Abstract
Observational studies have reported a correlation between Helicobacter pylori infection and colorectal cancer (CRC); however, the underlying cause has remained unclear. This research was aimed at determining whether there is a correlation between H. pylori infection and CRC by measuring the prevalence of H. pylori CagA antibodies and VacA antibodies. Using data from many genome-wide association studies (GWAS), we conducted a Mendelian randomization (MR) study with two sample GWAS. Then, we used bidirectional MR to evaluate the association between H. pylori infection and CRC for identifying causation. The most common method of analysis was the inverse variance-weighted technique. In addition, we performed supplementary analyses using the weighted median technique and MR-Egger regression. Horizontal pleiotropic outliers were identified and corrected using the MR Pleiotropy RESidual Sum and Outlier (MR-PRESSO) method. Genetically predicted anti-H. pylori IgG seropositivity was not causally associated with CRC [odds ratio (OR): 1.12; 95% confidence interval (CI): 0.98–1.27, P = 0.08] and neither were H. pylori VacA antibody levels (OR = 0.96, 95% CI: 0.90–1.02, P = 0.25) or H. pylori CagA antibody levels (OR = 1.00, 95% CI: 0.93–1.07, P = 0.92). Furthermore, reverse MR analysis did not reveal evidence for a causal effect of CRC on H. pylori infection. The weighted median, the MR-Egger method, and MR-PRESSO yielded identical results. Using genetic data, MR analysis showed there was no evidence for a causal association between seroprevalence of H. pylori infection and CRC. The relationship between H. pylori infection and CRC requires further research.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the third most common malignancy worldwide and the second greatest cause of cancer-related mortality. It has the highest occurrence rate of all malignancies in individuals aged under 50 years1. Approximately 1.9 million newly diagnosed cases of CRC were reported in 2020, accounting for 10% of all malignancies worldwide2. The prevalence of CRC causes serious economic and societal burdens. Available data on its etiology suggest that its prevalence is influenced by advanced age, sex, ethnicity, family history of colorectal cancer, familial polyposis syndrome, chronic inflammatory bowel diseases, physical inactivity, unhealthy diet, geographical variation, and consumption of alcohol and cigarettes3.
Helicobacter pylori is a gram-negative bacteria that colonizes the stomach mucosa of humans and causes gastritis in around half the world’s population4. The primary virulence pathogenic factors of H. pylori infection are vacuolar cytotoxin A (VacA) and cytotoxin-associated protein A (CagA). Gastritis, duodenal ulcers, gastric cancer, and other GI and non-GI problems have all been found to be associated with H. pylori infection5. Epidemiological studies have recently focused on the association between H. pylori infection and CRC6,7. There is also evidence that a high incidence of H. pylori seropositivity in patients with CRC8,9 despite several reports stating that H. pylori infection has no role in the genesis of CRC10,11. Furthermore, several studies have even confirmed the presence of H. pylori in CRC or colonic polyps12,13. Notably, the evidence available so far is based on observational research. Chronic inflammation plays a role in the development of both H. pylori-associated diseases, such as gastritis, and CRC. Inflammatory conditions, such as inflammatory bowel disease (IBD), can confound the genetic associations by influencing both H. pylori infection status and CRC risk14. It is difficult to conduct a randomized controlled trial to check for an association between H. pylori infection and CRC. Consequently, elucidating if H. pylori infection is associated with CRC is challenging. Owing to the high prevalence of H. pylori infections in the population and the relative ease and cost-effectiveness of anti-H. pylori therapy15, identifying any association between H. pylori infection and CRC risk is of great public health and clinical importance.
To quantify the association of single nucleotide polymorphisms (SNPs) and H. pylori infection with CRC risk, we used a Mendelian randomization (MR) technique16 based on genetic variance index exposures to establish causal relationships of disease-related risk factors. Using descriptive data from large-scale genome-wide association studies (GWAS) of H. pylori infection and CRC, we used two-sample MR to test the hypothesis that H. pylori infection is associated with increased risk of developing CRC.
Materials and methods
Mendelian randomization design
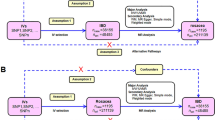
Figure 1 shows the overall setup for the current MR study. Large-scale GWAS summary data are now more readily available, making MR a valuable tool in epidemiological investigations17. As shown in Supplementary Fig. S1, three crucial instrumental variable (IV) assumptions were used as the foundation for the MR study’s assumed validity: (a) relevance assumption: genetic variations are assumed to be highly correlated with exposure; (b) independence assumption: there are no unobserved confounders associated with the genetic variants; and (c) exclusion restriction: genetic polymorphisms are not linked to the outcome when exposure and other confounding factors are present18. To further evaluate the efficacy of the IVs, the F-statistic was calculated for each SNP using the following formula:
where in R2 is the amount of exposure variation that can be explained by each genetic variant and N is the number of people in each sample19. An F-statistic of < 10 indicated that weak IVs may have skewed the direction of the confounded, observational relationship between phenotype and outcome20.
GWAS summary data
Genetic associations with CRC were acquired from FinnGen consortium (https://www.finngen.fi/en) data version 7 (R7 release version 1 June 2022); this GWAS included 4957 CRC cases and 304,197 controls. The GWAS analyses were adjusted in the FinnGen study in terms of sex, major genetic components, and the genotyping batch21. To further elucidate the presence of a causal association between H. pylori infection and CRC, two GWAS studies were used to derive genetic associations for colon cancer (CC) and rectal cancer (RC), and both these studies used data from the UK Biobank study (http://www.nealelab.is/uk-biobank/); one of these studies comprised 1697 cases and 359,444 controls and the other comprised 375 cases and 360,766 controls. Pooled GWAS data from anti-H. pylori IgG levels were determined from public data compiled in the EBI database22 (https://gwas.mrcieu.ac.uk/datasets/ieu-b-4905/), covering 1058 European cases and 3,625 European controls, as shown in Table 1.
Selection of the genetic instruments
Genetic IVs may be obtained via either a literature search or a GWAS summary data analysis. The Rotterdam studies RS-I, RS-II, and RS-III all yielded H. pylori-seropositive genetic IVs. Mayerle et al.23 performed a study on a European population that included 2763 cases and 8175 controls. As shown in Table 2, two SNPs (rs10004195 and rs368433) were identified to be independently and strongly associated with H. pylori seropositivity and have therefore been labeled as IVs. SNPs associated with anti-H. pylori VacA from the GWAS meta-analysis of EBI data (https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST90006916/) included 1,571 samples with 9,178,635 SNPs, of which 13 SNPs with P values < 5 × 10–6 were considered to be associated with VacA and used as IVs. Similarly, anti-H. pylori CagA from the EBI database (https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST90006911/) included 985 samples with 9,165,056 SNPs, of which ten SNPs with P values < 5 × 10–6 were considered to be associated with CagA and chosen as IVs.
To verify reverse causality between H. pylori infection and CRC risk. we obtained genetic IVs of CRC from the FinnGen study, and H. pylori IgG seropositivity from the EBI database (https://gwas.mrcieu.ac.uk/datasets/ieu-b-4905/) as the outcome for reverse causal inference. First, to search for the CRC genetic IVs that met the assumptions of MR relevance, the SNPs had to reach genome-wide significance with a P value of 5 × 10–8. Second, we then checked for linkage disequilibrium (r2 < 0.001 and clump distance > 10,000 kb). Third, when we harmonized the exposure and outcome datasets, we eliminated SNPs with intermediate allele frequencies and ambiguous SNPs with mismatched alleles to guarantee that effect alleles belonged to the same allele. Fourth, to avoid the effect of weak IVs on the causal effect, the F-statistic of each selected IV had to be greater than 10. Finally, PhenoScanner V2 (http://www.phenoscanner.medschl.cam.ac.uk/) was used to test the hypothesis that IVs are independent of confounders and outcomes by analyzing genome-wide significant relationships (P < 5 × 10–8). After the above criteria were satisfied, five SNPs associated with CRC were selected as IVs for subsequent analysis, as shown in Supplementary Table S1.
Ethics statement
This MR study was conducted using GWAS summary statistics or shared datasets. All of these data were anonymized, freely downloadable, and could be used without restriction. Therefore, a separate ethics statement or consent was not required. The study was approved by the hospital’s Institutional Review Board (Ethics 28 Committee of Chongqing General Hospital, ID: XJS S2022-052-01).
Statistical analyses
Using a two-sample Mendelian randomization (MR) approach, we determined the cause of H. pylori infection and its reverse causality with CRC. The inverse variance-weighted (IVW) technique has been used as the main statistical model24. We estimated genetic predictive associations for seropositive H. pylori using the IVW fixed-effects method as only two SNPs were available25. The weighted median26 and MR-Egger27 methods were used for the sensitivity analyses of MR analyses involving more than two IVs. After removing potentially misleading outliers, we used the MR-PRESSO test to analyze and compensate for horizontal pleiotropy and provide causal estimates28. Cochrane Q values were used to evaluate heterogeneity among the estimated s of SNPs in each analysis, with a Cochran Q-derived P value of < 0.05 indicating possible horizontal pleiotropy29. The results of multiple comparisons were recalculated using Bonferroni's procedure30, and a P value of < 0.005 (0.05/10) was considered to indicate a causal connection. Using the mRnd website (https://shiny.cnsgenomics.com/mRnd/), we determined the statistical power of the experiment31. All analyses were two-sided and implemented in R (version 4.2.1) through the TwoSampleMR package (version 0.5.6). Forest plots were created using the “forestplot” R package (version 3.1.1).
Results
Causal effect of H. pylori infection and subtypes on CRC
It was discovered that the two SNPs known as the IVs rs10004195A and rs368433C were strongly and independently associated with H. pylori seropositivity. The F-statistic for these IVs was 547.85 and 299.10, respectively, thus avoiding the impact of weak IVs on the causal effect.
We incorporated 15 independent SNPs with P < 5 × 10–6 as IV SNPs for anti-H. pylori VacA bodies and anti-CagA bodies, respectively. However, two VacA-associated SNPs and five CagA-associated SNPs were not available in the pooled statistics from CRC, as shown in Supplementary Table S3. According to the CC and RC summary statistics provided in Supplementary Table S4, one CagA-associated SNP was missing. There were no F-statistics below 10, and the variation explained by these IVs was close to 22% for VacA and 18.8% for CagA. Our MR analysis has significant power (over 80% to identify an OR of 1.2) to find moderate correlations of VacA and CagA with CRC, CC, and RC; however, it has restricted low power in assessing the effect of H. pylori infection on CRC, CC, and RC (66% power to detect an OR of 1.20). The results of MR estimates using several philosophies are presented in Fig. 2. There was no evidence of a causal connection between having a genetic predisposition to H. pylori infection and an increased risk of CRC. The primary IVW method showed that genetically predicted H. pylori seropositivity in the FinnGen GWAS was not causally associated with CRC [odds ratio (OR) = 1.12, 95% confidence interval (CI): = 0.98–1.27, P = 0.08], with CC (OR = 1.00, 95% CI: 0.99–1.00, P = 0.81), and with RC (OR = 1.00, 95% CI: 0.99–1.00, P = 0.83) in the UK Biobank GWAS. In addition, the MR estimates showed that the causal effect of VacA and CagA on CRC and its subtypes was the same as that of H. pylori infection on CRC. In addition, the sensitivity analyses of both MR-Egger and weighted median had comparable conclusions, as shown in Table 3. Scatter plots for effect sizes of SNPs for H. pylori infection, anti-VacA antibody, and anti-CagA antibody for CRC, CC, and RC are shown in Fig. 3 and Supplementary Fig. S7. The results of the Cochran Q test revealed no significant heterogeneity, indicating that there was no probable horizontal pleiotropy in any of the estimated s of SNPs in any study. The Cochran Q test showed no heterogeneity. Furthermore, the MR-Egger regression revealed no evidence of directional pleiotropy. The MR-PRESSO results were robust, and no outliers were detected (Supplementary Table S2). A single SNP did not drive the causative estimates of H. pylori infection, as shown by a leave-one-out analysis. Supplementary Figs. S2–S6 show the results of leave-one-out analyses and forest and funnel plots.
A scatter plot of the causal relationships between H. pylori and CRC using different MR methods. (A) Causal estimates for H. pylori seroprevalence on CRC. (B) Causal estimates for VacA on CRC. (C) Causal estimates for CagA on CRC. (D) Causal estimates for CRC on H. pylori infection. The slope of each line corresponds to the causal estimate for each method.
Causal effect of CRC on Helicobacter pylori infection
The IVs of CRC were identified using the FinnGen consortium GWAS. A total of five SNPs, namely rs11213823C, rs16969344G, rs2337113G, rs2735940G, and rs7897408A, were selected. rs11213823C maps to the colorectal cancer-associated 2 gene (COLCA2), which has been identified as being causally associated with the increased risk of CRC32. Shorter telomeres33 and increased cancer risk34 have been associated with the human telomerase reverse transcriptase gene and its related SNP, rs2735940G. Unfortunately, little information and few related publications are currently available on the SNPs rs16969344G, rs2337113G, and rs7897408G.
The IVW method revealed no evidence for a correlation between genetically predicted CRC and H. pylori infection. (OR = 1.03, 95% CI: 0.85–1.24, P = 0.74). The weighted median approach (OR = 1.06, 95% CI: 0.87–1.29, P = 0.50) and the MR-Egger method (OR = 0.25, 95% CI = 0.05–1.07, P = 0.15) generated identical results, as shown in Fig. 2. There was no heterogeneity, no directed polymorphism, and no outlier detected, as shown in Supplementary Table S2.
Discussions
Using four genetic data pools, our study did not directly compare H. pylori serology and colorectal cancer (CRC). Instead, we employed a Mendelian randomization approach using potential single nucleotide polymorphisms (SNPs) associated with H. pylori infection as instrumental variables to estimate the possible causality between H. pylori and CRC. Our results did not show any clear evidence of a causal effect of genetic predisposition to H. pylori infection on CRC risk. Multiple MR sensitivity analysis methods showed consistent results.
Regarding the association between H. pylori infection and CRC, we acknowledge that H. pylori infection is influenced by hygiene standards and family history. While our study focused on the potential genetic links between H. pylori and CRC, we cited previous research, such as the study by Mayerle et al.23, to provide additional context. We acknowledge that further replication and validation of the identified SNPs as surrogate biomarkers for predicting H. pylori infection are necessary. However, our study aimed to explore the potential genetic factors underlying the association, which can contribute to a better understanding of the biology of H. pylori-related CRC35.
To our knowledge, there is no inconsistent evidence on the association of H. pylori infection with an increased risk of CRC. A study on a large consortium of cohorts, which included ten prospective cohorts of American populations, conducted by Butt et al.8 in 2019 revealed that H. pylori VacA-specific seropositivity was associated with an increased risk of CRC (OR, 1.11; 95% CI, 1.01–1.22). In another study involving the same cohorts performed in 2020, Epplein et al.36 analyzed H. pylori VacA toxin levels and performed the Streptococcus gallolyticus pilus protein assay and found a significant association between H. pylori and CRC risk. A meta-analysis of 27 studies by Yang et al.37 conducted in 2019 and another meta-analysis of 48 studies by Choi et al.38 conducted in 2020 report similar conclusions. Additionally, the findings of another cohort study conducted by Wang et al.39 in 2020 in China support that H. pylori infection increases the risk of colorectal polyps and malignancy.
Nevertheless, a multi case–control research carried out by Fernández et al.40 in Spain in 2017 found that neither H. pylori seropositivity nor CagA and VacA were associated with a higher risk of CRC (OR, 0.91; 95% CI: 0.71–1.16). Regarding anti-CagA antibodies, their presence in H. pylori infection is not universal. Different studies have reported varying rates of anti-CagA antibody development. Nonetheless, our study did not solely rely on anti-CagA antibodies for the analysis but considered a broader perspective by investigating potential genetic associations. Moreover, in Blase et al.’s41 nested case–control study that involved individuals of Caucasian, they reported that H. pylori infection was not associated with CRC. A cross-sectional study by Boyuk et al.11 in 2019 also reported similar conclusions. In 2014, Guo et al.42 performed a meta-analysis of studies on the East Asian population to further investigate the association between H. pylori infection and colorectal neoplasm; their results showed that H. pylori infection had no apparent association with colorectal neoplasm. The studies of Patel et al.10 on the Hispanic population in the United States, Limburg et al.43 on Finnish population, and Machida-Montani et al.44 on Japanese population all support the absence of a correlation between H. pylori infection and colorectal malignancies.
According to the two large meta-analyses, one by Hooi et al.45 and the other by Ren et al.46, the prevalence of H. pylori infection is roughly 79.1% in Africa, 54.7% in Asia, 47% in Europe, 44.2% in the mainland of China, and 37.1% in North America. In a subgroup analysis of the cohort study conducted by Butt et al.8, they reported that H. pylori infection raises the risk of CRC in African Americans. Another study by Blase et al.41 refuted the notion that H. pylori infection was associated with a higher risk of CRC in elderly Caucasian population.
There are a number of possible explanations for the controversy surrounding H. pylori infection and the risk of CRC. First, there were no prospective, randomized, or blinded methods in any of these epidemiologic observational studies. The discrepancies in findings are most likely attributable to biases associated with improper confounding control, selection bias, or reverse causality. Second, different studies have used different diagnostic strategies for CRC and H. pylori infections. Based on the global guidelines of the World Gastroenterology Organization (WGO)47, the urine breath test is the most recommended non-invasive test to check for H. pylori infection. However, some studies measured H. pylori infection using serum or IgG antibodies to H. pylori or H. pylori DNA sequences, and therefore, the criteria for the diagnosis of H. pylori infection were inconsistent across studies8,12,48,49. Third, although biopsy is the standard method for diagnosing CRC, some studies obtained specimens via surgery, and these surgical specimens revealed inconsistent pathological classification. Fourth, these differences could be attributed to differences in the age, sex, geographic region, ethnicity, dietary habits, socioeconomic status, anti-H. pylori IgG titers, and anti-H. pylori antibody types and levels.
We acknowledge that CRC is a multifactorial disease influenced by various non-hereditary factors. However, genetic susceptibility can still play a role in disease development, including gene-environment interactions. By exploring the potential genetic links between H. pylori and CRC, our study aimed to contribute to the understanding of the complex etiology of CRC. As drawing conclusions from observational studies to avoid confounding risk factors has been difficult, it is important to investigate the mechanisms underlying the infection caused by H. pylori that lead to CRC. With MR methods, causality can be revealed reliably and without bias because MR is a natural randomized controlled trial performed during fertilization and has similar randomized controlled trials study design. Based on the rigorous assumptions regarding the relationship between the genetic score for exposure and the outcome, as underlying the principles of Mendelian Randomization, there is no compelling evidence suggesting a causal association between infection and colorectal cancer. Our study is the first attempt to elucidate the connection between H. pylori infection and CRC from the perspective of genetic variation.
The findings of the GWAS investigation yielded only two SNPs (rs10004195 and rs368433) that were significantly and independently linked to H. pylori infection. The SNP rs10004195 was found at the toll-like receptor (TLR) locus on chromosome 4p14, and the SNP rs368433 was found at the FCGR2A locus on chromosome 1q23. TLRs, the pattern recognition receptors with the most recognizable features, are critical for both innate and adaptive immune responses50. Tang et al.51 published their findings on TLR10 (rs10004195) and H. pylori infection in a Chinese population in 2015; they discovered that TLR10 (rs10004195) had a protective effect against H. pylori infection (OR = 0.83; 95% CI: 0.72–0.96). Another study conducted by EI-Omar et al.52 reported that low expression of the TLR rs10004195 was associated with increased protection against H. pylori infection, whereas high expression of the TLR rs10004195 was associated with H. pylori seropositivity. Two meta-analyses performed respectively by Karpiski et al.53 and Moradi-Marjaneh et al.54 revealed that gastrointestinal microorganisms play a significant role in colorectal carcinogenesis. Notably, most ongoing studies are at the stage of animal experiments55, and the potential association between TLRs and CRC signaling pathways still needs to be further investigated and confirmed in clinical trials. The SNP rs368433 is associated with the FCGR2A gene. Zhang et al.56 found that patients treated with cetuximab for CRC had a poorer outcome when they had the FCGR2A genotype. Geva et al.57 reported that no differences were found between FCGR polymorphisms when treated with cetuximab.
There have been some studies on virulence factors of H. pylori infection and their impact on the host immune response and physiology of gastric disease58,59. However, to our knowledge, no precise information on signaling pathways associating H. pylori infection with CRC pathogenesis is currently available, and the overall information on the contribution of H. pylori-related extraintestinal disorders to CRC is too limited to be fully understood. Thus far, there has been no report identifying a direct molecular signaling pathway leading to CRC through SNPs associated with H. pylori infection, and there is no evidence for the presence of H. pylori or corresponding proteins in CRC tissues. Further studies are warranted to confirm the presence of H. pylori in the colorectal epithelium and its potential direct effects in causing CRC59. Our data suggest no direct causative link between H. pylori infection and CRC incidence, and bidirectional MR analysis was performed to clarify the causality. Our study has several notable strengths. First, we could simulate randomized controlled trials in observational settings using the MR design. Although randomized controlled trials are widely considered the most reliable approach to identify the presence of a causal relationship, it is not always possible to carry one out due to its high cost. However, since SNPs are randomly allocated at conception, MR studies can successfully correct for this potential confounding bias. Unlike the approaches used in other observational studies, MR offers the advantage of being able to address the reverse causality problem. Second, our study results may have implications for healthcare policies regarding H. pylori infection and CRC. Given the high incidence of H. pylori infection in the general population, establishing causation between H. pylori infection and CRC can affect public health strategies for early prevention and appropriate intervention. Our results indicate that people with genetic predisposition to H. pylori infection may not benefit from CRC screening.
In summary, while we acknowledge the limitations and complexities of studying the association between H. pylori and CRC, we believe that our study provides valuable insights by employing a Mendelian randomization approach and considering potential genetic factors.
Conclusions
This MR study was aimed at investigating the causality of H. pylori infection with CRC. The findings do not lend credence to the notion that a higher seroprevalence of H. pylori infection is associated with a higher risk of CRC caused by genetic variation.
Data availability
The GWAS data related to CRC were sourced from the FinnGen website at https://www.finngen.fi/en. The summary-level data for CC and RC were obtained from the Neale Lab UK Biobank portal, available at http://www.nealelab.is/uk-biobank/. Additionally, the consolidated GWAS data can be found at the MRC IEU OpenGWAS database, located at https://gwas.mrcieu.ac.uk/datasets.
Abbreviations
- H. pylori :
-
Helicobacter pylori
- CRC:
-
Colorectal cancer
- CC:
-
Colon cancer
- RC:
-
Rectal cancer
- SNPs:
-
Single-nucleotide polymorphisms
- IVs:
-
Instrumental variants
- EBI:
-
European Bioinformatics Institute
- IVW:
-
Inverse variance weighted
- MR:
-
Mendelian randomization
- MR-PRESSO:
-
MR Pleiotropy Residual Sum and Outlier
- nSNPs:
-
The number of SNPs used in the analysis
- OR:
-
Odds ratio
- VacA:
-
Vacuolar cytotoxin A
- CagA:
-
Cytotoxin-associated protein A
- CI:
-
Confidence interval
References
Keum, N. & Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 16, 713–732. https://doi.org/10.1038/s41575-019-0189-8 (2019).
Xi, Y. & Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 14, 101174. https://doi.org/10.1016/j.tranon.2021.101174 (2021).
Lu, L., Mullins, C. S., Schafmayer, C., Zeißig, S. & Linnebacher, M. A global assessment of recent trends in gastrointestinal cancer and lifestyle-associated risk factors. Cancer Commun. (Lond.) 41, 1137–1151. https://doi.org/10.1002/cac2.12220 (2021).
Sonnenberg, A. Epidemiology of Helicobacter pylori. Aliment Pharmacol. Ther. 55(Suppl 1), S1-s13. https://doi.org/10.1111/apt.16592 (2022).
Wu, Y. et al. GWAS of peptic ulcer disease implicates Helicobacter pylori infection, other gastrointestinal disorders and depression. Nat. Commun. 12, 1146. https://doi.org/10.1038/s41467-021-21280-7 (2021).
Shen, L., Bian, R., Wang, W. & Zhao, J. Association of Helicobacter pylori infection with colorectal adenoma in the Chinese urban population: A cross-sectional study. Microb. Pathog. 158, 105111. https://doi.org/10.1016/j.micpath.2021.105111 (2021).
Zuo, Y. et al. Association between Helicobacter pylori infection and the risk of colorectal cancer: A systematic review and meta-analysis. Medicine (Baltimore) 99, e21832. https://doi.org/10.1097/md.0000000000021832 (2020).
Butt, J. et al. Serologic response to Helicobacter pylori proteins associated with risk of colorectal cancer among diverse populations in the United States. Gastroenterology 156, 175-186.e172. https://doi.org/10.1053/j.gastro.2018.09.054 (2019).
Kim, T. J. et al. Helicobacter pylori infection is an independent risk factor of early and advanced colorectal neoplasm. Helicobacter https://doi.org/10.1111/hel.12377 (2017).
Patel, S. et al. The association of H. pylori and colorectal adenoma: Does it exist in the US Hispanic population?. J. Gastrointest. Oncol. 5, 463–468. https://doi.org/10.3978/j.issn.2078-6891.2014.074 (2014).
Boyuk, B. et al. Helicobacter pylori infection coexisting with intestinal metaplasia is not associated with colorectal neoplasms. Prz. Gastroenterol. 14, 133–139. https://doi.org/10.5114/pg.2019.85897 (2019).
Grahn, N., Hmani-Aifa, M., Fransén, K., Söderkvist, P. & Monstein, H. J. Molecular identification of Helicobacter DNA present in human colorectal adenocarcinomas by 16S rDNA PCR amplification and pyrosequencing analysis. J. Med. Microbiol. 54, 1031–1035. https://doi.org/10.1099/jmm.0.46122-0 (2005).
Soylu, A. et al. Immunohistochemical testing for Helicobacter pylori existence in neoplasms of the colon. BMC Gastroenterol. 8, 35. https://doi.org/10.1186/1471-230x-8-35 (2008).
Epplein, M. et al. Helicobacter pylori protein-specific antibodies and risk of colorectal cancer. Cancer Epidemiol. Biomark. Prev. 22, 1964–1974 (2013).
Malfertheiner, P., Megraud, F., Rokkas, T., Gisbert, J.P., Liou, J.M., Schulz, C., Gasbarrini, A., Hunt, R.H., Leja, M., O'Morain, C. et al. Management of Helicobacter pylori infection: The Maastricht VI/Florence consensus report. Gut. https://doi.org/10.1136/gutjnl-2022-327745 (2022).
Czesnikiewicz-Guzik, M. et al. Causal association between periodontitis and hypertension: Evidence from Mendelian randomization and a randomized controlled trial of non-surgical periodontal therapy. Eur. Heart J. 40, 3459–3470. https://doi.org/10.1093/eurheartj/ehz646 (2019).
Davies, N. M., Holmes, M. V. & Davey Smith, G. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 362, k601. https://doi.org/10.1136/bmj.k601 (2018).
Lin, Z., Pan, I. & Pan, W. A practical problem with Egger regression in Mendelian randomization. PLoS Genet. 18, e1010166. https://doi.org/10.1371/journal.pgen.1010166 (2022).
Palmer, T. M. et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat. Methods Med. Res. 21, 223–242. https://doi.org/10.1177/0962280210394459 (2012).
Burgess, S. & Thompson, S. G. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 40, 755–764. https://doi.org/10.1093/ije/dyr036 (2011).
Kurki, M.I., Karjalainen, J., Palta, P., Sipilä, T.P., Kristiansson, K. & Donner, K. FinnGen: Unique genetic insights from combining isolated population and national health register data. medRxiv (2022).
Lyon, M. S. et al. The variant call format provides efficient and robust storage of GWAS summary statistics. Genome Biol. 22, 32. https://doi.org/10.1186/s13059-020-02248-0 (2021).
Mayerle, J. et al. Identification of genetic loci associated with Helicobacter pylori serologic status. Jama 309, 1912–1920. https://doi.org/10.1001/jama.2013.4350 (2013).
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665. https://doi.org/10.1002/gepi.21758 (2013).
Borenstein, M., Hedges, L. V., Higgins, J. P. & Rothstein, H. R. A basic introduction to fixed-effect and random-effects models for meta-analysis. Res. Synth. Methods 1, 97–111. https://doi.org/10.1002/jrsm.12 (2010).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314. https://doi.org/10.1002/gepi.21965 (2016).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525. https://doi.org/10.1093/ije/dyv080 (2015).
Verbanck, M., Chen, C. Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698. https://doi.org/10.1038/s41588-018-0099-7 (2018).
Cohen, J. F. et al. Cochran’s Q test was useful to assess heterogeneity in likelihood ratios in studies of diagnostic accuracy. J. Clin. Epidemiol. 68, 299–306. https://doi.org/10.1016/j.jclinepi.2014.09.005 (2015).
Curtin, F. & Schulz, P. Multiple correlations and Bonferroni’s correction. Biol. Psychiatry 44, 775–777. https://doi.org/10.1016/s0006-3223(98)00043-2 (1998).
Brion, M. J., Shakhbazov, K. & Visscher, P. M. Calculating statistical power in Mendelian randomization studies. Int. J. Epidemiol. 42, 1497–1501. https://doi.org/10.1093/ije/dyt179 (2013).
Zhou, X. et al. Alcohol consumption, DNA methylation and colorectal cancer risk: Results from pooled cohort studies and Mendelian randomization analysis. Int. J. Cancer 151, 83–94. https://doi.org/10.1002/ijc.33945 (2022).
Dratwa, M. et al. Relationship between telomere length, TERT genetic variability and TERT, TP53, SP1, MYC gene co-expression in the clinicopathological profile of breast cancer. Int. J. Mol. Sci. 23, 5164. https://doi.org/10.3390/ijms23095164 (2022).
Choi, J. E. et al. Polymorphisms in telomere maintenance genes and risk of lung cancer. Cancer Epidemiol. Biomark. Prev. 18, 2773–2781. https://doi.org/10.1158/1055-9965.epi-09-0323 (2009).
Wang, X. et al. Mendelian randomization analysis of C-reactive protein on colorectal cancer risk. Int. J. Epidemiol. 48, 767–780. https://doi.org/10.1093/ije/dyy244 (2019).
Epplein, M. et al. Association of combined sero-positivity to Helicobacter pylori and Streptococcus gallolyticus with risk of colorectal cancer. Microorganisms 8, 1698. https://doi.org/10.3390/microorganisms8111698 (2020).
Yang, F., Xu, Y.L. & Zhu, R.F. Helicobacter pylori infection and the risk of colorectal carcinoma: A systematic review and meta-analysis. Minerva Med. 110, 464–470. https://doi.org/10.23736/s0026-4806.19.05942-1 (2019).
Choi, D.S., Seo, S.I., Shin, W.G. & Park, C.H. Risk for colorectal neoplasia in patients with Helicobacter pylori infection: A systematic review and meta-analysis. Clin. Transl. Gastroenterol. 11, e00127. https://doi.org/10.14309/ctg.0000000000000127 (2020).
Wang, M. et al. Association of Helicobacter pylori infection with colorectal polyps and malignancy in China. World J. Gastrointest. Oncol. 12, 582–591. https://doi.org/10.4251/wjgo.v12.i5.582 (2020).
Fernández de Larrea-Baz, N. et al. Helicobacter pylori antibody reactivities and colorectal cancer risk in a case–control study in Spain. Front. Microbiol. 8, 888. https://doi.org/10.3389/fmicb.2017.00888 (2017).
Blase, J. L. et al. Prediagnostic Helicobacter pylori antibodies and colorectal cancer risk in an elderly, Caucasian population. Helicobacter 21, 488–492. https://doi.org/10.1111/hel.12305 (2016).
Guo, Y. & Li, H. Y. Association between Helicobacter pylori infection and colorectal neoplasm risk: A meta-analysis based on East Asian population. J. Cancer Res. Ther. 10(Suppl), 263–266. https://doi.org/10.4103/0973-1482.151482 (2014).
Limburg, P. J. et al. Helicobacter pylori seropositivity and colorectal cancer risk: A prospective study of male smokers. Cancer Epidemiol. Biomark. Prev. 11, 1095–1099 (2002).
Machida-Montani, A. et al. Atrophic gastritis, Helicobacter pylori, and colorectal cancer risk: A case–control study. Helicobacter 12, 328–332. https://doi.org/10.1111/j.1523-5378.2007.00513.x (2007).
Hooi, J. K. Y. et al. Global prevalence of Helicobacter pylori infection: Systematic review and meta-analysis. Gastroenterology 153, 420–429. https://doi.org/10.1053/j.gastro.2017.04.022 (2017).
Ren, S. et al. Prevalence of Helicobacter pylori infection in China: A systematic review and meta-analysis. J. Gastroenterol. Hepatol. 37, 464–470. https://doi.org/10.1111/jgh.15751 (2022).
Katelaris, P. et al. Helicobacter pylori World Gastroenterology Organization global guideline. J. Clin. Gastroenterol. 57, 111–126. https://doi.org/10.1097/mcg.0000000000001719 (2023).
Ponzetto, A. & Figura, N. Colon cancer risk and VacA toxin of Helicobacter pylori. Gastroenterology 156, 2356. https://doi.org/10.1053/j.gastro.2018.11.083 (2019).
Leja, M., Grinberga-Derica, I., Bilgilier, C. & Steininger, C. Review: Epidemiology of Helicobacter pylori infection. Helicobacter 24(Suppl 1), e12635. https://doi.org/10.1111/hel.12635 (2019).
Bae, M. et al. Akkermansia muciniphila phospholipid induces homeostatic immune responses. Nature 608, 168–173. https://doi.org/10.1038/s41586-022-04985-7 (2022).
Tang, F. B. et al. Toll-like receptor 1 and 10 polymorphisms, Helicobacter pylori susceptibility and risk of gastric lesions in a high-risk Chinese population. Infect. Genet. Evol. 31, 263–269. https://doi.org/10.1016/j.meegid.2015.02.005 (2015).
El-Omar, E. M. Helicobacter pylori susceptibility in the GWAS era. Jama 309, 1939–1940. https://doi.org/10.1001/jama.2013.5590 (2013).
Karpiński, T. M., Ożarowski, M. & Stasiewicz, M. Carcinogenic microbiota and its role in colorectal cancer development. Semin. Cancer Biol. 86, 420–430. https://doi.org/10.1016/j.semcancer.2022.01.004 (2022).
Moradi-Marjaneh, R. et al. Toll like receptor signaling pathway as a potential therapeutic target in colorectal cancer. J. Cell Physiol. 233, 5613–5622. https://doi.org/10.1002/jcp.26273 (2018).
Yang, Y. et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21. Gastroenterology 152, 851-866.e824. https://doi.org/10.1053/j.gastro.2016.11.018 (2017).
Zhang, W. et al. FCGR2A and FCGR3A polymorphisms associated with clinical outcome of epidermal growth factor receptor expressing metastatic colorectal cancer patients treated with single-agent cetuximab. J. Clin. Oncol. 25, 3712–3718. https://doi.org/10.1200/jco.2006.08.8021 (2007).
Geva, R. et al. FCGR polymorphisms and cetuximab efficacy in chemorefractory metastatic colorectal cancer: An international consortium study. Gut 64, 921–928. https://doi.org/10.1136/gutjnl-2014-307234 (2015).
Hathroubi, S., Servetas, S. L., Windham, I., Merrell, D. S. & Ottemann, K. M. Helicobacter pylori biofilm formation and its potential role in pathogenesis. Microbiol. Mol. Biol. Rev. 82, e00001-00018. https://doi.org/10.1128/mmbr.00001-18 (2018).
Morey, P. et al. Helicobacter pylori depletes cholesterol in gastric glands to prevent interferon gamma signaling and escape the inflammatory response. Gastroenterology 154, 1391-1404.e1399. https://doi.org/10.1053/j.gastro.2017.12.008 (2018).
Acknowledgements
We would like to acknowledge the participants and investigators of the FinnGen, EBI, and UK biobank consortiums.
Funding
This work was supported by the Chongqing Pharmaceutical Vocational Education Group Teaching and Research Project (CQZJ202043), Chongqing Science and Health Joint Medical Research Project (2022MSXM037), and by Basic Research and Frontier Exploration Project of Science and Technology Commission of Yuzhong District, Chongqing (201901026).
Author information
Authors and Affiliations
Contributions
(I) Conception and design: F.L., P.Z., M.G., S.Z.; (II) Administrative support: M.G., S.Z.; (III) Provision of study materials or patients: F.L., P.Z., X.R.; (IV) Collection and assembly of data: S.Z., F.L., X.R.; (V) Data analysis and interpretation: F.L., P.Z., S.Z., M.G.; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Luo, F., Zhou, P., Ran, X. et al. No evident causal association between Helicobacter pylori infection and colorectal cancer: a bidirectional mendelian randomization study. Sci Rep 13, 18544 (2023). https://doi.org/10.1038/s41598-023-45545-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-45545-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.