Abstract
Squamous cell carcinoma of the external auditory canal (EACSCC) is an extraordinarily rare and aggressive malignant disease. Establishment of EACSCC cell line with robust molecular characteristics is essential for the basic and translational research of EACSCC. Here, we show the newly established EACSCC cell line SCEACono2, derived from a patient with well-to-moderately differentiated EACSCC. We analyzed histologic and genetic features of SCEACono2 hiring multiple experiments, including next-generation sequencing (NGS). Immunocytochemical staining of SCEACono2 showed positivity of p53 and SCC1/2. Furthermore, SCEACono2 exhibited a unique characteristic that cytokeratin, vimentin as well as cancer stem cell markers (CD44, CD133, ALP and Oct3/4) were positive. SCEACono2 had an ability to form tumors at the temporal lesion xenograft nude mice model. NGS revealed that SCEACono2 harbored the somatic mutations of TP53 (p.G245S) and NOTCH1 (p.A465T). RNA-seq and downstream bioinformatics analysis revealed significant enrichment of genes involved in inflammation and cell adhesion in SCEACono2 compared to SCC-9 and HSC-4. STR profiling indicated no evidence of cross-contamination. In conclusion, SCEACono2 could serves as a promising and robust research resource of EACSCC in vitro and in vivo.
Similar content being viewed by others
Introduction
Squamous cell carcinoma of the external auditory canal is extraordinarily rare1,2,3,4. The external auditory canal is divided into bony and cartilaginous parts. Both parts are covered with cutaneous tissue, and the tympanic membrane is at the most medial part. Chronic inflammation produced especially by frequent ear-picking can be caused carcinogenesis in the skin covering the bone and cartilaginous part of the external auditory canal5. However, its rarity makes it difficult to set up prospective clinical studies and complicates retrospective meta-analysis, preventing the accumulation of high-quality evidence. Furthermore, the delay in establishing the cell line causes the lack of basic research evidence. So far, few reports of molecular biological studies on external auditory canal carcinoma have been reported. Almost studies have mainly used the histopathological/immunohistochemistrical method on clinical samples6,7,8,9,10,11.
The establishment of the stable cell line of the external auditory canal carcinoma, which can be the most crucial tool for the experiments both in vivo and in vitro, has been desired to accumulate the evidence of molecular biology study. A stable cell line of this carcinoma is mandatory to reveal the precise molecular characterization of external auditory canal squamous cell carcinoma and proceed with the molecular biological experiment both in vivo and in vitro. Now reported cell line of external auditory canal squamous cell carcinoma was established by Mario A. Hermsen’s group in Spain12. Until now, there has not been a report on the cell line derived from the Asian population. In Japan, ear picking, also called “mimikaki”, is a popular habit and a unique culture, which has been considered a risk factor for chronic inflammation leading to the carcinogenesis of the external auditory canal carcinoma5. Thus, we have tried to establish the stable cell line of the external auditory canal from Japanese people. Sekino et al.13 established the method to maintain the primary culture of the external auditory SCC derived from Japanese. We finally established the stable cell line of this carcinoma derived from a Japanese patient using this protocol. Furthermore, we analyze the biological and genetic characteristics of this stable cell line in this study to demonstrate the value of this cell line as a tool for molecular biology research.
Results
Establishment of SCEACono2 derived from a primary external auditory canal carcinoma tissue
The patient was a 71-year-old woman with well-to-moderately differentiated SCC of the right external auditory canal (clinical stage: cT4N0M0). Tumor tissue was collected from surgically resected primary site. Then, the primary cells derived from the tumor tissue cultured following the previously reported method13. We yielded a clonally proliferating tumor colony on a feeder layer of fibroblast cells (MMC-TIG-1-20). Subsequently, the tumor colonies were picked up and seeded on culture dishes without the feeder cells. Among the picked-up colonies, we have selected the colony that survived and grew up as a monolayer on a dish, which is presumably suitable for establishment of a cell line. To date, these cells have been passaged in vitro for more than 50 passages and exhibited stable growth and thus we named this stable cell line as SCEACono2. The population doubling time of this cell line was about 24 h (Fig. 1A). SCEACono2 adhered firmly to culture flask surface, forming island-like colonies, and had epithelial-like morphology (Fig. 1B). Importantly, the established cell line did not exhibit signs of senescence, which is considered immortalized. Any signs of contamination such as fungus, bacteria or mycoplasma was not evident in the cultured SCEACono2 cells.
SCEACono2 is positive for SCC markers and cancer stem cell markers
We next validated the characteristics of SCEACono2 as a SCC-derived cell line. For this purpose, we performed immunocytochemical staining of SCC marker proteins. SCEACono2 exhibited positivity for p53 and SCCA1/2 (Fig. 2A). Furthermore, both cytokeratin and vimentin were positive, suggesting a hybrid state of epithelial and mesenchymal characteristics (Fig. 2B). In order to examine stemness-like features of SCEACono2 cells, the presence of cancer stem cell markers (CD44, CD133, Oct3/4, ALP) was examined14,15,16,17. SCEACono2 cells showed positive for all of these cancer stem cell markers (Fig. 3A–D).
Immunostaining of SCEACono2 cell. (A) Fluorescence immunostaining of SCEACono2 cells with cancer-specific antibodies. The cells were double-stained with [anti-p53-Alexa 488/green] and [anti-SCCA1/2 + Alexa Fluor® 555 anti-mouse IgG/red]. The SCEACono2 cells have double positivity for p53 and SCC. (B) Fluorescence immunostaining of SCEACono2 cells with anti-cytokeratin-Alexa 488 (green) and anti-vimentin-PE (red). SCEACono2 cells were positive for both [anti-pan-Cytokeratin-Alexa 488/green] and [anti-Vimentin-PE/red]. Cytokeratin and vimentin are located in the cytoplasm but not in nuclei.
Immunofluorescence and alkaline phosphatase staining in SCEACono2 cell. (A–C) SCEACono2 cell was tested with anti-CD44-PE (A), anti-CD133-APC (B), and anti-Oct-3/4 (C) + Alexa Fluor® 555 anti-mouse IgG. DAPI (blue) was used to identify the location of cell nuclei for immunofluorescence staining. SCEACono2 cells have positivity of CD44, CD133, or Oct3/4. (C) ALP activity is partially observed in SCEACono2 cells.
Chromosomal instability of SCEACono2 cell line
The complicated karyotype and abnormal chromosome number of the SCEACono2 cells were revealed by chromosome analysis (Fig. 4A). Chromosome number analysis was performed on 20 cells. The number of chromosomes varied from 74 to 81, with 81 being the most common (6 cells) (Fig. 4B). Among these cells, karyotyping analysis was also performed on 8 cells that could be analyzed. Eight cells showed abnormal karyotypes, indicating chromosomal instability. We observed the five most common chromosomal aberrations in these cells as follows;
-
(1)
A chromosome with an additional fragment of unknown origin on the short arm of chromosome 9.
-
(2)
A chromosome with an additional fragment of unknown origin on the short arm of chromosome 14.
-
(3)
A chromosome with an additional fragment of unknown origin on the long arm of chromosome 18.
-
(4)
Marker chromosome 1 (mer 1).
-
(5)
Marker chromosome 3 (mer 3).
The karyograms of each cell are shown in Supplementary Fig. 1.
The short tandem repeat (STR) profile
To rule out the possibility of cross-contaminations with available cell lines, we performed STR analysis of SCEACono2. We have extracted the genomic DNA from the SCEACono2 cells after 100 times passaging. Although the STR profile of the cultured SCEACono2 cells did not exactly match that of the original patient tumor sample, their STR similarity was 0.96, indicating that the cultured SCEACono2 highly resembles the original tumor cells. (Fig. 5 and Table 1) We, therefore, considered these cells to be identical cell strains. In addition, we confirmed that there were no other identical cell lines in the Japanese Collection of Research Bioresources (JCRB).
Short tandem repeat (STR) profiles. Short tandem repeat values of cell line and tumor tissue for fifteen gene loci and amelogenin (X and Y chromosomes). The evaluation value between the cell culture and host tumor tissue was 0.96, which was high enough that the STR profile of the cell line was the same as those of the host tumor sample.
Tumorigenicity in Nude mice
We next examined the tumorigenic capacity of SCEACono2 in vivo. One week after the injection of SCEACono2 cells at the right temporal region in nude mice, subcutaneous tumor size was macroscopically increased, and we resected the tumors 16 days after inoculation. (Fig. 6A, B) We examined various injection sites to validate tumorigenicity of SCEACono2, including subcutaneous groin, knee, flank, and temporal sites. Importantly, the tumor formation rate of SCEACono2 exhibited 100% (n = 5), indicating that this cell line is suitable for cell line-derived xenograft models. Immunohistochemical staining showed that the xenografted tumor tissues were positive for the proliferation marker Ki 67 and p53 (Fig. 6C–E).
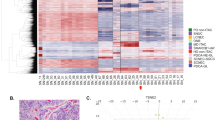
Tumorigenicity in BALB/c mice and histological analysis of the Cell line-derived Xenograft tumors in Nude mice. (A, B) Subcutaneous tumor mass (black arrow in A and red circle in B) in BALB/c mice which inoculated with 5 × 106 cells at 14 days post-injection. (C) Hematoxylin and Eosin (H&E) stain of the paraffin-embedded tumor sample confirmed the tumor formation of the differentiated squamous cell carcinoma. (D, E) Immunohistochemistry (IHC) analysis of SCEACono2 xenografts. Human Ki 67 and p53 were detected by IHC. The slide IHC image under low magnification and representative IHC image under high magnification of p53 and Ki 67 stain were shown on the left and right, respectively.
Whole exome sequencing (WES)
To clarify the genetic alterations of SCEACono2, we performed WES on genomic DNA derived from the cultured SCEACono2 cells, the original tumor tissues and the patient-matched PBMC as the non-cancerous control. The sequencing data had an average sequencing depth of 126.16 (range; 124.39–127.89), suggesting sufficient quality for mutation calling. Cultured SCEACono2 cells harbor several non-synonymous somatic mutations such as FREM3(p.S61R), GLIS3(p.S24R), COL15A1(p.V1265E), NOTCH1(p.A465T), FSCB(p.A310V), INF2(p.R87H) and TP53(p.G245S) (Table 2). Among these mutations, TP53 was also found as a significant mutation in the primary tumor. Although other mutations were excluded upon filtering, we confirmed that all of them were included in the primary tumor sample with low variant read frequencies (data not shown), suggesting that the cells harboring these mutations were selected among the heterogeneous cancer cell population. Taken together, SCEACono2 resembles genetic characteristics of the primary tumor.
Transcriptomic characteristics of SCEACono2
Finally, we performed RNA-seq and downstream bioinformatics analysis to explore the gene expression signature of SCEACono2 cells. Publicly available HNSCC cell lines HSC-4 and SCC-9 were subjected to this analysis simultaneously in order to compare their transcriptomic characteristics. Principal component analysis showed the clear separation of these three cell lines in transcriptomic levels, suggesting the unique transcriptional programs activated in SCEACono2 compared to HNSCC cell lines (Fig. 7A). We identified 1864 and 2077 upregulated genes in SCEACono2 compared to HSC-4 and SCC-9, respectively (Fig. 7B, C). In order to identify a unique transcriptional signature characterizing SCEACono2, we have extracted overlapped genes among these upregulated genes. 829 genes (27%) were found to be overlapped, whereas 1035 (33%) and 1248 (40%) genes were upregulated only in the comparison to HSC-4 and SCC-9 cell, respectively (Fig. 7D). Gene ontology analysis showed significant enrichment of genes involved in the regulation of MAPK cascade (GO: 0043408), regulation of cell adhesion (GO: 0030155), response to cytokine (GO: 0034097) and inflammatory response (GO: 0006954) in these overlapping upregulated genes. (Fig. 7E).
Comparison of gene expression among the newly established cell line, HSC-4, and SCC-9. (A) Principal component analysis showed the clear separation of these three cell lines in transcriptomic levels. (B) Volcano plot showing gene expression (FDR < 0.05; Log2FC > 1) altered (highlighted in red color) on the comparison between the SCEACono2 cell and HSC-4 cell. (C) Volcano plot showing gene expression (FDR < 0.05; Log2FC > 1) altered (highlighted in red color) on the comparison between the SCEACono2 cell and SCC-9 cell. (D) Venn diagram showing commonly upregulated genes in both comparisons of gene expression between the SCEACono2 cell line and HSC-4 and between the SCEACono2 cell line and SCC-9. Genes highlighted in pink are observed to be commonly upregulated in both comparisons. (E) Gene ontology (GO) enrichment analysis of upregulated genes. [− log10(p value)] of the corresponding biological process is shown. A dark orange bar indicates a small p value (p value < 0.1).
Discussion
Squamous cell carcinomas of the head and neck include the tongue, pharyngeal, laryngeal, and sinonasal carcinomas. The development of treatment strategies for head and neck squamous cell carcinoma has been supported by building evidence from clinical and basic research. Temporal bone squamous cell carcinoma is included as a type of temporal bone squamous cell carcinoma. However, temporal bone squamous cell carcinoma of the external auditory canal and middle ear is extremely rare compared to other head and neck SCC, and the frequency of this pathogen has been reported to account for < 0.2% of head and neck squamous cell carcinoma1,4,18. Due to its rarity, the accumulation of molecular biological evidence and establishing of treatment guidelines have been delayed. The immortalized cell lines for each pathogen have played a major role in establishing molecular biological evidence, elucidating molecular biological mechanisms of carcinogenesis and metastasis, or verifying therapeutic resistance and radiosensitivity. The establishment of cell lines for squamous cell carcinoma of the external auditory canal has lagged far behind that of other head and neck SCC. We have previously reported a method for creating a primary culture of cancer cells isolated from patient cancer tissue. Using this method, we have established a new cell line of squamous cell carcinoma of the external auditory canal. Therefore, in this report, we verified and reported the molecular and biological characteristics of the new cell line.
According to the Cellosaurus (https://web.expasy.org/cellosaurus/), excluding derivative cell lines, only one cell line, CAE606, is available as a cell line derived from the external auditory canal squamous cell carcinoma so far12. As we have previously reported, maintaining cancer cells isolated from squamous cell carcinoma of the external auditory canal is still challenging; the rarity of the disease and the difficulties in maintenance of the primary cells may explain why the establishment of immortalized cells from this disease has rarely been reported. Furthermore, the process of cell line establishment from fresh tumor tissues is energy- and time-consuming, and difficult due to contamination, out-growth of cancer-associated fibroblasts and cellular senescence, which causes the low success rate of establishment19. In this study, we tried to establish the new cell line from external auditory carcinoma based on the method that our group had reported previously13. We did not use either SV40 T-antigen, HPV E6/E7 or telomerase reverse transcriptase to immortalize cells20,21,22,23,24,25. During the primary cell culture, we could obtain a colony with enough mutations of the intrinsic gene to result in long-term survival, and our newly established cell line without the transfer of exogenous genes will be more suitable for studying EAC-SCC26.
The shape of SCEACono2 cells was quite similar to the cobblestone-like epithelioid morphology. The population doubling time was about 24 h, and this proliferative capacity can be comparable to other cells, including the Hela and SCC-9, commonly used in scientific research. Our newly established cell line harbors the typical markers of squamous cell carcinoma, including the SCCA1/2 and p53. Furthermore, this cell line expresses both vimentin and cytokeratin. This is implicated in the epithelial mesenchymal transition, associated with invasive behavior, poor differentiation, and poor prognosis. Several cancer stem cell markers, CD44, CD133, and Oct3/4, were positive, and ALP was partially positive14,15,16,17,27,28,29. These findings mean that this cell line has the characteristic of the stem cell partially. The SCEACono2 cells also displayed chromosomal instability, including the changes in chromosome number and the generation of hybrid DNA, which is related to the promotion of tumorigenesis and development30,31,32,33.
We confirmed that this new cell line could be a tool for scientific research in vivo. Injection of the newly derived cells under the temporal skin of a nude mouse resulted in tumor formation. The immunohistochemistry staining of xenograft shows the positive of Ki 67 and p53. This cell-line-derived xenograft could be used to elucidate the tumor's anatomical metastatic mechanism and select therapeutic agents for EACSCC.
In this study, we analyzed the transcriptome of the newly established cell line with RNA sequencing compared to two cell lines of tongue squamous cell carcinoma: HSC-4 and SCC-9. We found that the genes related to inflammation and cell adhesion are more expressed in the newly established cell line than the other two. Furthermore, newly established cell line harbor several somatic mutations: FREM(p.S62R), GLIS3(p.S24R), COLI5A1(p.V1265E), NOTCH1(p.A465T), FSCB(p.A310V), INF2(p.R87H), and TP53(p.G245S). The somatic mutation of TP53 is the most frequently mutated tumor suppressor gene in human squamous cell carcinoma34. Furthermore, The somatic mutation of NOTCH1 is also commonly found in human squamous cell carcinoma35. These results suggest that the major drive mutations of EACSCC are similar to those of other SCC. In addition to these somatic mutations, missense mutations in FREM3, GLIS3, COLI5A1, FSCB, and INF2 are identified. However, the molecular significance of these missense mutations in EACSCC remains unclear. The previously established cell line, CAE606, harbors inactivating mutation in CDKN2A and TP53 and high-level amplification of CCND1, affecting the cell cycle pathway. Our cell line also harbors the major drive mutations of EACSCC and has the transcriptome profiling related to inflammation and cell adhesion. These analyses with next-generation analysis suggested that this newly established cell line with these genetic backgrounds is an appropriate cell line for EACSCC research.
In conclusion, we established new cell lines derived from human squamous cell carcinoma of the external auditory meatus and named SCEACono2. This newly established cell line could be a useful tool for the pathological analysis of EACSCC to provide further insight into the mechanism of tumorigenesis, metastasis, and selection of the antitumor-drug.
Methods
Ethic statement
The Clinical Research Ethics Review Committee of Kyushu University Hospital approved the study (permit no. 29-43, 30-268, and 700-00). Written informed consent for the current research project was obtained before the tumor tissue, and a blood sample were harvested. This study was also conducted according to the principles of the Declaration of Helsinki. The animal experiment was also reviewed and approved by the Committee of. Ethics of Animal Experiments, Kyushu University Graduate School of Medical Sciences. The animals were treated according to the “Guidelines for Proper Conduct of Animal Experiments” (Science Council of Japan). The study was carried out in compliance with the ARRIVE guidelines.
Growth medium
The growth medium consisted of Dulbecco’s Modified Eagle Medium (D-MEM)/Ham’s F12 (Sigma-Aldrich, St. Louis, MO), supplemented with 10% fetal bovine serum (FBS: Sigma-Aldrich), 0.1 mM MEM Non-Essential Amino Acids Solution (Thermo Fisher Scientific, Waltham, MA), 1 mM sodium pyruvate (Thermo Fisher Scientific), 2 mM L-glutamine (Thermo Fisher Scientific) and 1% Antibiotic–Antimycotic Mixed Stock Solution (Nacalai Tesque, Inc., Kyoto, Japan).
Immunofluorescence and alkaline phosphatase staining
SCEACono2 cell for immunostaining was prepared as follows. SCEACono2 suspension for staining the Vimentin, Cytokeratin, p53, SCCA1/2, CD133, Oct3/4, and CD44 were seeded on eight-chamber slides and allowed to attach overnight. The cultures were fixed with methanol: acetone 1:1 (v/v) for 20 min at − 20 °C. These preparations for cytoplasmic antigen staining were additionally permeabilized with PBS containing 0.5% Triton X-100 and 0.05% NaN3 for 10 min at r.t. Following fixation, for cell surface marker staining, preparations were incubated with anti-CD44-PE or anti-CD133-APC for 1 h at r.t. For cytoplasmic marker staining, samples were incubated with anti-pan-Cytokeratin-Alexa Fluor® 488 and Vimentin-PE at r.t. for 1 h and with anti-p53-Alexa Fluor® 488 and anti-SCCA1/2 (mouse IgG) at 4 °C overnight. For SCCA1/2 and Oct3/4 antigens, the samples were stained with goat anti-mouse IgG (H + L) highly cross-adsorbed secondary antibody conjugated Alexa Fluor® 555 after the primary antibody reaction. Slides were then mounted with DAPI Fluoromount-G® (SouthernBiotech, Birmingham, AL) for nuclear staining. Unfixed cytospin preparations were tested with the Vector® Blue Alkaline Phosphatase Substrate kit (Vector Labs, Burlingame, CA) to check for alkaline phosphatase (ALP) activity.
Chromosome analysis: chromosomes of SCEACono2 cell
After 48 h of incubation in a culture medium, Semiconfluent and actively dividing cells were incubated in 0.5 μg/ml Colcemid for 20 min, and cells were harvested. Harvested cells were pelleted out and treated with hypotonic solution (0.075 M KCL) for 10 min before fixation in methanol-acetic acid (3:1). The suspension of fixed cells was dropped onto microscope slides. Chromosomes of SCEACono2 cells were prepared using a standard air-drying method, metaphase spreads were counted to determine the modal number, and the G-banding technique analyzed karyotypes. The metaphase chromosome spreads slide was evaluated using Ikaros software (MetaSystems, Altussheim, Germany).
Cell-line-derived xenograft
In vivo, the tumorigenicity of the SCEACono2 cells was assessed by the ability to form tumors subcutaneously in Nude mice. SCEACono2 cells were harvested and re-suspended to prepare a cell suspension of 5 × 107 cells/ml. BALB/c nude mice (5 weeks, body weight 16–18 g) were subcutaneously injected in a 0.1 ml suspension in each region. When the apparent tumor formation was macroscopically confirmed, the tumor was separated from the mice and fixed in 4% formaldehyde for pathological examination. The tumorigenesis experiment was repeated using 3 Nude mice. BALB/c nude mice were purchased from The Jackson Laboratory Japan, Inc. The xenograft tumors were examined with immunohistochemistry measure. Following deparaffinization, hydration, and blockage of endogenous peroxidase activity by 3% hydrogen peroxide, the specimen sections underwent antigen retrieval process in citrate buffer (pH6.0) and were incubated overnight at 4 °C with antibodies against p53(1:800, NCL-L-p53-DO7, Leica Camera AG, Wetzlar Germany) and Ki 67(1:100 M7240, Dako, Carpinteria, CA, USA). Then the sections were incubated with poly-peroxidase-anti-mouse/rabbit IgG and stained with diaminobenzidine (Fujifilm Wako Pure Chemical, Osaka, Japan), followed by counterstaining with hematoxylin and dehydration.
Short tandem repeat analysis
Short tandem repeat analysis Genomic DNA was extracted from the cell line and the tumor samples from the patients using a DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA). The Gene Print PowerPlex 16HS System (Promega, Madison, WI, USA) was used to amplify the extracted DNA. The amplified fragments were detected using an Applied Biosystems 3500xL Genetic Analyzer (Thermo Fisher Scientific).
DNA/RNA extraction, library preparation
The extraction of genomic DNA from the SCEACono2 and the patient-matched PBMC was conducted using DNeasy Blood and Tissue Kits (Qiagen, Chatsworth, CA, USA). The extraction of RNA from SCEACono2, HSC-4, and SCC-9 cell lines were performed using a NucleoSpin™ RNA plus kit (MachereyHagel, Düren, Germany).
Whole exome sequencing (WES)
Library preparation from DNA extracted from cultured SCEACono2 cells, the parent EACSCC tissue as well as patient-matched PBMC was conducted using an Agilent SureSelect All Exon V6 exome capture kit (Agilent Technologies, Santa Clara, CA, USA). Library preparation and WES were performed at the Beijing Genomics Institute (Shenzhen, China). The captured libraries were sequenced using DNBSEQ-G400 sequencer with the paired-end 100-bp sequence read option according to the manufacturer’s protocols.
Alignment and detection of somatic variants of SCEACono2 cells
The WES data was processed in the Genomon 2.6.2 pipeline (http://genomon.hgc.jp). Briefly, the sequencing reads were aligned to the NCBI Human Reference Genome Build 37 (hg19) with BWA version 0.7.8 using default parameters (http://bio-bwa.sourceforge.net/). PCR duplicate reads were removed using Picard (http://www.picard.sourceforge.net). Detection of somatic mutations in SCEACono2 cells and the parent EACSCC tissue was performed using the EBCall algorithm (Shiraishi Y, Sato Y, Chiba K, et al. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res. 2013;41:e89.) with the following parameters: (a) mapping quality score ≥ 30; (b) base quality score ≥ 15; (c) sequencing depths ≥ 8; (d) variant reads in SCEACono2 and EACSCC ≥ 4; (e) variant allele frequency (VAF) in SCEACono2 and EACSCC ≥ 0.05; (f) VAF in PBMC < 0.1; (g) minus logarithm of P-value of Fisher’s exact test ≥ 1.3; and (h) minus logarithm of the P value of EBCall ≥ 5.
RNA sequencing and downstream analysis
Library preparation from RNA extracted from cultured cells and RNA sequencing were performed using a DNBSEQ-G400 sequencer at the Beijing Genomics Institute (Shenzhen, China). The sequenced reads were aligned to the human reference genome GRCh38 using STAR v2.7.9a with Gencode v38 annotations. Gene count tables were generated with RSEM v1.3.1. Differentially expressed genes (DEGs) between SCEACono2, HSC-4 and SCC-9 was detected using R package DEseq2 v1.10.1. Cut-off values of DEGs were determined as absolute log2 fold change > 2 and FDR-adjusted p value < 0.01. Gene ontology analysis was performed using Metascape.
Informed consent
Informed consent was obtained from all subjects involved in the study.
Data availability
The RNA-Seq datasets (accession no. GSE240311) and the whole exome sequencing data (accession no. JGAS000645) have been deposited to the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) and the Japanese Genotypephenotype Archive (https://www.ddbj.nig.ac.jp/jga/index.html), respectively.
References
Allanson, B. M., Low, T. H., Clark, J. R. & Gupta, R. Squamous cell carcinoma of the external auditory canal and temporal bone: An update. Head Neck Pathol. 12, 407–418. https://doi.org/10.1007/s12105-018-0908-4 (2018).
Bacciu, A., Clemente, I. A., Piccirillo, E., Ferrari, S. & Sanna, M. Guidelines for treating temporal bone carcinoma based on long-term outcomes. Otol. Neurotol. 34, 898–907. https://doi.org/10.1097/MAO.0b013e318281e0a9 (2013).
Prasad, S. C., D’Orazio, F., Medina, M., Bacciu, A. & Sanna, M. State of the art in temporal bone malignancies. Curr Opin Otolaryngol Head Neck Surg 22, 154–165. https://doi.org/10.1097/MOO.0000000000000035 (2014).
Seligman, K. L., Sun, D. Q., Ten Eyck, P. P., Schularick, N. M. & Hansen, M. R. Temporal bone carcinoma: Treatment patterns and survival. Laryngoscope https://doi.org/10.1002/lary.27877 (2019).
Tsunoda, A., Sumi, T., Terasaki, O. & Kishimoto, S. Right dominance in the incidence of external auditory canal squamous cell carcinoma in the Japanese population: Does handedness affect carcinogenesis?. Laryngoscope Investig. Otolaryngol. 2, 19–22. https://doi.org/10.1002/lio2.43 (2017).
Alessandrini, L. et al. Temporal bone squamous cell carcinoma: Molecular markers involved in carcinogenesis, behavior, and prognosis: A systematic review. Int. J. Mol. Sci. https://doi.org/10.3390/ijms23094536 (2022).
Hongo, T. et al. Programmed death-ligand 1 expression and tumor-infiltrating lymphocytes in temporal bone squamous cell carcinoma. Laryngoscope https://doi.org/10.1002/lary.29689 (2021).
Hongo, T. et al. High-risk HPV-related squamous cell carcinoma in the temporal bone: A rare but noteworthy subtype. Virchows Arch. 482, 539–550. https://doi.org/10.1007/s00428-023-03497-7 (2023).
Miyazaki, M. et al. Poorly differentiated clusters predict a poor prognosis for external auditory canal carcinoma. Head Neck Pathol. 13, 198–207. https://doi.org/10.1007/s12105-018-0939-x (2019).
Miyazaki, M. et al. Highly expressed tumoral emmprin and stromal CD73 predict a poor prognosis for external auditory canal carcinoma. Cancer Sci. https://doi.org/10.1111/cas.14508 (2020).
Ungar, O. J. et al. Invasion patterns of external auditory canal squamous cell carcinoma: A histopathology study. Laryngoscope 131, E590–E597. https://doi.org/10.1002/lary.28676 (2021).
Garcia-Marin, R. et al. A novel external auditory canal squamous cell carcinoma cell line sensitive to CDK4/6 inhibition. Otolaryngol. Head Neck Surg. https://doi.org/10.1177/01945998221089186 (2022).
Sekino, Y. et al. Establishment and characterization of a primary cell culture derived from external auditory canal squamous cell carcinoma. FEBS Open Bio https://doi.org/10.1002/2211-5463.13225 (2021).
Baillie, R., Tan, S. T. & Itinteang, T. Cancer stem cells in oral cavity squamous cell carcinoma: A review. Front. Oncol. 7, 112. https://doi.org/10.3389/fonc.2017.00112 (2017).
Chen, X. et al. p53 positively regulates the expression of cancer stem cell marker CD133 in HCT116 colon cancer cells. Oncol. Lett. 16, 431–438. https://doi.org/10.3892/ol.2018.8619 (2018).
Kamal, A. F. et al. Isolation, culture and characterization of cancer stem cells from primary osteosarcoma. Open Stem Cell J. 5, 1–13 (2018).
Major, A. G., Pitty, L. P. & Farah, C. S. Cancer stem cell markers in head and neck squamous cell carcinoma. Stem Cells Int. 2013, 319489. https://doi.org/10.1155/2013/319489 (2013).
Moody, S. A., Hirsch, B. E. & Myers, E. N. Squamous cell carcinoma of the external auditory canal: An evaluation of a staging system. Am. J. Otol. 21, 582–588 (2000).
Bian, X. et al. Establishment and characterization of a new human colon cancer cell line, PUMC-CRC1. Sci. Rep. 11, 13122. https://doi.org/10.1038/s41598-021-92491-7 (2021).
Dalerba, P. et al. Reconstitution of human telomerase reverse transcriptase expression rescues colorectal carcinoma cells from in vitro senescence: Evidence against immortality as a constitutive trait of tumor cells. Cancer Res. 65, 2321–2329. https://doi.org/10.1158/0008-5472.CAN-04-3678 (2005).
Maqsood, M. I., Matin, M. M., Bahrami, A. R. & Ghasroldasht, M. M. Immortality of cell lines: Challenges and advantages of establishment. Cell Biol. Int. 37, 1038–1045. https://doi.org/10.1002/cbin.10137 (2013).
Pavez Lorie, E. et al. Characterisation of the novel spontaneously immortalized and invasively growing human skin keratinocyte line HaSKpw. Sci. Rep. 10, 15196. https://doi.org/10.1038/s41598-020-71315-0 (2020).
Tsao, S. W. et al. Establishment of two immortalized nasopharyngeal epithelial cell lines using SV40 large T and HPV16E6/E7 viral oncogenes. Biochim. Biophys. Acta 1590, 150–158. https://doi.org/10.1016/s0167-4889(02)00208-2 (2002).
Wang, R. et al. Establishment and characterization of a prostate cancer cell line from a prostatectomy specimen for the study of cellular interaction. Int. J. Cancer 145, 2249–2259. https://doi.org/10.1002/ijc.32370 (2019).
Zhan, K., Lin, M., Zhao, Q. M., Zhan, J. S. & Zhao, G. Q. Biological characterization of bovine mammary epithelial cell lines immortalized by HPV16 E6/E7 and SV40T. In Vitro Cell Dev. Biol. Anim. 52, 906–910. https://doi.org/10.1007/s11626-016-0063-8 (2016).
Feng, H. et al. Intrinsic gene changes determine the successful establishment of stable renal cancer cell lines from tumor tissue. Int. J. Cancer 140, 2526–2534. https://doi.org/10.1002/ijc.30674 (2017).
Al-Mudamgha, Z. A., Rassam, M. B., Al-Salihi, A. R. & Al-Sammeraie, F. T. Alkaline phosphatase of cancerous larynx tissue in comparison with the placental enzyme. Biochemical and histochemical studies. Acta Oncol. 36, 213–218. https://doi.org/10.3109/02841869709109232 (1997).
Noguchi, K. et al. Susceptibility of pancreatic cancer stem cells to reprogramming. Cancer Sci. 106, 1182–1187. https://doi.org/10.1111/cas.12734 (2015).
O’Connor, M. D. et al. Alkaline phosphatase-positive colony formation is a sensitive, specific, and quantitative indicator of undifferentiated human embryonic stem cells. Stem Cells 26, 1109–1116. https://doi.org/10.1634/stemcells.2007-0801 (2008).
Bakhoum, S. F. et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472. https://doi.org/10.1038/nature25432 (2018).
Johnson, S. C. & McClelland, S. E. Watching cancer cells evolve through chromosomal instability. Nature 570, 166–167. https://doi.org/10.1038/d41586-019-01709-2 (2019).
Li, J., Dallmayer, M., Kirchner, T., Musa, J. & Grunewald, T. G. P. PRC1: Linking cytokinesis, chromosomal instability, and cancer evolution. Trends Cancer 4, 59–73. https://doi.org/10.1016/j.trecan.2017.11.002 (2018).
McClelland, S. E. Role of chromosomal instability in cancer progression. Endocr. Relat. Cancer 24, T23–T31. https://doi.org/10.1530/ERC-17-0187 (2017).
Cancer Genome Atlas, N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582. https://doi.org/10.1038/nature14129 (2015).
Dotto, G. P. & Rustgi, A. K. Squamous cell cancers: a unified perspective on biology and genetics. Cancer Cell 29, 622–637. https://doi.org/10.1016/j.ccell.2016.04.004 (2016).
Acknowledgements
The super-computing resource was provided by Human Genome Center, the Institute of Medical Science, the University of Tokyo. We would like to thank Uni-edit (http://uni-edit.net/) for editing and proof-reading this manuscript.
Funding
This work was supported in part by JSPS KAKENHI (Grant Numbers JP 22H03236 and 22K09745) and the Soda Toyoji Memorial Foundation. Institutional Review Board Statement: Our study was approved by the ethics committee of Kyushu University Hospital (permit no. 29-43, approval date: 17 April 2017, no. 30-268, approval date: 4 October 2018, and no. 700-00, approval date: 26 December 2016).
Author information
Authors and Affiliations
Contributions
N.K. designed the study. R.K., H.O. and N.T. supervised this project. K.S., M.O., A.I., S.M. and S.I. performed the experiments and analyzed the results in cell culture and characterization. T.M., R.K. and T.H. performed the histological examination. K.S. performed genetic analysis. M.O. and A.I. performed the experiments and analyzed data in cell quality control. N.K. and K.S. prepared the manuscript. N.K. and N.T. provided the funding. All authors reviewed the results and the manuscript and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Komune, N., Sato, K., Ono, M. et al. Biological and genetic characterization of a newly established human external auditory canal carcinoma cell line, SCEACono2. Sci Rep 13, 19636 (2023). https://doi.org/10.1038/s41598-023-46926-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-46926-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.