in d, the counterclockwise rotation of the actin filament gliding in the forward translocation direction. (f) The xy- and xz-trajectories of the QD bound to the actin filament shown in d. Rotation handedness is confirmed by referring to the xy- and xz-trajectories of the QD because the sinusoidal oscillation phases of the two traces shift with a quarter of the wavelength as the gliding actin filaments rotate around their longitudinal axis. The numbered labels ❶–❹ and thin black dotted lines indicate the peaks and midpoints for a given period of the sinusoidal-like xz-trajectory. Schematic of the left-handed corkscrewing motion of the actin filament over time (bottom). Red cylinder and yellow sphere represent the actin filament and QD, respectively. The blue and black curved arrows indicate the gliding and rotation directions of the actin filament, respectively. The numbers ❶–❹ correspond to those indicated above. Optimally, the z- and y-coordinates of the QD bound to the actin filament have maximum values at ❶ and ❷ and minimum values at ❸ and ❹, respectively. In case actin filaments are gliding along a curved path, the y-coordinate accordingly shifts.
To determine the xyz-positions of streptavidin-coated QDs bound to sparsely biotinylated actin filaments gliding across the PI(4,5)P2-containing SLB-bound myosin IC-coated surface, we used a tPOT microscope (Fig. 1a–c). The tPOT microscope, originally developed to track single fluorescent molecules35, QDs31, microbeads36, or cells37, achieved nanometer-order accuracy in the 3D positioning of actin filament-bound QDs fixed on a glass coverslip38 (Supplementary Fig. 2). Using this method, we observed that the 3D trajectory of the QDs bound to the gliding actin filaments exhibited helical motion (Fig. 1d, Supplementary Fig. 3a, and Supplementary Movie 1); this indicates that myosin IC causes the rotation of gliding actin filaments around their longitudinal axis (corkscrewing motion). These corkscrewing motions have not been previously observed for myosin I family members and demonstrate the torsional and axial gliding force production characteristics of single-headed myosin IC. The 3D trajectory (Fig. 1d) and yz-trajectory (Fig. 1e) of the SLB-bound myosin IC-driven QD-tagged actin filament revealed the left-handed direction of rotation of the actin filament. Furthermore, the xy- and xz-trajectories of the QDs showed oscillations; however, the xy-oscillatory trajectories of the QDs were often irregular, indicating that the QD-bound gliding actin filaments often bent and drastically changed their gliding direction (Fig. 1f and Supplementary Fig. 3b). A phase lag was often observed between the sinusoidal oscillations of the xy- and xz-trajectories, with the xy-trajectory lagging slightly behind the xz-trajectory; this also indicates the left-handed corkscrewing motion of the actin filament (Fig. 1f). The torsional force by myosin IC that caused the rotation of the gliding actin filaments was effective even when bound to the diffusive lipid membrane. The handedness of the myosin IC-driven corkscrewing motion in the actin filaments was consistent with those reported in a previous study on dimeric myosin II and V25.
Quantification of corkscrewing actin filaments driven by membrane-bound myosin IC
In the in vitro actin filament corkscrewing assay, myosin IC-driven gliding actin filaments often changed their direction of motion in the xy-plane owing to their flexibility. Thus, the xy-trajectories of the QDs bound to gliding actin filaments also changed accordingly. In contrast, most of the xz-trajectories of the QDs exhibited constant oscillations (Figs. 1f and 2a). We selected the actin filament-bound QDs that glided reasonably straight and further analyzed their 3D positions. From the xz-trajectories that showed ≥ 2 oscillations, the corkscrewing pitches (that is the gliding distance of the actin filament per rotation around its longitudinal axis) of each actin filament were determined by fitting the xz-trajectory of the QD with sinusoidal function (Fig. 2a). The mean corkscrewing pitch of myosin IC was 0.58 ± 0.16 μm (mean ± standard deviation, SD) under 6% PI(4,5)P2-containing SLBs (Supplementary Table 2). This corkscrewing pitch was markedly longer than the intrinsic helical pitch of the actin filament (approximately 72 nm), suggesting that the helical trajectories of QDs occurred owing to the intrinsic properties of myosin IC. The gliding and rotational velocities of the corkscrewing motion were determined by linear approximation of the plots of x-displacements (Fig. 2b) and revolutions (Fig. 2c) versus time for individual QDs with ≥ 2 revolutions. The mean gliding and rotational velocities were 9.5 ± 4.0 nm s−1 (mean ± SD) and + 0.016 ± 0.003 rev s−1 (mean ± SD, the plus ( +) sign represents left-handed revolution), respectively (Supplementary Table 2).
Individual traces of corkscrewing actin filaments on SLBs. (a) xz-trajectories of the QDs bound to corkscrewing actin filaments. Each xz-trajectory of the QDs (0.72 µM myosin IC, 6% PI(4,5)P2, n = 26) exhibited a robust corkscrewing motion with a relatively constant pitch. Individual traces of the QDs in (a) were fitted with a sinusoidal function to obtain the corkscrewing pitch. Time courses of the x-displacements (b) and revolutions (c) of the QDs bound to corkscrewing actin filaments. Individual traces of the QDs in b and c were fitted with linear functions to obtain the gliding and rotational velocities, respectively. Average gliding and rotational velocities were 9.5 ± 4.0 μm s−1 and + 0.016 ± 0.003 rev s−1, respectively.
Effect of PI(4,5)P2 concentration in SLBs on the corkscrewing motion
The PH domain in the myosin IC tail has a high affinity for the anionic phospholipid PI(4,5)P23. Although relatively low, LCBD also has an affinity for anionic phospholipids such as PI(4,5)P2 and phosphatidylserine3,39. In a previous study, the gliding velocity of myosin IC-driven actin filaments on fluid SLBs in an in vitro gliding assay decreased with increasing concentrations of anionic phospholipids in SLB14. It has been suggested that the gliding velocity may be affected by the inhibition of motor domain dynamics caused by the interaction of lever arm-containing LCBD with anionic phospholipids in SLB. To investigate the effect of anionic phospholipids in SLBs on torque generation, we performed the corkscrewing assay using SLBs with varying PI(4,5)P2 concentrations (2–10 mol%) and Drosophila myosin IC. The gliding velocity of actin filaments at 2% PI(4,5)P2 concentration was faster than that at 4–10% PI(4,5)P2 concentrations (Fig. 3a); this was consistent with the finding of a previous study on mouse myosin IC14. The mean gliding velocity at 2% PI(4,5)P2 concentration was approximately twice that at 10% PI(4,5)P2 concentration; no significant differences in gliding velocity were observed at 4%, 6%, and 10% PI(4,5)P2 concentration (Supplementary Table 2). The rotational velocity was the slowest at 4% PI(4,5)P2 concentration and approximately two-thirds of the fastest velocity at 2% PI(4,5)P2 concentration (Fig. 3b and Supplementary Table 2). Moreover, we examined the effect of myosin IC concentration on gliding velocity and found that gliding velocity was not affected by myosin IC concentration (Supplementary Fig. 4). Furthermore, we found that the corkscrewing pitch was sensitive to changes in PI(4,5)P2 concentrations; the pitch at low PI(4,5)P2 concentration was longer than that at high PI(4,5)P2 concentration (Fig. 3c and Supplementary Table 2). The plots of gliding velocity versus corkscrewing pitch at all PI(4,5)P2 concentrations were approximated linearly, indicating a strong correlation (correlation coefficient = 0.71) between them within the range of the experimental conditions (Fig. 3d). Because the corkscrewing pitch value was determined by dividing gliding velocity by rotational velocity, the longest pitch value at 2% PI(4,5)P2 concentration indicates that the ratio of gliding velocity to rotational velocity is the highest. A change in the corkscrewing pitch implies a change in the ratio of gliding to rotational velocities; therefore, PI(4,5)P2 concentrations in SLBs may have different effects on the torsional and longitudinal forces generated by myosin IC.
Dependence of the corkscrewing motion on PI(4,5)P2 concentration. Gliding velocity (a), rotational velocity (b), and pitch (c) versus PI(4,5)P2 concentration of individual QDs bound to corkscrewing actin filaments at an applied myosin IC concentration of 0.72 µM. The box represents the 75th–25th percentiles, and the median and mean values are indicated by black lines and red diamonds, respectively [n = 16 (2%), 18 (4%), 26 (6%), and 18 (10%)]. ***, p < 0.001; **, p < 0.01; *, p < 0.05; and ns, nonsignificant in the Wilcoxson rank-sum test. (d) Correlation of the gliding velocity and pitch of the corkscrewing motion on SLBs containing 2% (black filled circle), 4% (black filled rhombus), 6% (black filled triangle), and 10% ( ×) PI(4,5)P2. Red line indicates a linear approximation of the plots. The correlation coefficient between the gliding velocity and corkscrew pitch was 0.71 (n = 78).
Discussion
To investigate the motility of myosin IC on SLBs mimicking the cell membrane, 3D nanometry using the tPOT microscope was applied to quantify the movement of actin filaments driven by full-length, single-headed Drosophila myosin IC anchored to the PI(4,5)P2-containing fluidic SLBs. The QDs bound to gliding actin filaments were tracked in 3D and the corkscrewing motions of actin filaments driven by nonprocessive myosin IC were observed. The direction of the off-axis torque generated by SLB-bound myosin IC was consistent with that reported for myosin II, V, and X anchored to solid substrates such as glass coverslips, microbeads, and QDs25,30. Our present experimental system requires the binding of QDs to actin filaments as tracers; therefore, we cannot rule out the possibility that the presence of QDs bound to actin filaments promotes or inhibits the myosin IC-driven corkscrewing motion. Quantifying corkscrewing motion using a single fluorescent dye molecule25,35 as a tracer instead of QD will help elucidate the effect of QDs on the corkscrewing motion.
The mechanisms underlying the filament rotation by myosins remain unclear; however, several factors have been proposed to contribute to the corkscrewing motion of cytoskeletal filaments by nonprocessive motor proteins, including the off-axis component of motor domain conformational change, motor filament interaction-induced molecular drag, or directionally biased selection of the motor to the filament binding site25,40,41,42; it can be inferred that the corkscrewing motion is the result of a combination of different mechanisms. Moreover, it has been proposed that the rotational motion of processive myosins, such as myosin V and VI, results from the interaction between the stride length of myosin and the helical disposition of actin monomers26,27. Based on this approach, the stride length of processive myosin slightly differs from that of the actin helical structure, resulting in helical motions of processive myosins. However, nonprocessive myosins, such as myosin IC and skeletal muscle myosin II, have short stride lengths; thus, the abovementioned mechanism that factors the stride length is insufficient to clearly explain the observed corkscrewing pitch that reaches the submicrometer level (Fig. 2a)24,25. If myosin IC followed the monomers on the filaments, the corkscrewing pitch should match the pitch of the actin helical structure (approximately 72 nm); however, our study revealed that the corkscrewing pitch by monomeric myosin IC on SLB was approximately 0.5 µm, suggesting the involvement of a mechanism of corkscrewing motion that is independent of myosin stride length.
A previous theoretical study explained the corkscrewing motion of actin filaments gliding across a surface covered with multiple nonprocessive myosin monomers. In the model, myosin monomers were anchored to the cover glass and preferentially bound to the approaching specific actin monomers within the filaments, inducing the rotation of gliding actin filaments43. The actin monomers, termed “target zone,” to which myosin motor domains selectively bind, are periodically located in the filament44,45. The left-handed corkscrew motion of the actin filament around its longitudinal axis arises from the selective binding of myosin motor domains to the target zone located to the left of the longitudinal axis of the filament, pulling in the direction of the long axis of the filament, that is, rotating the filament around the longitudinal axis during the stroke of the lever arm along the longitudinal axis of the filament. Herein, the handedness of the myosin IC-induced corkscrewing motion (Fig. 1d) is consistent with that of the rotation estimated by the aforementioned model. Furthermore, the model predicts that the stiffness of the myosin motor domain affects the magnitude of the torque and corkscrewing pitch, as greater stiffness pulls the target zone more strongly43. It is likely that at high PI(4,5)P2 concentrations in fluidic SLBs, the mechanical properties of the lever arm, which weakly interacts with PI(4,5)P23, are altered, resulting in a shorter corkscrewing pitch (Fig. 3c). The selectivity of myosin heads regarding actin monomers is also predicted to be affected by the gliding velocity of the actin filament43. A strong correlation was observed between the gliding velocity and corkscrewing pitch (Fig. 3d), suggesting that the corkscrewing pitch by myosin IC can exhibit sensitivity toward their gliding velocity. Herein, the results indicate that the corkscrewing motion can be explained by the selective binding of myosin motor domains to target zone actin filaments, although the possibility of oblique power strokes of the lever arms cannot be ruled out.
We demonstrated the chiral corkscrewing motion of gliding actin filament driven by SLB-bound myosin IC, containing varying concentrations of PI(4,5)P2, indicating that the lipid composition of the membrane, a factor not previously considered, may regulate torque generation by myosin IC. A possible mechanism by which the PI(4,5)P2-containing membrane may directly regulate the myosin IC-generated torque is the electrostatic interaction of PI(4,5)P2 with LCBD14, which acts as a mechanical lever arm. If the interaction between PI(4,5)P2 and LCBD inhibits conformational changes in the motor domain during the mechanochemical cycle and alters the ratio of the translational force component to the rotational force component in response to PI(4,5)P2 concentration (Fig. 3a,b), the corkscrew pitch can be altered (Fig. 3c).
Although the mechanism of torque generation by myosin IC remains unclear, the torque component in the motor domain may be efficient for myosin IC to transport vesicles around obstacles in a crowded intracellular environment46, similar to that proposed for other myosin30 and microtubule-based motor proteins, such as kinesins and dyneins47,48. In addition to being an intracellular transporter, myosin IC functions as an antagonist of myosin ID, another member of the myosin I family, during the chirality formation of the epidermal cells of Drosophila melanogaster organs. Various studies have shown that myosin IC overexpression results in an inversion of the organ phenotype, and the knockdown of myosin IC results in left–right asymmetry defects in the organ9,49. In particular, whole-body twisting induced by myosin IC overexpression in the larval epidermal tissue, which is devoid of left–right asymmetry, implies that myosin IC plays a crucial role in establishing the left–right asymmetry in organisms15. Myosin IC is responsible for regulating the effect of the myosin ID-induced left–right asymmetry of Drosophila; the motor domain determines organ chirality15. There are notable differences in myosin IC and ID motilities because myosin ID propels the actin filaments in leftward circles (circular motion) in the in vitro gliding assay, whereas myosin IC does not15. Furthermore, vesicles coated with multiple myosin IC and ID molecules can move processively along actin filaments, with myosin ID having a longer run length than myosin IC11. However, the mechanism by which these motile properties contribute to the establishment of cell and organ chirality remains unknown. Although the rotation of gliding actin filaments around their longitudinal axis (corkscrewing motion) driven by myosin IC observed in the present study does not directly address the question of the molecular mechanism underlying the circular motion of the actin filament, myosin IC, which exhibits the left-handed corkscrewing motion (Fig. 1), should be a left-to-right force component in the direction perpendicular to the longitudinal axis of actin filaments; in contrast, myosin ID, which exhibits a circular left-rotational motion15, should be a right-to-left force component. Considering that the actin-attachment lifetime of myosin IC is ninefold longer than that of myosin ID11, myosin IC may suppress the circular left-rotational motion of myosin ID-driven actin filaments and be involved in establishing left–right asymmetry in cells and organs. Quantifying the direction of the myosin ID-induced corkscrewing motion may provide further insights into the underlying molecular mechanism.
Myosin IC may participate in establishing cell chirality, along with cadherin and β-catenin, which are responsible for cell–cell adhesion10. Myosin IC, which binds to the actin filaments longer than myosin ID, may serve as a tether that holds actin filaments to the cell membrane11. Formin, a factor that promotes actin polymerization is also essential for establishing left–right asymmetry in Drosophila50. Formin drives the left-handed rotation of actin filaments around their longitudinal axis while processively polymerizing actin filaments51. It has been speculated that the rotation of polymerizing actin filament by formin induces the formation of counterclockwise cell chirality52. The torque generated by formin may be transmitted to the cell membrane via myosin IC bound to the actin filaments, resulting in cell chirality11. Because the torque of the left-handed corkscrewing motion produced by myosin IC is in the same direction as that exerted by formin on the actin filaments, the two torque components may reinforce each other. Furthermore, the tension in the actin filament network near the cell membrane may be altered by the torque exerted by myosin IC molecules tethered to the actin filaments, which may be the direct role of the torque generated by myosin IC in this process.
Reportedly, myosin IC directly regulates the cell morphology in neuronal growth cone53. Asymmetric inhibition of myosin IC in the growth cone causes the rearrangement of the actin cytoskeleton network and changes the cell shape in the direction of the inhibition area. Torque generation around the longitudinal axis of actin filaments generated by myosin V54 and X55, which interact with the cell membrane and actin cytoskeleton, has suggested to be involved in the formation of chiral-shaped filopodia. Considering that myosin IC is bound to the cell membrane via its tail domain and to the cortical actin cytoskeleton via its motor domain, it is conceivable that the torque force generated by myosin IC at the cell membrane surface may also be involved in regulating cell membrane morphology and cortical actin cytoskeleton organization. Future studies with higher spatial resolution observations of the in vivo 3D dynamics of the cortical actin filament and myosin IC are necessary to reveal the biological role of torque generation by myosin motor proteins.
Materials and methods
Construction of plasmids for myosin IC and calmodulin
The Drosophila melanogaster myosin IC gene was inserted into the pFastBac vector (Invitrogen, Carlsbad, CA, USA) to generate the pFastBac Drosophila myosin IC baculovirus transfer vector. Myosin IC (UniProt ID: Q23979) cDNA cloned from Drosophila melanogaster (a gift from Dr. K. Matsuno) was mutated to create an NcoI site in the upstream region of the nucleotide sequence encoding residue 1 and an AgeI site in the downstream region of the nucleotide sequence encoding residue 1035 of myosin IC. This DNA was cut with NcoI and AgeI and the fragment was ligated with the NcoI-AgeI fragment of the pFastBac XI-K Full vector56. The resulting construct, Drosophila full-length myosin IC (pFastBac Drosophila myosin IC), encodes a FLAG-tag (DYKDDDDK) in the N-terminus and a Myc tag (EQKLISEEDL) and His8-tag (HHHHHHHH) in the C-terminus. The Drosophila melanogaster calmodulin gene was inserted into the pFastBac vector to generate the pFastBacCaM baculovirus transfer vector. The calmodulin cDNA cloned from Drosophila melanogaster (a gift from Dr. K. Matsuno) was mutated to create an XbaI site upstream of the nucleotide sequence encoding residue 1 and an XhoI site downstream of the nucleotide sequence encoding 149 residues of Drosophila calmodulin. The BamHI-KpnI digest fragment was ligated with the BamHI-KpnI digest fragment of pFastBac. DH10Bac was transformed with pFastBacMyo1c and pFastBacCaM, respectively, to obtain the bacmid of baculovirus. Transfection of the bacmid into SF9 resulted in baculoviruses with myosin IC and calmodulin constructs.
Preparation of biological samples
Drosophila myosin IC was co-expressed with Drosophila calmodulin in High Five™ cell culture (Invitrogen). Cells were harvested and washed with 10 mM HEPES, pH 8.0, 150 mM NaCl, and 1 mM MgCl2. The pelleted cells were suspended with 3 vol/g cells of buffer A (30 mM HEPES, pH 8.0, 200 mM NaCl, 50 mM KCl, 5 mM MgCl2, 10% glycerol, 10 mM β-mercaptoethanol, 0.5 µM Chara corallina calmodulin, and a mixture of protease inhibitors) containing 7 mM ATP. Then, 3 vol/g cells of buffer A containing 1% Nonidet P-40 was added, and the ingredients were mixed. Chara calmodulin, which was added to supplement Drosophila calmodulin during purification, has 88% amino acid identity with Drosophila calmodulin. After incubation on ice for 15 min, the lysate was centrifuged at 228,000×g for 30 min at 4 °C. The supernatant was mixed with 0.3 mL of nickel-nitrilotriacetic acid-agarose (Qiagen) in a 50-mL tube on a rotating wheel for 1 h at 4 °C. The resin suspension was then loaded on a column and washed with 30 mL of buffer A containing 30 mM imidazole, followed by 10 mL of buffer B (30 mM HEPES, pH 8.0, 50 mM KCl, 5 mM MgCl2, 30 mM imidazole, 10% glycerol, 10 mM β-mercaptoethanol, approximately 0.5 µM Chara corallina calmodulin, and a mixture of protease inhibitors) containing 2 mM ATP. Drosophila myosin IC was eluted with buffer B containing 250 mM imidazole. The eluted Drosophila myosin IC was mixed with 0.3 mL of anti-FLAG M2 affinity resin (Sigma-Aldrich) in a 50-mL tube on a rotating wheel for 1 h at 4 °C. The resin suspension was then loaded on a column and washed with 10 mL of buffer C (25 mM HEPES, pH 7.4, 150 mM KCl, 4 mM MgCl2, 10% glycerol, 0.5 mM DTT, approximately 0.5 µM Chara corallina calmodulin, and a mixture of protease inhibitors) containing 2 mM ATP, followed by washing with 30 mL of buffer D (25 mM HEPES, pH 7.4, 25 mM KCl, 4 mM MgCl2, 10% glycerol, 0.5 mM DTT, Chara corallina calmodulin, and a mixture of protease inhibitors) containing 0.2 mM ATP. Drosophila myosin IC was eluted with buffer D containing 0.2 mg/mL FLAG peptide (Sigma-Aldrich). The purity and homogeneity of protein samples were confirmed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) (Supplementary Fig. 5). Chara corallina calmodulin (Q9LDQ9) was expressed in E. coli strain BL21 (DE3) and purified using the trichloroacetic acid method and affinity chromatography using Phenyl Sepharose CL-4B57. Purified myosin IC and calmodulin were rapidly frozen in liquid nitrogen and stored in a −80 °C freezer. G-actin was purified by a standardized protocol from acetone powder prepared from rabbit skeletal muscle using a previously described method58. G-actin was polymerized at a total actin concentration of 1.2 μM in F buffer (10 mM HEPES–KOH, pH 7.5, 100 mM KCl, 1 mM ATP, 1 mM MgCl2) and stabilized with 4 µM Alexa 647-labeled phalloidin (Invitrogen). Then, actin filaments were labeled with 1 mM Biotin-(AC5)2 Sulfo-OSu (Dojindo, Kumamoto, Japan) in the buffer (10 mM HEPES–KOH, pH 7.5, 0.2 mM ATP, 0.2 mM CaCl2, 0.2 mM DTT) for 1 h at 4 °C. The actin filament mixture was ultracentrifuged, and excess biotin was removed by washing with F buffer before suspension. The precipitate of biotinylated actin filaments was suspended in F buffer.
SLB preparation
Glass coverslips (24 mm × 36 mm, 130–170 μm thickness; Matsunami Glass Ind., Japan) were washed with a 5 M KOH solution and then rinsed with ultrapure water. The coverslips were dried and plasma cleaned (PDC-32D; Harrick Plasma) for 30 min. The flow chamber was assembled with 24 mm × 36 mm and 18 mm × 18 mm glass coverslips using two thin strips of tape (810-1-18, Scotch 3 M) that were placed approximately 3 mm apart. The volume of the flow chamber was approximately 5 µL. Giant unilamellar vesicles (GUVs) were prepared via the inverted emulsion method59. The lipid-in-oil mixture was prepared by dissolving the desired phospholipids in medium-chain triglyceride oil (Nisshin OilliO Group, Japan) by heating the mixture at 60 °C for 3 h in the water bath. For the corkscrewing assay, the lipid-in-oil mixture contained 90–98% DOPC (molecular weight 786, Avanti Polar Lipids) and 2–10% PI(4,5)P2 (molecular weight 1096, Avanti Polar Lipids). For FRAP and Avi-488 single-molecule tracking, the lipid-in-oil mixture contained 0.05% ATTO647N–DOPE (molecular weight 855, ATTO-TEC GmbH) and 1% DSPE–PEG-2000-biotin (molecular weight 3016, Avanti Polar Lipids). To form water-in-oil droplets, the lipid-in-oil mixture was suspended in an inner solution (20 mM HEPES–KOH, pH 7.5, 75 mM NaCl, 80 mM sucrose, 300 mM glucose). The emulsion was gently loaded onto the outer solution (20 mM HEPES–KOH, pH 7.5, 75 mM NaCl, 380 mM glucose). To form the GUVs, the solutions were centrifuged for 15 min, the supernatant was removed, and the precipitated GUVs were suspended. SUV dispersions were prepared by sonicating the GUV solution for 1 h. CaCl2 was added to the SUV dispersion to a final concentration of 2.5 mM to facilitate SLB formation immediately before infusion into the glass flow chamber. The SUV dispersions were then added to the chambers and incubated for 30 min. The chamber was then washed with 10 volumes of the H20S75 buffer (20 mM HEPES–KOH, pH 7.5, 75 mM KCl) before use.
FRAP experiment
The FRAP experiment was performed by opening/closing the path of a 488 nm wavelength laser for bleaching while observing SLBs with a 632 nm wavelength laser. The 632 nm wavelength laser (05-LHP-991, Melles Griot) was used at 10 mW laser power to observe ATTO647N–DOPE in SLBs, and the 488 nm wavelength laser (488 LS FP, Coherent OBIS) was used at 40 mW laser power for bleaching. The FRAP experiment was performed as follows: the prebleached SLB was recorded for 2 s and then bleached for 5 s with a 488 nm wavelength laser, followed by the recovery of the intensity of the recorded SLB. Images were acquired with a 100 ms exposure for 600 frames using a modified TIRF microscope (IX71, Olympus) fitted with a UPlan-Apochromat TIRF 100×/1.45 NA objective lens and an electron-multiplying (EM)-charge-coupled device (CCD) camera (iXon DV887DCS, Andor Technology) using the Solis software (Andor Technology). The DLIPID value was calculated by fitting the recovery curve of the ATTO647N–DOPE fluorescence intensity60.
Three-dimensional Prismatic Optical Tracking (tPOT) microscopy
The principle of 3D tracking has been described previously31,61. The back-focal-plane (BFP) of the objective was focused outside the camera port of a modified inverted microscope (Eclipse Ti-PFS, Nikon) with an achromatic lens-1 to create an equivalent BFP (eBFP) (Fig. 1b). A custom-made wedge prism (3.5°) coated with an antireflective layer was precisely positioned at the eBFP to split beam path. Two split images of the sample were focused on the camera with another lens-2, which was used to determine the magnification of the optical system. The tPOT microscope was equipped with a 100×/1.49 NA, plan-apochromat TIRF objective lens (Nikon), either a Cy5 filter set (Semrock) for Alexa 647 labeled actin filaments or a GFP-3035C filter set (Semrock) for QDs, a stable stage (KS-N, ChuukoushaSeisakujo, Tokyo, Japan), a LED light source (D-LEDI, Nikon), a pulse motor (SGSP-13ACT, Sigma Koki, Tokyo, Japan), and controller (QT-CM2, Chuo Precision Ind., Tokyo, Japan). Images were acquired with 500 or 1000 ms exposure for 600 or 300 frames by an EM-CCD camera (iXon X3 DU897, Andor Technology) via Solis software (Andor Technology). The position of the two optically separated images of a QD was determined using the 2D Gaussian fitting as (x1, y1) and (x2, y2), and x, y, and z were calculated using (x1 + x2)/2, (y1 + y2)/2 and y1 − y2, respectively, using Igor software. To calibrate the real z-axis position, the objective was moved vertically at a constant speed using the stage equipped with the pulse motor and controller while observing the QDs bound to the glass surface. The calculated and real z-axis position (as defined by the pulse motor) corresponded linearly over a range of ±0.4 µm from the focal plane. The temperature of the sample stage was controlled and maintained at 25 °C using a custom-built water-circulating temperature control system (F-12, Julabo, Germany)62. The temperature was measured using a micro thermos sensor (HD-1100E, Anritsu Meter, Japan).
Actin filament corkscrewing assays
The corkscrewing assays were performed using QD-labeled actin filaments. QD-labeled actin filaments were prepared by adding 50 nM streptavidin-coated QDs (Qdot525 streptavidin conjugate, Thermo Fisher Scientific) diluted in KMg25 buffer (10 mM MOPS, pH 7.0, 25 mM KCl, 1 mM DTT, 1 mM EGTA, and 1 mM MgCl2), mixed with an equal volume of 800 nM sparsely biotinylated and Alexa Fluor 647 phalloidin-labeled actin filament, and incubated for 30 min at room temperature. After incubation, the QD-labeled actin filament solution was diluted 1/50th of the original concentration with the KMg25 buffer. QD-labeled actin filaments were prepared for each day of the assay. Before the assay, 2 mg/mL BSA was infused into the SLB chamber and incubated for a minimum of 30 min for blocking. QD-labeled actin filaments (0.5 µL) were mixed with 1 μL of 3.25 μM myosin IC in a total of 4.5 μL of KMg25 buffer containing 1 mM ATP, 9.7 μM Chara calmodulin, 3 mg/mL glucose, 50 U/mL glucose oxidase (Sigma-Aldrich), 40 μg/mL catalase (Sigma-Aldrich), 0.3 mg/mL creatine kinase (Roche), and 5 mM creatin phosphate (Roche) immediately before each experiment to prepare the assay mixture. Into the SLB-coated glass flow chamber, one chamber volume of the assay mixture was infused and the chamber was then sealed using vacuum grease. In the preliminary screening for a condition that allows us to observe the stable motion of the actin filaments, we realized that high myosin concentrations should be introduced into the flow chamber. Therefore, we used a sufficient myosin concentration (0.72 µM in the flow chamber) that allowed a stable gliding movement, although we were not aware of exactly how many myosins simultaneously interact with an actin filament at any given time. The assays were performed at 25 °C. The number of QDs bound to the actin filaments was one in most cases; if more than one QD was bound, further analysis was not performed. The actin filaments were introduced into the flow chamber at a concentration such that approximately one actin filament was observed in one field of view (a circle with a radius of 7.8 μm); the mean length of the QD-bound actin filament was 2.1 ± 1.5 μm (mean ± SD, n = 102) (Supplementary Fig. 6). However, the actual length of the actin filament in the corkscrewing motion remains unknown because our optical system does not allow the simultaneous observation of actin filaments and QDs.
Quantification of the corkscrewing motion of actin filaments
The acquired images of the QDs were averaged every 4 frames using ImageJ software. Based on the averaged sequential images, fluctuating (rapid forward or backward movement relative to the direction of travel), wiggling (frequent small movements to the left or right relative to the direction of travel), or crossing QDs were ignored with no further analysis. Using Igor software, we tracked the QDs bound to actin filaments that exhibited stable directional movement. For further analysis, x-, y-, and z-positions in the individual trajectories were averaged every 20 frames. The 3D trajectories were displayed using the MATLAB software, which has an easy-to-use graphical display, to determine if the trajectories exhibited the corkscrewing motion (Fig. 1d and Supplementary Fig. 3). Among the 17, 21, 32, and 22 QD movements for 2%, 4%, 6%, and 10% PI(4,5)P2 concentrations, respectively, that were analyzed to obtain the 3D trajectories, cases that were not suitable for further analyses were observed; in these cases, the QDs did not exhibit a clear corkscrewing motion [n = 1, 2% PI(4,5)P2; n = 2, 4% PI(4,5)P2; n = 3, 6% PI(4,5)P2; and n = 3, 10% PI(4,5)P2] or QDs exhibited frequent stops (n = 1, 4% PI(4,5)P2; n = 1, 6% PI(4,5)P2); and n = 1, 10% PI(4,5)P2]. Furthermore, the amplitude of projection was too small owing to some unknown reason (< 25 nm, half as large as the sum of the radii of the actin filament and QD, n = 2, 6% PI(4,5)P2). Finally, 16, 18, 26, and 18 trajectories for 2%, 4%, 6%, and 10% PI(4,5)P2 concentrations, respectively, which clearly exhibited left-handed corkscrewing motions, were obtained. To determine the individual corkscrewing pitch, gliding velocity, and rotational velocity, we selected the xz-trajectory that showed at least 2 continuous oscillations. The individual corkscrewing pitch was determined by fitting the xz-trajectory with a sine function. Individual gliding and rotational velocities were determined by fitting the t-x and t-rev trajectories, respectively, with linear functions.
Statistics and reproducibility
Tracking data for QDs were obtained from at least two independent experiments, and sample sizes are indicated in detail in the text or the “Materials and methods” section.
Data availability
All samples used in this study are available from the corresponding authors upon request. The source data for graphs in the main figures are provided as Supplementary Data 1.
Code availability
Custom scripts for tracking QDs were written using Igor Pro 5.05A. Trajectories were analyzed using MATLAB R2023a. These scripts are available upon reasonable request.
References
McConnell, R. E. & Tyska, M. J. Leveraging the membrane—cytoskeleton interface with myosin-1. Trends Cell Biol. 20, 418–426 (2010).
McIntosh, B. B. & Ostap, E. M. Myosin-I molecular motors at a glance. J. Cell Sci. 129, 2689–2695 (2016).
Hokanson, D. E. & Ostap, E. M. Myo1c binds tightly and specifically to phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate. Proc. Natl. Acad. Sci. USA 103, 3118–3123 (2006).
Boguslavsky, S. et al. Myo1c binding to submembrane actin mediates insulin-induced tethering of GLUT4 vesicles. Mol. Biol. Cell 23, 4065–4078 (2012).
Papadopulos, A., Tomatis, V. M., Kasula, R. & Meunier, F. A. The cortical acto-myosin network: From diffusion barrier to functional gateway in the transport of neurosecretory vesicles to the plasma membrane. Front. Endocrinol. 4, 20–24 (2013).
Bose, A. et al. Glucose transporter recycling in response to insulin is facilitated by myosin Myo1c. Nature 420, 821–824 (2002).
Brandstaetter, H., Kendrick-Jones, J. & Buss, F. Myo1c regulates lipid raft recycling to control cell spreading, migration and Salmonella invasion. J. Cell Sci. 125, 1991–2003 (2012).
Gillespie, P. G. Myosin I and adaptation of mechanical transduction by the inner ear. Philos. Trans. R. Soc. B Biol. Sci. 359, 1945–1951 (2004).
Hozumi, S. et al. An unconventional myosin in Drosophila reverses the default handedness in visceral organs. Nature 440, 798–802 (2006).
Petzoldt, A. G. et al. DE-Cadherin regulates unconventional myosin ID and myosin IC in Drosophila left-right asymmetry establishment. Development 139, 1874–1884 (2012).
Báez-Cruz, F. A. & Ostap, E. M. Drosophila class-I myosins that can impact left-right asymmetry have distinct ATPase kinetics. J. Biol. Chem. 299, 104961 (2023).
Saarikangas, J., Zhao, H. & Lappalainen, P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 90, 259–289 (2010).
Sciortino, A. & Bausch, A. R. Pattern formation and polarity sorting of driven actin filaments on lipid membranes. Proc. Natl. Acad. Sci. USA 118, e2017047118 (2021).
Pyrpassopoulos, S., Feeser, E. A., Mazerik, J. N., Tyska, M. J. & Ostap, E. M. Membrane-bound Myo1c powers asymmetric motility of actin filaments. Curr. Biol. 22, 1688–1692 (2012).
Lebreton, G. et al. Molecular to organismal chirality is induced by the conserved myosin 1D. Science 362, 949–952 (2018).
Ma, G. J., Ferhan, A. R., Jackman, J. A. & Cho, N. J. Elucidating how different amphipathic stabilizers affect BSA protein conformational properties and adsorption behavior. Langmuir 36, 10606–10614 (2020).
Richter, R. P., Maury, N. & Brisson, A. R. On the effect of the solid support on the interleaflet distribution of lipids in supported lipid bilayers. Langmuir 21, 299–304 (2005).
Zot, H. G., Doberstein, S. K. & Pollard, T. D. Myosin-I moves actin filaments on a phospholipid substrate: Implications for membrane targeting. J. Cell Biol. 116, 367–376 (1992).
Grover, R. et al. Transport efficiency of membrane-anchored kinesin-1 motors depends on motor density and diffusivity. Proc. Natl. Acad. Sci. USA 113, E7185–E7193 (2016).
Pernier, J. et al. Myosin 1b is an actin depolymerase. Nat. Commun. 10, 5200 (2019).
Lopes, J. et al. Membrane mediated motor kinetics in microtubule gliding assays. Sci. Rep. 9, 9548 (2019).
Giese, S. et al. Mechanochemical properties of human myosin-1C are modulated by isoform-specific differences in the N-terminal extension. J. Biol. Chem. 296, 100128 (2021).
Nishizaka, T., Toshiki Yagi, Y. T. & Lshiwata, S. Right-handed rotation of an actin filament in an in vitro motile system. Nature 45, 1998–2001 (1993).
Sase, I., Miyata, H., Ishiwata, S. & Kinosita, K. Axial rotation of sliding actin filaments revealed by single-fluorophore imaging. Proc. Natl. Acad. Sci. USA 94, 5646–5650 (1997).
Beausang, J. F., Schroeder, H. W., Nelson, P. C. & Goldman, Y. E. Twirling of actin by myosins II and V observed via polarized TIRF in a modified gliding assay. Biophys. J. 95, 5820–5831 (2008).
Ali, M. Y. et al. Myosin v is a left-handed spiral motor on the right-handed actin helix. Nat. Struct. Biol. 9, 464–467 (2002).
Ali, M. Y. et al. Unconstrained steps of myosin VI appear longest among known molecular motors. Biophys. J. 86, 3804–3810 (2004).
Arsenault, M. E., Sun, Y., Bau, H. H. & Goldman, Y. E. Using electrical and optical tweezers to facilitate studies of molecular motors. Phys. Chem. Chem. Phys. 11, 4834–4839 (2009).
Sun, Y., McKenna, J. D., Murray, J. M., Ostap, E. M. & Goldman, Y. E. Parallax: High accuracy three-dimensional single molecule tracking using split images. Nano Lett. 9, 2676–2682 (2009).
Sun, Y. et al. Single-molecule stepping and structural dynamics of myosin X. Nat. Struct. Mol. Biol. 17, 485–491 (2010).
Yajima, J., Mizutani, K. & Nishizaka, T. A torque component present in mitotic kinesin Eg5 revealed by three-dimensional tracking. Nat. Struct. Mol. Biol. 15, 1119–1121 (2008).
Yamagishi, M., Fujimura, S., Sugawa, M., Nishizaka, T. & Yajima, J. N-terminal β-strand of single-headed kinesin-1 can modulate the off-axis force-generation and resultant rotation pitch. Cytoskeleton 77, 351–361 (2020).
Knight, J. D., Lerner, M. G., Marcano-Velázquez, J. G., Pastor, R. W. & Falke, J. J. Single molecule diffusion of membrane-bound proteins: Window into lipid contacts and bilayer dynamics. Biophys. J. 99, 2879–2887 (2010).
Seu, K. J. et al. Effect of surface treatment on diffusion and domain formation in supported lipid bilayers. Biophys. J. 92, 2445–2450 (2007).
Fujimura, S. et al. Dissection of the angle of single fluorophore attached to the nucleotide in corkscrewing microtubules. Biochem. Biophys. Res. Commun. 485, 614–620 (2017).
Maruyama, Y. et al. CYK4 relaxes the bias in the off-axis motion by MKLP1 kinesin-6. Commun. Biol. 4, 180 (2021).
Marumo, A., Yamagishi, M. & Yajima, J. Three-dimensional tracking of the ciliate Tetrahymena reveals the mechanism of ciliary stroke-driven helical swimming. Commun. Biol. 4, 1209. https://doi.org/10.1038/s42003-021-02756-0(2021).
Yamaguchi, S. et al. Torque generation by axonemal outer-arm dynein. Biophys. J. 108, 872–879 (2015).
Tang, N., Lin, T. & Ostap, E. M. Dynamics of Myo1c (Myosin-Iβ) lipid binding and dissociation. J. Biol. Chem. 277, 42763–42768 (2002).
Yajima, J. & Cross, R. A. A Torque component in the kinesin-1 power stroke. Nat. Chem. Biol. 1, 338–341 (2005).
Nitzsche, B. et al. Working stroke of the kinesin-14, ncd, comprises two substeps of different direction. Proc. Natl. Acad. Sci. USA. 113, E6582–E6589 (2016).
Mitra, A. et al. A Brownian ratchet model explains the biased sidestepping of single-headed kinesin-3 KIF1A. Biophys. J. 116, 2266–2274 (2019).
Vilfan, A. Twirling motion of actin filaments in gliding assays with nonprocessive myosin motors. Biophys. J. 97, 1130–1137 (2009).
Steffen, W., Smith, D., Simmons, R. & Sleep, J. Mapping the actin filament with myosin. Proc. Natl. Acad. Sci. USA 98, 14949–14954 (2001).
Wu, S. et al. Electron tomography of cryofixed, isometrically contracting insect flight muscle reveals novel actin-myosin interactions. PLoS ONE 5, e12643 (2010).
Nakamori, Y. et al. Myosin motor Myo1c and its receptor NEMO/IKK-γ promote TNF-α-induced serine307 phosphorylation of IRS-1. J. Cell Biol. 173, 665–671 (2006).
Brunnbauer, M. et al. Torque generation of kinesin motors is governed by the stability of the neck domain. Mol. Cell 46, 147–158 (2012).
Can, S., Lacey, S., Gur, M., Carter, A. P. & Yildiz, A. Directionality of dynein is controlled by the angle and length of its stalk. Nature 566, 407–410 (2019).
Hozumi, S. et al. Head region of unconventional myosin I family members is responsible for the organ-specificity of their roles in left-right polarity in Drosophila. Dev. Dyn. 237, 3528–3537 (2008).
Chougule, A. et al. The Drosophila actin nucleator DAAM is essential for left-right asymmetry. PLoS Genet. 16, e1008758 (2020).
Mizuno, H. et al. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science 331, 80–83 (2011).
Tee, Y. H. et al. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat. Cell Biol. 17, 445–457 (2015).
Wang, F. S., Liu, C. W., Diefenbach, T. J. & Jay, D. G. Modeling the role of myosin 1c in neuronal growth cone turning. Biophys. J. 85, 3319–3328 (2003).
Tamada, A. & Igarashi, M. Revealing chiral cell motility by 3D Riesz transform-differential interference contrast microscopy and computational kinematic analysis. Nat. Commun. 8, 2194 (2017).
Li, W. et al. Chiral growth of adherent filopodia. Biophys. J. 2, 3704–3721 (2023).
Haraguchi, T. et al. Functional diversity of class XI myosins in Arabidopsis thaliana. Plant Cell Physiol. 59, 2268–2277 (2018).
Awata, J., Saitoh, K., Shimada, K., Kashiyama, T. & Yamamoto, K. Effects of Ca2+ and calmodulin on the motile activity of characean myosin in vitro. Plant Cell Physiol. 42, 828–834 (2001).
Spudich, J. A. & Watt, S. The regulation of rabbit skeletal muscle contraction. J. Biol. Chem. 246, 4866–4871 (1971).
Pautot, S., Frisken, B. J. & Weitz, D. A. Production of unilamellar vesicles using an inverted emulsion. Langmuir 19, 2870–2879 (2003).
Soumpasis, D. M. Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys. J. 41, 95–97 (1983).
Yamagishi, M., Sumiyoshi, R., Drummond, D. R. & Yajima, J. Anchoring geometry is a significant factor in determining the direction of kinesin-14 motility on microtubules. Sci. Rep. 12, 15417. https://doi.org/10.1038/s41598-022-19589-4 (2022).
Sugawa, M., Maruyama, Y., Yamagishi, M., Cross, R. A. & Yajima, J. Motor generated torque drives coupled yawing and orbital rotations of kinesin coated gold nanorods. Commun. Biol. 5, 1368 https://doi.org/10.1038/s42003-022-04304-w (2022).
Acknowledgements
We thank Y.Y. Toyoshima and M. Sugawa for the experimental techniques and fruitful discussion; K. Matsuno for providing Drosophila myosin IC and calmodulin cDNA; and T. Toyota for the experimental methods. This work was supported by MEXT KAKENHI (Grant number JP23K18135) and MEXT KAKENHI Grant-in-Aid for Transformative Research Areas (Grant number JP23H04401 to J.Y.) and for Scientific Research on Innovative Areas (Grant number JP22H04833 to K.I.).
Author information
Authors and Affiliations
Contributions
J.Y. designed the project; Y.S. performed the experiments and data analysis; Y.S., K.Y., K.M., T.H., M.Y., and S.S. prepared biological samples; A.M. and M.Y. contributed new analytical tools; J.Y. and K.I. supervised the project; Y.S. and J.Y. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sato, Y., Yoshimura, K., Matsuda, K. et al. Membrane-bound myosin IC drives the chiral rotation of the gliding actin filament around its longitudinal axis. Sci Rep 13, 19908 (2023). https://doi.org/10.1038/s41598-023-47125-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-47125-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.