Abstract
Regional lymph node metastasis is an important predictor for survival outcome and an indicator for postoperative adjuvant chemotherapy in patients with colorectal cancer. Even with advances in adjuvant chemotherapeutic regimens, 5-year distant metastasis and survival rates are still unsatisfactory. Here, we evaluate the clinical significance of polymorphisms in receptors for HMGB1, which is the hallmark of chemotherapy-induced immunogenic cell death, in patients with stage II–III colon carcinoma (COAD). We found that high cytosolic HMGB1 is elicited in stage III COAD patients who received adjuvant chemotherapy. Patients with the TLR1-N248S polymorphism (rs4833095), which causes loss-of-function in HMGB1-mediated TLR1–TLR2 signaling, may influence the therapeutic efficacy of adjuvant chemotherapy, leading to a high risk of distant metastasis within 5 years [HR = 1.694, 95% CI = 1.063–2.698, p = 0.027], suggesting that TLR1-N248S is an independent prognostic factor for locally advanced colon carcinoma patients. We found that defective TLR1 impaired TLR1/2 signaling during dendritic cell (DC) maturation for the antitumor immune response under immunogenic chemotherapy oxaliplatin (OXP) treatment. Defective TLR1 on DCs impaired their maturation ability by HMGB1 and reduced the secretion of IFNγ from T cells to eradicate tumor cells in vitro. Moreover, systemic inhibition of TLR1/2 dramatically reduced the tumor-infiltrating immune cells by OXP treatment, leading to poor therapeutic response to OXP. In contrast, administration of a TLR1/2 agonist synergistically increased the benefit of OXP treatment and triggered a high density of tumor-infiltrating immune cells. We also observed that fewer tumor-infiltrating cytotoxic T lymphocytes were located within the tumor microenvironment in patients bearing the TLR1-N248S polymorphism. Overall, our results suggest that dysfunctional TLR1 may reduce the therapeutic response to adjuvant chemotherapy by impairing HMGB1-mediated DC maturation and attenuating the antitumor immune response in locally advanced colon carcinoma patients.
Similar content being viewed by others
Introduction
Lymph node metastasis is a crucial predictor of survival outcome and an indicator for postoperative adjuvant chemotherapy in patients with colorectal cancer (CRC)1,2,3. Despite advances in therapeutic approaches such as combinational chemotherapy regimens, targeted therapy and radiotherapy, metastatic disease remains the leading factor in CRC mortality. Not all patients benefit from standard chemotherapy regimens, making it critical to stratify patients using predictive and prognostic biomarkers to identify those who will benefit the most from chemotherapy, such as immune-related genes4.
Toll-like receptors (TLRs) play a crucial role in intestinal mucosal innate and acquired immunity to maintain gut homeostasis5. TLRs not only recognize exogenous pathogen-associated molecular patterns (PAMPs) but also sensitize endogenous damage-associated molecular patterns (DAMPs) that are released from injured or dying cells6. These DAMPs shape adaptive anticancer immunity through the activation of antigen-presenting cells (APCs) for tumor antigen processing and cross-presentation to CD8+ T cells. Therefore, several groups have established prognostic biomarkers based on immunogenic cell death (ICD)-associated DAMP signatures to estimate prognosis and immunotherapy effectiveness in several malignancies, such as triple-negative breast cancer (TNBC) and head and neck squamous cell carcinoma (HNSCC)7,8. Several immunogenic chemotherapeutic drugs, such as oxaliplatin (OXP) and doxorubicin (DOX), promote ICD, releasing DAMPs, including high-mobility group box 1 (HMGB1), heat shock protein 70 (Hsp70), ATP, annexin A1 (ANXA1) and calreticulin (CRT)9,10. HMGB1 can activate TLR2 and TLR4 on dendritic cells (DCs) to initiate antigen presentation and cytokine secretion, activating antigen‐specific CD4+ T cells and CD8+ T cells to recognize and eradicate tumor cells. Furthermore, TLR2 agonists have significantly inhibited cancer growth by stimulating CD8+ T cells and natural killer (NK) cells11,12. TLR2 interacts with TLR1 or TLR6 as a functional heterodimer to activate downstream signaling in regulating the mucosal immune response within the gut13. Several single nucleotide polymorphisms (SNPs) in the TLR1 gene impair the response to several bacterial agonists for this receptor and affect T helper 1 cytokine production, including P315L (rs5743613), N248S (rs4833095) and I602I (rs5743618)14. These polymorphisms of TLR1 are associated with the response to FOLFIRI plus bevacizumab in metastatic CRC15. Additionally, the expression of TLR1 and TLR2 serves as prognostic factors in breast cancer and pancreatic ductal adenocarcinoma (PDAC)16,17. Recent studies have shown that the activation of TLR1/2 signaling enhances the antitumor efficacy of CTLA-4 blockade, suggesting that TLR1/2 signaling participates in not only innate immunity against bacterial infection but also adaptive immunity to eradicate tumor cells. Therefore, several TLR immunomodulators have been linked to anticancer therapies in preclinical studies and clinical trials18,19, including the TLR2 agonist SMU-Z1 and polysaccharide krestin (PSK)11,20, TLR3 agonist AMP‐51621, TLR5 agonist entolimod (CBLB502)22, TLR7 agonist imiquimod23 and TLR9 agonist CpG‐790924.
In this study, we investigated the impact of genetic variation in TLR1 and TLR2, which are the receptors for HMGB1, on the clinical outcomes of colon carcinoma (COAD) patients who underwent postoperative adjuvant chemotherapy, specifically those with regional lymph node metastatic COAD. Our findings suggest that stage III COAD patients carrying the TLR1-N248 polymorphism exhibit poorer distant metastasis-free survival (DMFS) and disease-free survival (DFS). Defective TLR1 signaling decreases HMGB1-mediated dendritic cell (DC) maturation induced by immunogenic chemotherapy. Blocking TLR1/2 signaling significantly reduced the therapeutic efficacy of the immunogenic chemotherapeutic agent, resulting in reduced infiltration of DCs, CD4, and CD8 within the tumor microenvironment. However, administering a TLR1/2 agonist significantly increased the therapeutic efficacy of OXP by boosting the antitumor immune response. In conclusion, our results indicate that dysfunctional TLR1 reduces the extent of antitumor immunity elicited by immunogenic chemotherapy, leading to poor response to OXP and worse survival outcomes in locally advanced CRC patients.
Material and methods
Patient characteristics
From 2006 to 2014, 869 stage II–III colon carcinoma patients who were histologically and clinically diagnosed underwent surgery, followed by postoperative adjuvant chemotherapy (high-risk stage II and stage III patients), and were followed-up once every 6 months postoperatively for 5 years at China Medical University Hospital (CMUH). Adjuvant chemotherapy regimens were administered as previously described5,14. All study protocols were approved by the Institutional Review Board (IRB) of CMUH [Protocol number: CMUH105-REC2-073].
Tissue microarray (TMA) construction and immunohistochemistry
Stage II–III colon carcinoma patients who were diagnosed and treated between 2006 and 2014 at China Medical University Hospital were enrolled in our cohort25,26. The TMA included resected primary tumor tissue and their corresponding normal mucosa specimens, which was approved by the Institutional Review Board (IRB) of China Medical University Hospital [Protocol number: CMUH105-REC2-073]. The collection of specimens and methods was carried out in accordance with the approval guidelines from the committee. Briefly, central areas of the resected primary tumor were marked on hematoxylin/eosin-stained slides by pathology, and the corresponding area on the matching formalin-fixed, paraffin-embedded tissue (FFPE) was then identified by punching 2-mm-diameter tissue cylinders. The criteria for TMA spots were at least 50% tumor cells27.
Immunohistochemistry (IHC) was performed using 3-μm-thick TMA sections with the indicated antibodies, followed by an HRP-conjugated avidin biotin complex (ABC) Kit (Vector Laboratories, CA, USA), incubation with HRP substrate DAB chromogen (Vector Laboratories) and counterstaining in hematoxylin28,29. The immune cell markers CD8 (ab217344, Abcam) were counted at 400× magnification under a microscope. The average number of TILs in five high-power fields (400× magnification) was scored by two pathologists. Cytosolic HMGB1 (ab18256, Abcam) expression was based on the H-score25,30, according to the intensity and percentage of positive cells for histoscore (H-score), which was calculated by a semiquantitative assessment of both the intensity of the staining (0: negative staining; 1: weak; 2: moderate; and 3: strong) and the percentage of immunopositive cells. The range of the H-score was from 0 to 300. The cytosolic HMGB1expression status was categorized as low or high according to the median value of the H-score.
Genomic DNA extraction and SNP genotyping
Genomic DNA from nontumor tissues of rectal cancer patients was extracted from two 5-μm-thick FFPE slides using a QIAamp® DNA FFPE Extraction Kit (QIAGEN GmbH, Hilden, Germany). For SNP genotyping, 10 ng of total genomic DNA was used for PCR amplification and was performed using the iPLEX® HS panel on the MassARRAY® System (Agena Bioscience, San Diego, CA, USA), which employs matrix-assisted laser desorption/ionization time-of-fight mass spectrometry for amplicon detection (MALDITOF-MS; SpectroACQUIRE, Agena Bioscience). Primers designed (Supplementary Table S1) for PCR amplification of specific polymorphisms and extension reactions were prepared using MassARRAY® Assay Design Version 3.1 software (Agena Bioscience, San Diego, CA, USA). Following PCR, SAP addition, and the iPLEX HS® extension reaction, the samples were desalted by resin treatment for 15 min, spotted onto SpectroCHIP® Arrays (Agena Bioscience, San Diego, CA), analyzed by mass spectrometry, and ultimately interpreted using SpectroTYPER v4.0 software (Agena Bioscience, San Diego, CA). The characteristics of the selected polymorphisms are shown in Table S1.
Cell line and reagents
Two human colorectal cancer cell lines, HT29 and HCT116, the monocytic leukemia cell line THP1 and human T-cell leukemia Jurkat cells, were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (HyClone, MA, USA) at 37 °C with a humidified atmosphere of 5% CO2 and 95% air. CU-CPTT22 (TLR1/2 antagonist), TLR4-IN-C34 (TLR4 antagonist), FPS-ZM1 (RAGE antagonist) and Pam3CSK4 (TLR1/2 agonist) were purchased from MedChemExpress (MCE, NJ, USA).
Western blot analysis
Total cell lysates (20–40 μg) were separated via 6–12% SDS‒PAGE and transferred onto a PVDF membrane (Millipore, MA, USA)31. The membranes were blocked with BlockPRO™ Blocking buffer (BioLion Biotech, Taipei, Taiwan), probed with specific antibodies overnight at 4 °C, and then incubated with HRP-conjugated secondary antibodies for 1 h. After antibody incubation, the membranes were incubated with Immobilon Western Chemiluminescent HRP Substrate (Millipore) and analyzed by an ImageQuant™ LAS 4000 biomolecular imager (GE Healthcare, Amersham, UK). The digital data were processed using Adobe Photoshop and quantified using ImageJ software (NIH, MD, USA). Each blot was stripped by immunoblotting stripping buffer (BioLion Tech.) before reprobing with the other antibodies. The antibodies used in western blot analysis were as follows: anti-p-IKKα/β (AP0546, Abclonal), anti-p-JNK (AP0631, Abclonal) and anti-TLR1 (A0997, Abclonal)28,29.
THP1-derived immature DCs and colorectal cancer cell line coculture
The human monocytic leukemia cell line THP1 was cultured and maintained in RPMI 1640 medium supplemented with 10% FCS (Life Technologies, Grand Island, New York, USA), 2 mM glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 mg/ml streptomycin at 37 °C in a humidified incubator with 95% air and 5% CO2. After the gene was silenced by lentivirus carrying shRNA against the indicated genes, immature DCs were generated from THP1 cells as previously described32. Briefly, THP1 cells were differentiated into immature DCs by adding 1500 IU/ml rhIL-4 (Sino Biological, Beijing, China) and 1500 IU/ml rhGM-CSF (Sino Biological, Beijing, China) in culture medium for at least 7 days, with cytokine-supplemented culture medium changed every 2–3 days at 37 °C in a humidified incubator with 95% air and 5% CO2. These THP1-iDC cells were treated with recombinant human HMGB1 (Sino Biological, Beijing, China) for 24 h. DC maturation was measured by flow cytometry. These THP1-iDC cells were also cocultured with OXP-treated HT29-GFP cells to analyze the DC maturation marker CD86 by flow cytometry. In addition, the human T-cell leukemia Jurkat cell line was coincubated with OXP-treated HT29/THP1-iDCs for 15 h to analyze the level of IFNγ.
Animal model
BALB/c mice (female, 5 weeks old) were maintained according to the institutional guidelines approved by the Institutional Animal Care and Use Committee of China Medical University [Protocol number: CMUIACUC-2020-057]. CT26 cells (2 × 105 cells/mouse) were suspended in 100 μL 20% Matrigel and subcutaneously inoculated into the right flank of each mouse. After 10 days, mice were administered oxaliplatin (OXP, 6 mg/kg/mouse, intraperitoneal injection) 4 times at 3-day intervals on Days 13, 16, 19, 22 and 25. TLR1/2 antagonist CU-CPT22 (2.5 mg/kg, HY-108471, MCE) and TLR1/2 agonist Pam3CSK4 (2.5 mg/kg, HY-P1180, MCE). The tumor volume was measured at 3-day intervals in the study, and tumor volumes were calculated according to the formula (width2 × length)/2. All animals were sacrificed appropriately at the end of the experiments with CO2, and the tumor tissues were collected for further analysis, including immunoblotting analysis and immunohistochemistry33,34. The study was performed with the protocols in the Institutional Animal Care and Use Committee of China Medical University and carried out in compliance with the ARRIVE guidelines35.
Tumor processing to isolate tumor-infiltrating lymphocytes (TILs) and lymph nodes for flow cytometry
For analysis of tumor-infiltrating immune cells, mice from each treatment group were sacrificed on Day 28, and their tumors and tumor-draining lymph nodes were isolated. Isolated tumors were weighed, and mechanically dissected into small pieces (1–2 mm) by a beaver blade, filtered through a 70-μm nylon cell strainer, and then resuspended in blank RPMI medium. Thereafter, the cell suspensions were layered over Ficoll-Paque media and centrifuged at 1025×g for 20 min. The layer of mononuclear cells was transferred into a conical tube, 20 ml of complete RPMI media was added, and then the cells were gently mixed and centrifuged at 650×g for 10 min twice. Finally, the supernatant was removed, and the TILs were resuspended in complete RPMI medium36.
For Treg staining, these cells were fixed and permeabilized with a FoxP3 Fix/Perm buffer kit from Biolegend according to the manufacturer’s instructions and then stained with intracellular antibodies for further analysis by flow cytometry. For quantification, the absolute numbers of different cell types per gram of tumor were measured by flow cytometric analysis. For surface marker staining, TILs were resuspended in 500 μL of staining buffer (2% BSA, 0.1% NaN3 in PBS). The cells were stained with different surface marker panels: (1) T cells phenotype: CD3-FITC (100,204, Biolegend, CA, USA), CD4-APC/FireTM750 (116,020, Biolegend, CA, USA), CD8a-PerCPCy5.5 (100,734, Biolegend, CA, USA), CD44-PE (103,008, Biolegend, CA, USA), CD45-PECy7 (103,114, Biolegend, CA, USA), CD62L-APC (104,412, Biolegend, CA, USA) and their isotype; (2) Regulatory T lymphocyte (Treg): FoxP3-Alexa488 (126,406, Biolegend, CA, USA), CD25-PE (101,904, Biolegend, CA, USA), CD3-PerCPCy5.5 (100,218, Biolegend, CA, USA), CD45-PECy7 (103,114, Biolegend, CA, USA), CD127-Alexa647 (135,020, Biolegend, CA, USA), CD4-APC/FireTM750 (116,020, Biolegend, CA, USA), and their isotype; (3) IFNγ+CD8+ T cells: CD3-FITC (100,204, Biolegend, CA, USA), IFNγ-PE (505,808, Biolegend, CA, USA), CD45-PECy7 (103,114, Biolegend, CA, USA), CD8-APC/FireTM750 (100,766, Biolegend, CA, USA) and their isotype. Data were acquired on a Guava easyCyte flow cytometer and analyzed by FlowJo Software37,38.
Statistical analysis
The statistical analysis was performed by SPSS (IBM SPSS Statistics 22, WA, USA), and GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA) was utilized. A two-sided p < 0.05 was used as the significance level for all tests, such as Student’s t test, Pearson chi-square test and Fisher’s exact test. Univariate and multivariate analyses were performed by Cox regression analysis to estimate the hazard ratios (HRs) and 95% confidence intervals (CIs). These influential factors were adjusted in the Cox models, including sex (male vs. female), age (≥ 65 vs. < 65), pT stage (pT 3–4 vs. pT 1–2), tumor location (proximal vs. distal), tumor differentiation (poor vs. well to moderate), LVI (present vs. absent), PNI (present vs. absent) and TLR1-N248 (variant vs. WT) and TLR2 promoter (variant vs. WT). Kaplan‒Meier analysis was used to assess distant metastasis-free survival (DMFS) and disease-free survival (DFS) with the survival time between surgery and events such as tumor relapse and death.
Ethical approval
This study was reviewed and approved by the Internal Review Board (IRB) of China Medical University Hospital [Protocol number: CMUH105-REC2-073]. The method was carried out in accordance with the committee’s approved guidelines.
Informed consent
Informed consents were obtained from all participants in the study.
Results
Clinical characteristics and genotype of TLR1/TLR2, which are HMGB1 receptors, in stage II–III patients
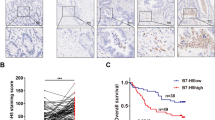
Our previous studies have demonstrated that the density and cytosolic pattern of HMGB1 (cyto-HMGB1) is an independent prognostic factor in locally advanced rectal cancer patients who underwent preoperative concurrent chemoradiotherapy, suggesting that chemoradiotherapy elicits the release of HMGB1 to promote ICD and antitumor immunity26. In this study, we investigated whether cyto-HMGB1 is associated with survival outcomes in patients with locally advanced colon adenocarcinoma (COAD) who received adjuvant chemotherapy. As shown in Fig. 1A–C, high cyto-HMGB1 levels in tumor tissues of stage III COAD patients were associated with poor distant metastasis-free survival (DMFS) and disease-free survival (DFS). Patients with low cyto-HMGB1 had a negative correlation with higher nuclear HMGB1 expression and were associated with a more favorable survival outcome, which was consistent with previous studies39. These results suggest that environment-driven inflammation for cyto-HMGB1 before chemotherapy may promote tumor progression and inhibit antitumor immunity with other mechanisms, such as autophagy39,40,41,42,43. However, chemotherapy-induced ICD signatures may be associated with antitumor immunity and influence the risk of distant metastasis and the therapeutic efficacy of chemotherapy8. Our results demonstrated that cyto-HMGB1 was significantly increased in advanced-stage CRC patients after chemotherapy treatment (Fig. 1D, p = 0.0017).
The association between distant metastasis-free survival (DMFS), disease-free survival (DFS) and TLR1 genotypes in locally advanced stage III COAD patients. (A) Representative images of cyto-HMGB1 expression within the tumor microenvironment in advanced colorectal cancers. (B) Kaplan–Meier curves showed that inflammation-driven cyto-HMGB1 expression was associated with poor DMFS in locally advanced COAD patients (n = 410, p = 0.0125). (C) Kaplan–Meier curves showed that inflammation-driven cyto-HMGB1 expression was associated with poor DFS in locally advanced COAD patients (n = 410, p = 0.0164). (D) The intensity of cyto-HMGB1 (H-score) within the TME in colorectal cancer patients before and after chemotherapy treatment was evaluated by IHC (n = 19, p = 0.0017, unpaired t test). (E) Kaplan–Meier curves showed that TLR1-WT (AA genotype) is associated with favorable DMFS in locally advanced COAD patients compared to TLR1-N248S (AG and GG genotypes). (F) Kaplan–Meier curves showed that TLR1-WT (AA genotype) is associated with favorable DFS in locally advanced COAD patients compared to TLR1-N248S (AG and GG genotypes).
To evaluate whether genetic defects in TLR1, TLR2 and TLR4 may influence HMGB1-mediated antitumor immunity and be associated with distant metastasis and tumor relapse in advanced-stage CRC patients after chemotherapy treatment. We evaluated the genetic polymorphisms of TLR1/2, namely, TLR1 (N248S/rs4833095), TLR1 (S602I/rs5743618), TLR2 (N729S/rs61735278), TLR2 promoter (− 196 to − 174 deletion/rs111200466), TLR4 (D299G/rs4986790) and TLR4 (T399I/rs4986791), in stage II–III COAD patients (n = 869). The individual genotypes and allele frequencies were shown and were consistent with the global allele frequencies in Table S1, which were classified based on the NCBI dbSNP. The genotype GG of TLR1 (N248S/rs4833095) and promoter deletion of TLR2 (− 196 to − 174 deletion/rs111200466) were prevalent among COAD patients (Table 1, n = 869). The mean age at diagnosis was 65.14 ± 13.65 years (range, 13–92 years). The majority of the patients were men (53.9%). After surgery, 60 patients (6.9%) exhibited local recurrence, and 147 patients (16.9%) exhibited distant metastasis within 10 years.
Association of SNPs with clinical outcome in stage II–III COAD patients
Because the prevalence of TLR1 (S602I/rs5743618), TLR2 (N729S/rs61735278) and TLR4 (D299G/rs4986790 and T399I/rs4986791) is lower, we only evaluated the relationship of TLR1-N248S and TLR2 promoter (− 196 to − 174 deletion) to DMFS and DFS. As shown in Table 2, older age (59% vs. 80%, p < 0.001) and presence of lymphovascular invasion (LVI, 61% vs. 71%, p = 0.01) exhibited a significantly higher risk in DMFS and DFS, and presence of perineural invasion (PNI, 59% vs. 70%, p = 0.033) was significantly associated with poor 5-year DFS in stage II COAD patients by Kaplan–Meier survival analysis. The presence of TLR1-N248S and the TLR2 promoter (− 196 to − 174 deletion) was not associated with DMFS and DFS in stage II COAD patients. However, the variants of TLR1-N248 (AG/GG genotype) had a notably higher risk in DMFS (52% vs. 68%, p = 0.019) and DFS (51% vs. 66%, p = 0.026) in stage III COAD, suggesting that the TLR1-N248S polymorphism may be associated with regional lymph node metastasis.
In the univariate analysis of DMFS in locally advanced stage III COAD patients, the following parameters were associated with patient survival rate: age, pT stage, LVI and PNI. Moreover, patients with the TLR1-N248 polymorphism had an increased risk for lower DMFS (HR = 1.725, 95% CI = 1.084–2.745, p = 0.0215, Fig. 1E) and DFS (HR = 1.632, 95% CI = 1.121–2.395, p = 0.0296, Fig. 1F) compared with patients carrying wild-type TLR1 (Table 3). These results indicate that the TLR1-N248 polymorphism has significant prognostic value for locally advanced stage III COAD patients. Subsequently, we examined whether the inclusion of other variables was significantly associated with the survival of locally advanced COAD. Our results showed that the TLR1-N248 polymorphism (HR = 1.694, 95% CI = 1.063–2.698, p = 0.027) was an independent prognostic factor of DMFS for locally advanced COAD patients (Table 3), indicating that the TLR1-N248 polymorphism has significant prognostic value for COAD patients with regional lymph node metastasis. These results suggested that loss-of-function in TLR1 may impair the immunogenic effect of released HMGB1 on antitumor immunity elicited by immunogenic chemotherapeutic agents such as oxaliplatin (OXP).
Defective TLR1 signaling attenuated OXP-elicited TLR1/TLR2 signaling activation
To demonstrate that defective TLR1 impairs HMGB1-mediated TLR1/2 signaling for DC maturation by immunogenic chemotherapy, we first treated HT29 and HCT116 cells with OXP and analyzed the level of released HMGB1 (Fig. 2A). Indeed, we found that released HMGB1 was clearly detected in conditioned medium (CM) after OXP treatment (Fig. 2A). These OXP-treated CM from HT29 and HCT116 cells significantly triggered the activation of the TLR-mediated downstream, MYD88, ERK and IRF3 signaling pathway in THP1-iDC cells (Fig. 2B). However, the addition of anti-HMGB1 neutralizing antibodies clearly diminished the activation of the TLR-mediated signaling pathway, suggesting that HMGB1 released by OXP was critical to enhance DC maturation by TLR receptors. The released HMGB1 promoting DC maturation was mainly dependent on TLR2, TLR4 and RAGE for antitumor immunity26,44,45. Therefore, we combined several small molecule inhibitors, including FPS-ZM1 (RAGE antagonist), TLR4-IN-C34 (TLR4 antagonist) and CU-CPTT22 (TLR1/2 antagonist), with HMGB1 recombinant protein (rhHMGB1) to evaluate DC maturation. We found that blockade of TLR1/2, TLR4 and RAGE significantly decreased HMGB1-enhanced DC maturation (Fig. 2C). The level of DC maturation markers CD80 and CD86 were significantly decreased (Fig. 2C). But we found that TLR1/2 and TLR4 mainly participate in HMGB1-mediated DC maturation, compared to RAGE signaling pathway.
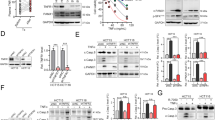
Knockdown of TLR1 reduced TLR1/2 signaling for DC maturation. (A) The level of HMGB1 in conditioned medium (OXP: 25 μM) after 24 h treatment was evaluated by an ELISA kit (n = 3). One-way ANOVA t test, ***p < 0.001. (B) The conditioned medium from OXP (25 μM)-treated HT29 and HCT116 cells was collected after 24 h of treatment. These CMs were preincubated with anti-HMGB1 antibodies (1 μg/mL) for 0.5 h and then added to THP1-iDCs, which were differentiated into immature DCs (iDCs) by IL-4 (1500 IU/ml) and GM-CSF (1500 IU/ml) for 7 days. (C) THP1-iDCs were treated with different small molecules for 1 h, and then rhHMGB1 was added for 24 h to analyze the maturation of DCs (CD86 and CD80) by qRT‒PCR (n = 3). One-way ANOVA t test, **p < 0.01. (D) TLR1 was knocked down in THP1 cells by lentivirus carrying shRNA with a spinoculation protocol. Then, THP1-iDCs were treated with rhHMGB1 for 24 h for analysis by immunoblotting. (E) THP1shNC-iDCs and THP1shTLR1-iDCs were treated with rhHMGB1 for 24 h to analyze the maturation of DCs (CD86 and CD80) by qRT‒PCR (n = 3). One-way ANOVA t test, **p < 0.01. (F) THP1shNC-iDCs and THP1shTLR1-iDCs were treated with rhHMGB1 protein for 24 h. DC maturation CD86 was analyzed by flow cytometry (n = 3). One-way ANOVA t test, **p < 0.01. (G) The diagram scheme of coculture analysis. HT29-GFP cells were treated with OXP (20 μM) for 24 h, and then the drugs were washed to coculture with THP1shNC-iDCs and THP1shTLR1-iDCs for 24 h. DC maturation of CD86 was analyzed by flow cytometry (n = 3). One-way ANOVA t test, **p < 0.01. (H) HT29 cells were treated with OXP (20 μM) for 24 h, and then the drugs were washed out and cocultured with THP1shNC-iDCs and THP1shTLR1-iDCs for 24 h. After coculture with iDCs for 24 h, the Jurkat T cells were coincubated for 15 h to analyze the IFNγ+ T cells by flow cytometry (n = 3). One-way ANOVA t test, *p < 0.05 and **p < 0.01.
We assume that dysfunctional TLR1 may attenuate HMGB1-activated TLR1/TLR2 signaling for DC maturation by OXP to influence its therapeutic efficacy. Therefore, we generated TLR1-silenced THP1 monocytes (Fig. 2D) and differentiated them into immature DCs (THP1-iDCs) by the cytokines IL4 and GM-CSF. These THP1-iDC cells were treated with rhHMGB1 protein to analyze the downstream effects of TLR1/2 signaling (Fig. 2D). We found that treatment with HMGB1 markedly activated TLR1/TLR2 signaling. However, silencing TLR1 led to a significant impact on the activation of TLR1/2 signaling in THP1-iDC cells, especially ERK activation (Fig. 2D). Additionally, the DC maturation markers CD80 and CD86 were significantly decreased (Fig. 2E). The surface DC maturation marker CD86 was also decreased in THP1shTLR1-iDC cells, suggesting that defective TLR1 directly impacts TLR1/2 signaling for DC maturation (Fig. 2F).
We then cocultured OXP-treated HT29-GFP cancer cells with THP1shNC-iDCs and THP1shTLR1-iDCs for 24 h (Fig. 2G). We found that the DC maturation marker CD86 was significantly decreased in THP1shTLR1-iDC cells after coculture with OXP-treated HT29-GFP cells compared to THP1shNC-iDC cells (Fig. 2G). Then, we cocultured Jurkat T lymphocytes with OXP-treated HT29/THP1-iDCs to evaluate the level of the cytotoxic marker IFNγ (Fig. 2H). As shown in Fig. 2H, we found that the level of IFNγ in Jurkat T lymphocytes was also significantly decreased in THP1shTLR1-iDCs (Fig. 2H). These results suggested that defective TLR1 attenuated TLR2 signaling, leading to less DC maturation and T-cell activation by immunogenic chemotherapy.
TLR1/2 signaling influences the therapeutic efficacy of chemotherapy by boosting antitumor immunity
To further validate that the therapeutic efficacy of chemotherapy was influenced by TLR1/2 signaling via antitumor immunity, we combined either the TLR1/2 antagonist CU-CPT22 or the TLR1/2 agonist Pam3CSK4 with OXP (Fig. 3A). Mice were given a subcutaneous tumor challenge with CT26 for 13 days and treated with OXP with or without CU-CPT22 and Pam3CSK4. We found that tumor volume was decreased in mice receiving 5 cycles of OXP (Fig. 3B). By pharmacologic inhibition of TLR1/2 signaling, we found that the therapeutic efficacy of OXP was significantly inhibited by administration of CU-CPT22 (Fig. 3B–D). In contrast, tumor regression was markedly reduced in the OXP/Pam3CSK4 group compared to mice that received OXP only (Fig. 3B–D), suggesting that activation of TLR1/2 signaling may enhance the therapeutic efficacy of OXP.
Systemic inhibition of TLR1/2 signaling reduced the therapeutic efficacy of chemotherapy in vivo. (A) Diagram of the animal experiment. (B) CT26 cells were subcutaneously inoculated into BALB/c mice for 13 days as the tumor volume reached 100 mm3. After randomization into 6 groups, OXP (6 mg/kg/mouse, i.p.) was administered on Days 13, 16, 19, 22 and 25. The TLR1/2 antagonist CU-CPT22 (2.5 mg/kg/mouse) and TLR1/2 agonist Pam3CSK4 (2.5 mg/kg/mouse) were administered 3 h before OXP administration. Tumor volume was measured every 3 days (n = 5–6). Two-way ANOVA t test, *p < 0.05, **p < 0.01 and ***p < 0.001. (C) The tumor volume was recorded on the end-point day (Day 28, n = 5–6). One-way ANOVA t test, *p < 0.05, **p < 0.01 and ***p < 0.001. (D) The resected tumor weight was recorded (n = 5–6). One-way ANOVA t test, *p < 0.05, **p < 0.01 and ***p < 0.001. (E) The densities of CD11c+ DCs and CD8+ T cells in resected tumors were analyzed by immunohistochemistry (n = 3–4). (F) The quantification of CD11c+ DCs in resected tumors is shown (n = 5). One-way ANOVA t test, *p < 0.05 and **p < 0.01. (G) The quantification of GzmB+CD8+ T cells in resected tumors is shown (n = 5). One-way ANOVA t test, *p < 0.05 and **p < 0.01. (H) Gating strategy of the tumor-infiltrating immune cell profile. (I) The phenotype of CD3+CD45+ tumor-infiltrating immune cells was analyzed by flow cytometry (n = 4–5). The percentage of CD4+ T cells (CD4+CD3+CD45+ T cells) and CD8+ T cells (CD8+CD3+CD45+ T cells) in total T cells (CD3+CD45+ T cells) was examined. One-way ANOVA t test, *p < 0.05.
We then evaluated the immune cell profile within the tumor microenvironment (TME) by flow cytometry and IHC. The density of tumor-infiltrating CD11c+ DCs and GzmB+CD8+ T cells was markedly decreased in the OXP/CU-CPT22 group (Fig. 3E–G). However, there was an increase in the OXP/Pam3CSK4 group compared to the OXP group (Fig. 3E–G). By flow cytometric analysis, we found that tumor-infiltrating CD8+ T cells were also significantly increased in the OXP/Pam3CSK4 group compared to the other groups (Fig. 3H,I). Blockade of TLR1/2 significantly reduced the infiltration of CD8 T cells elicited by OXP treatment (Fig. 3H,I), suggesting that activation of TLR1/2 signaling may enhance the therapeutic efficacy of OXP by boosting antitumor immunity. But we found that the density of tumor-infiltrating CD4+ T cells were significantly decreased in the OXP/Pam3CSK4 group compared to the other groups (Fig. 3I).
By evaluating the density of effector/memory CD8 cells, we found that there was a remarkable decrease in the OXP/CU-CPT22 group but an increase in the OXP/Pam3CSK4 group (Fig. 4A). The tumor-infiltrating IFNγ+ CD8 cells were also increased in the OXP/Pam3CSK4 group (Fig. 4B,C). However, we found no significant change in immunosuppressive Foxp3+ regulatory T cells within TME (Fig. 4D). Taken together, these results showed that OXP can induce infiltration of effector/memory and cytotoxic immune cells to recognize and eradicate residual tumor cells via TLR1/2-mediated signaling, indicating that TLR1/2 is critical to enhance the therapeutic efficacy of immunogenic chemotherapy. Additionally, the density of tumor-infiltrating CD8+ T cells was significantly lower in locally advanced stage III COAD patients bearing the TLR1-N248S polymorphism (Fig. 4E). Taken together, these results showed that defective TLR1 may reduce the therapeutic efficacy of chemotherapy in locally advanced COAD patients by attenuating HMGB1-mediated antitumor immunity.
Inhibition of TLR1/2 signaling remarkably impacts the infiltration of immune cells after immunogenic chemotherapy treatment in vivo. (A) The effector/memory cells of CD3+CD45+ tumor-infiltrating immune cells were analyzed by flow cytometry (n = 4–5). The percentage of CD8+ TEM cells (CD44+CD62L-CD8+CD3+CD45+ T cells) in total T cells (CD3+CD45+ T cells) was examined. One-way ANOVA t test, *p < 0.05 and **p < 0.01. (B) Gating strategy of the IFNγ+CD8+ tumor-infiltrating immune cell profile. (C) IFNγ+CD8+ tumor-infiltrating immune cells were analyzed by flow cytometry (n = 4–5). The percentage of IFNγ+CD8+ T cells (IFNγ+CD8+CD3+CD45+ T cells) in total T cells (CD3+CD45+ T cells) was examined. One-way ANOVA t test, *p < 0.05 and **p < 0.01. (D) The Foxp3+ T regulatory cells were analyzed by flow cytometry (n = 4–5). The percentage of Foxp3+Treg cells (Foxp3+CD4+CD25+ T cells) in total T cells (CD3+CD45+ T cells) was examined. One-way ANOVA t test. (E) The density of tumor-infiltrating CD8 cells within the tumor microenvironment in patients with locally advanced colon carcinoma was analyzed by immunohistochemistry. The association between CD8 density and TLR1-N248 polymorphism was analyzed (n = 410, p = 0.0455). Unpaired t test.
Discussion
Our data showed for the first time that TLR1-N248S was associated with the clinical outcome of patients with locally advanced stage III COAD, suggesting that this genetic defect might be associated with the risk of lymph node metastasis and poor DMFS. We found that defective TLR1 might attenuate HMGB1-mediated ICD to decrease the response to immunogenic chemotherapy, leading to a reduced antitumor response and poor therapeutic efficacy in vitro and in vivo. Furthermore, patients carrying the TLR1-N248S polymorphism had fewer tumor-infiltrating CD8+ lymphocytes within the TME in locally advanced COAD and associated with poor DFS. These findings indicate that TLR1 signaling plays a critical role in the therapeutic efficacy of chemotherapy by modulating antitumor immunity.
Our previous studies reported that the extent of HMGB1 release and immune cell infiltration were associated with favorable survival outcomes in locally advanced rectal cancer patients after preoperative chemoradiotherapy treatment26, suggesting that the HMGB1-TLR2/4 signaling pathway may participate in chemotherapy-induced antitumor immunity. TLR2 can form heterodimers with TLR1 or TLR6 to recognize different TLR ligands, resulting in different functions46. Previous studies showed that activation of TLR1/2 signaling could activate the immune response, but activation of TLR2/6 signaling could suppress T-cell immunity47,48. Moreover, recent studies have shown that TLR1/2 agonists enhance the antitumor efficacy of CTLA-4 and PD-L1 blockade by increasing intratumoral Treg depletion12,49 and stimulating cytotoxic T lymphocytes against tumor cells11. Here, our studies showed that TLR1 is essential for TLR1/2 signaling activation in DCs in response to immunogenic chemotherapy. We assumed that defective TLR1 may attenuate TLR1/2 signaling but enhance TLR2/6 signaling to suppress T-cell immunity. Therefore, HMGB1 released by immunogenic chemotherapy may activate TLR2/6 signaling in DCs to suppress T-cell-mediated immunity rather than promote the T-cell-mediated immune response in COAD patients with defective TLR1. Indeed, by silencing TLR1 expression on immature dendritic cells, we found that TLR2 signaling was less activated by HMGB1 compared to wild-type immature dendritic cells. Moreover, direct coculture of OXP-treated colorectal cancer cells with defective TLR1-iDCs showed that DC maturation and T-cell activation were remarkably attenuated in vitro, suggesting that TLR1 is essential for the HMGB1-TLR2 axis to boost antitumor immunity. Furthermore, systemic inhibition of TLR1/2 signaling markedly reduced the response to chemotherapy, leading to tumor regrowth. However, systemic administration of a TLR1/2 agonist significantly delayed tumor growth and increased the response to chemotherapy. Within the tumor microenvironment, we found that inhibition of TLR1/2 by the small molecule CU-CPT22 dramatically decreased the effect of OXP, leading to fewer tumor-infiltrating immune cells, including CD4+, CD8+ and IFNγ+ CD8+ cells. However, coadministration of a TLR1/2 agonist boosted the influence of immunogenic chemotherapy, increasing antitumor immunity to reshape the TME and eradicate residual tumors. Consistent with these observations, we found a lower density of tumor-infiltrating CD8+ lymphocytes within the tumor microenvironment in locally advanced colon carcinoma bearing the TLR1-N2418S polymorphism. Notably, we found no significant change in regulatory T (Treg) lymphocytes in these groups, which is inconsistent with previous studies47. Zhang et al. found that the TLR1/2 ligand not only promoted cytotoxic T lymphocyte infiltration but also reduced the infiltration of Tregs47. Sharma et al. also indicated that a TLR1/2 agonist enhanced the efficacy of an anti-CTLA-4 antibody by depleting regulatory T cells and increasing IFN-γ secretion from T cells to reshape the TME in a syngeneic melanoma model12. We assumed that the tumor immunophenotype might be different in these models. Our syngeneic colon model CT26 had more infiltration of Tregs than the melanoma Model B16F1050,51, which may influence the impact of TLR1/2 agonists on the immune status within the TME. These divergences need to be investigated in different syngeneic animal models in the future.
Previous reports have shown that several SNPs in TLR1 genes may modify the activation of its protein, implying that the genetic variation in the TLR1 gene might affect the innate immune response and clinical susceptibility to a wide spectrum of pathogens and confer a high risk of cancer52, including N248S and I602S. Here, we found that the TLR1-N248S polymorphism was not only associated with poor survival outcomes but also influenced the response to adjuvant chemotherapy. Consistent with our studies, Okazaki et al. also indicated that TLR1-I602S (rs5743618) could serve as a predictor of clinical response to FOLFIRI plus bevacizumab in patients with mCRC15. Moreover, Deng et al. showed that TLR1/2 expression had a positive correlation with lung cancer patient survival46, suggesting that TLR1/2 signaling may influence the therapeutic response to chemotherapy by modulating antitumor immunity. Therefore, low TLR1/2 expression or defective TLR1/2 signaling may result in poor clinical benefit of chemotherapy, especially immunogenic chemotherapeutic agents.
Taken together, our studies showed that the TLR1-N248S polymorphism is not only a prognostic but also a predictive factor for the clinical response to adjuvant chemotherapy in patients with locally advanced colon carcinoma. The TLR1-N248S polymorphism might reduce the ability of immunogenic chemotherapeutic agents to reshape the tumor microenvironment, leading to an insufficient antitumor immune response and tumor recurrence.
Data availability
All datasets generated or analyzed during the current study are included in this published article.
References
Yeom, S. S., Lee, S. Y., Kim, C. H., Kim, H. R. & Kim, Y. J. The prognostic effect of adjuvant chemotherapy in the colon cancer patients with solitary lymph node metastasis. Int. J. Colorectal. Dis. 34(8), 1483–1490. https://doi.org/10.1007/s00384-019-03346-7 (2019).
Hashiguchi, Y. et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int. J. Clin. Oncol. 25(1), 1–42. https://doi.org/10.1007/s10147-019-01485-z (2020).
Wang, L. et al. Prognostic utility of apical lymph node metastasis in patients with left-sided colorectal cancer. In Vivo 34(5), 2981–2989. https://doi.org/10.21873/invivo.12129 (2020).
Chiang, S. F. et al. Polymorphism of formyl peptide receptor 1 (FPR1) reduces the therapeutic efficiency and antitumor immunity after neoadjuvant chemoradiotherapy (CCRT) treatment in locally advanced rectal cancer. Cancer Immunol. Immunother. https://doi.org/10.1007/s00262-021-02894-8 (2021).
Abreu, M. T. Toll-like receptor signalling in the intestinal epithelium: How bacterial recognition shapes intestinal function. Nat. Rev. Immunol. 10(2), 131–144. https://doi.org/10.1038/nri2707 (2010).
Sato, Y., Goto, Y., Narita, N. & Hoon, D. S. Cancer cells expressing toll-like receptors and the tumor microenvironment. Cancer Microenviron. 2(Suppl 1), 205–214. https://doi.org/10.1007/s12307-009-0022-y (2009).
Xu, M. et al. Immunogenic cell death-relevant damage-associated molecular patterns and sensing receptors in triple-negative breast cancer molecular subtypes and implications for immunotherapy. Front. Oncol. 12, 870914. https://doi.org/10.3389/fonc.2022.870914 (2022).
Wang, X. et al. An immunogenic cell death-related classification predicts prognosis and response to immunotherapy in head and neck squamous cell carcinoma. Front. Immunol. 12, 781466. https://doi.org/10.3389/fimmu.2021.781466 (2021).
Huang, K. C. et al. Decitabine augments chemotherapy-induced PD-L1 upregulation for PD-L1 blockade in colorectal cancer. Cancers https://doi.org/10.3390/cancers12020462 (2020).
Huang, K. C. et al. Immunogenic cell death by the novel topoisomerase I inhibitor TLC388 enhances the therapeutic efficacy of radiotherapy. Cancers https://doi.org/10.3390/cancers13061218 (2021).
Cen, X. et al. TLR1/2 specific small-molecule agonist suppresses leukemia cancer cell growth by stimulating cytotoxic T lymphocytes. Adv. Sci. 6(10), 1802042. https://doi.org/10.1002/advs.201802042 (2019).
Sharma, N., Vacher, J. & Allison, J. P. TLR1/2 ligand enhances antitumor efficacy of CTLA-4 blockade by increasing intratumoral Treg depletion. Proc. Natl. Acad. Sci. USA 116(21), 10453–10462. https://doi.org/10.1073/pnas.1819004116 (2019).
Sugiura, Y. et al. TLR1-induced chemokine production is critical for mucosal immunity against Yersinia enterocolitica. Mucosal Immunol. 6(6), 1101–1109. https://doi.org/10.1038/mi.2013.5 (2013).
Yang, C. A. & Chiang, B. L. Toll-like receptor 1 N248S polymorphism affects T helper 1 cytokine production and is associated with serum immunoglobulin E levels in Taiwanese allergic patients. J. Microbiol. Immunol. Infect. 50(1), 112–117. https://doi.org/10.1016/j.jmii.2015.01.004 (2017).
Okazaki, S. et al. Clinical significance of TLR1 I602S polymorphism for patients with metastatic colorectal cancer treated with FOLFIRI plus bevacizumab. Mol. Cancer Ther. 15(7), 1740–1745. https://doi.org/10.1158/1535-7163.MCT-15-0931 (2016).
Wang, Y., Liu, S., Zhang, Y. & Yang, J. Dysregulation of TLR2 serves as a prognostic biomarker in breast cancer and predicts resistance to endocrine therapy in the luminal B subtype. Front. Oncol. 10, 547. https://doi.org/10.3389/fonc.2020.00547 (2020).
Lanki, M., Seppanen, H., Mustonen, H., Hagstrom, J. & Haglund, C. Toll-like receptor 1 predicts favorable prognosis in pancreatic cancer. PLoS ONE 14(7), e0219245. https://doi.org/10.1371/journal.pone.0219245 (2019).
Jinushi, M., Yagita, H., Yoshiyama, H. & Tahara, H. Putting the brakes on anticancer therapies: Suppression of innate immune pathways by tumor-associated myeloid cells. Trends Mol. Med. 19(9), 536–545. https://doi.org/10.1016/j.molmed.2013.06.001 (2013).
Pradere, J. P., Dapito, D. H. & Schwabe, R. F. The Yin and Yang of Toll-like receptors in cancer. Oncogene 33(27), 3485–3495. https://doi.org/10.1038/onc.2013.302 (2014).
Lu, H. et al. Polysaccharide krestin is a novel TLR2 agonist that mediates inhibition of tumor growth via stimulation of CD8 T cells and NK cells. Clin. Cancer Res. 17(1), 67–76. https://doi.org/10.1158/1078-0432.CCR-10-1763 (2011).
Jasani, B., Navabi, H. & Adams, M. Ampligen: A potential toll-like 3 receptor adjuvant for immunotherapy of cancer. Vaccine 27(25–26), 3401–3404. https://doi.org/10.1016/j.vaccine.2009.01.071 (2009).
Yang, H. et al. The Toll-like receptor 5 agonist entolimod suppresses hepatic metastases in a murine model of ocular melanoma via an NK cell-dependent mechanism. Oncotarget 7(3), 2936–2950. https://doi.org/10.18632/oncotarget.6500 (2016).
Panelli, M. C. et al. Sequential gene profiling of basal cell carcinomas treated with imiquimod in a placebo-controlled study defines the requirements for tissue rejection. Genome Biol. 8(1), R8. https://doi.org/10.1186/gb-2007-8-1-r8 (2007).
Hopkins, R. J. et al. Randomized, double-blind, active-controlled study evaluating the safety and immunogenicity of three vaccination schedules and two dose levels of AV7909 vaccine for anthrax post-exposure prophylaxis in healthy adults. Vaccine 34(18), 2096–2105. https://doi.org/10.1016/j.vaccine.2016.03.006 (2016).
Huang, C. Y. et al. Clinical significance of programmed death 1 ligand-1 (CD274/PD-L1) and intra-tumoral CD8+ T-cell infiltration in stage II-III colorectal cancer. Sci. Rep. 8(1), 15658. https://doi.org/10.1038/s41598-018-33927-5 (2018).
Huang, C. Y. et al. Cytosolic high-mobility group box protein 1 (HMGB1) and/or PD-1+ TILs in the tumor microenvironment may be contributing prognostic biomarkers for patients with locally advanced rectal cancer who have undergone neoadjuvant chemoradiotherapy. Cancer Immunol. Immunother. 67(4), 551–562. https://doi.org/10.1007/s00262-017-2109-5 (2018).
Huang, K. C. et al. ATAD3A stabilizes GRP78 to suppress ER stress for acquired chemoresistance in colorectal cancer. J. Cell Physiol. 236(9), 6481–6495. https://doi.org/10.1002/jcp.30323 (2021).
Lin, T. Y., Fan, C. W., Maa, M. C. & Leu, T. H. Lipopolysaccharide-promoted proliferation of Caco-2 cells is mediated by c-Src induction and ERK activation. Biomedicine 5(1), 5. https://doi.org/10.7603/s40681-015-0005-x (2015).
Wang, X. et al. RSF-1 overexpression determines cancer progression and drug resistance in cervical cancer. Biomedicine 8(1), 4. https://doi.org/10.1051/bmdcn/2018080104 (2018).
Chiang, S. F. et al. Upregulation of tumor PD-L1 by neoadjuvant chemoradiotherapy (neoCRT) confers improved survival in patients with lymph node metastasis of locally advanced rectal cancers. Cancer Immunol. Immunother. 68(2), 283–296. https://doi.org/10.1007/s00262-018-2275-0 (2019).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347(6220), 1260419. https://doi.org/10.1126/science.1260419 (2015).
Berges, C. et al. A cell line model for the differentiation of human dendritic cells. Biochem. Biophys. Res. Commun. 333(3), 896–907. https://doi.org/10.1016/j.bbrc.2005.05.171 (2005).
Huang, K.C.-Y. et al. DNMT1 constrains IFNβ-mediated anti-tumor immunity and PD-L1 expression to reduce the efficacy of radiotherapy and immunotherapy. OncoImmunology 10(1), 1989790. https://doi.org/10.1080/2162402X.2021.1989790 (2021).
Huang, K. C. et al. A novel engineered AAV-based neoantigen vaccine in combination with radiotherapy eradicates tumors. Cancer Immunol. Res. 11(1), 123–136. https://doi.org/10.1158/2326-6066.CIR-22-0318 (2023).
Percie du Sert, N. et al. The ARRIVE guidelines 20: Updated guidelines for reporting animal research. PLoS Biol. 18(7), e3000410. https://doi.org/10.1371/journal.pbio.3000410 (2020).
Lin, Y. S. et al. Targeting CD73 increases therapeutic response to immunogenic chemotherapy by promoting dendritic cell maturation. Cancer Immunol. Immunother. 72(7), 2283–2297. https://doi.org/10.1007/s00262-023-03416-4 (2023).
Huang, K. C. et al. Engineered sTRAIL-armed MSCs overcome STING deficiency to enhance the therapeutic efficacy of radiotherapy for immune checkpoint blockade. Cell Death Dis. 13(7), 610. https://doi.org/10.1038/s41419-022-05069-0 (2022).
Chen, T. W. et al. Dual inhibition of TGFbeta signaling and CSF1/CSF1R reprograms tumor-infiltrating macrophages and improves response to chemotherapy via suppressing PD-L1. Cancer Lett. 543, 215795. https://doi.org/10.1016/j.canlet.2022.215795 (2022).
Wang, C. Q. et al. Subcellular localization of HMGB1 in colorectal cancer impacts on tumor grade and survival prognosis. Sci. Rep. 10(1), 18587. https://doi.org/10.1038/s41598-020-75783-2 (2020).
Chen, R., Kang, R. & Tang, D. The mechanism of HMGB1 secretion and release. Exp. Mol. Med. 54(2), 91–102. https://doi.org/10.1038/s12276-022-00736-w (2022).
Cheng, K. J., Mohamed, E. H. M., Syafruddin, S. E. & Ibrahim, Z. A. Interleukin-1 alpha and high mobility group box-1 secretion in polyinosinic:polycytidylic-induced colorectal cancer cells occur via RIPK1-dependent mechanism and participate in tumourigenesis. J. Cell Commun. Signal. https://doi.org/10.1007/s12079-022-00681-3 (2022).
Lu, H. et al. Oncogenic role of HMGB1 as an alarming in robust prediction of immunotherapy response in colorectal cancer. Cancers https://doi.org/10.3390/cancers14194875 (2022).
Peng, R. Q. et al. Co-expression of nuclear and cytoplasmic HMGB1 is inversely associated with infiltration of CD45RO+ T cells and prognosis in patients with stage IIIB colon cancer. BMC Cancer 10, 496. https://doi.org/10.1186/1471-2407-10-496 (2010).
Huang, C. Y. et al. HMGB1 promotes ERK-mediated mitochondrial Drp1 phosphorylation for chemoresistance through RAGE in colorectal cancer. Cell Death Dis. 9(10), 1004. https://doi.org/10.1038/s41419-018-1019-6 (2018).
Dumitriu, I. E., Baruah, P., Bianchi, M. E., Manfredi, A. A. & Rovere-Querini, P. Requirement of HMGB1 and RAGE for the maturation of human plasmacytoid dendritic cells. Eur. J. Immunol. 35(7), 2184–2190. https://doi.org/10.1002/eji.200526066 (2005).
Deng, Y. et al. TLR1/TLR2 signaling blocks the suppression of monocytic myeloid-derived suppressor cell by promoting its differentiation into M1-type macrophage. Mol. Immunol. 112, 266–273. https://doi.org/10.1016/j.molimm.2019.06.006 (2019).
Zhang, Y. et al. TLR1/TLR2 agonist induces tumor regression by reciprocal modulation of effector and regulatory T cells. J. Immunol. 186(4), 1963–1969. https://doi.org/10.4049/jimmunol.1002320 (2011).
Skabytska, Y. et al. Cutaneous innate immune sensing of Toll-like receptor 2–6 ligands suppresses T cell immunity by inducing myeloid-derived suppressor cells. Immunity 41(5), 762–775. https://doi.org/10.1016/j.immuni.2014.10.009 (2014).
Wang, Y. et al. Adjuvant effect of the novel TLR1/TLR2 agonist Diprovocim synergizes with anti-PD-L1 to eliminate melanoma in mice. Proc. Natl. Acad. Sci. USA 115(37), E8698–E8706. https://doi.org/10.1073/pnas.1809232115 (2018).
Yu, J. W. et al. Tumor-immune profiling of murine syngeneic tumor models as a framework to guide mechanistic studies and predict therapy response in distinct tumor microenvironments. PLoS ONE 13(11), e0206223. https://doi.org/10.1371/journal.pone.0206223 (2018).
Taylor, M. A. et al. Longitudinal immune characterization of syngeneic tumor models to enable model selection for immune oncology drug discovery. J. Immunother. Cancer 7(1), 328. https://doi.org/10.1186/s40425-019-0794-7 (2019).
la Trejo-de, O. A., Hernandez-Sancen, P. & Maldonado-Bernal, C. Relevance of single-nucleotide polymorphisms in human TLR genes to infectious and inflammatory diseases and cancer. Genes Immun. 15(4), 199–209. https://doi.org/10.1038/gene.2014.10 (2014).
Acknowledgements
We are grateful for the tissue microarray support from the Translation Research Core, China Medical University Hospital. This study was supported in part by China Medical University Hospital (DMR-111-156, Taiwan) and the Ministry of Science and Technology (MOST110-2628-B-039-005 and MOST110-2314-B-039-032, Taiwan). This study was partially based on clinical information from the China Medical University Hospital Cancer Registry. Experiments and data analysis were performed in part through the use of the Medical Research Core Facilities Center, Office of Research & Development at China medical University, Taichung, Taiwan, R.O.C.
Author information
Authors and Affiliations
Contributions
Data curation, K.C.-Y.H. and T.-W.C.; Formal analysis, S.-F.C., W.T.-L.C., K.C.-Y.H., J.-A.L., W.-Z.H., C.-Y.L., L.-C.C. and J.-Y.C.; Funding acquisition, K.S.C.C. and K.C.-Y.H.; Investigation, W.T.-L.C., T.-W.K. and P.-C.Y.; Methodology, K.C.-Y.H., J.-Y.C.; Project administration, K.C.-Y.H.; Resources, W.T.-L.C., T.-W.K. and K.S.C.C.; Supervision, K.S.C.C.; Writing—original draft, K.C.-Y.H.; Writing—review & editing, K.S.C.C.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, K.CY., Ke, TW., Chen, JY. et al. Dysfunctional TLR1 reduces the therapeutic efficacy of chemotherapy by attenuating HMGB1-mediated antitumor immunity in locally advanced colorectal cancer. Sci Rep 13, 19440 (2023). https://doi.org/10.1038/s41598-023-46254-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-46254-1
This article is cited by
-
Activation of STING by the novel liposomal TLC388 enhances the therapeutic response to anti-PD-1 antibodies in combination with radiotherapy
Cancer Immunology, Immunotherapy (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.