Abstract
Advanced head and neck squamous cell carcinoma (HNSCC) patients have been treated with cisplatin (CDDP) chemoradiation, and the variability of treatment effects has been attributed to single nucleotide variants (SNVs) in genes of metabolic pathways. This study investigated the roles of GSTM1, GSTT1, GSTP1 c.313A>G, XPC c.2815A>C, XPD c.934G>A and c.2251A>C, XPF c.2505T>C, ERCC1 c.354C>T, MLH1 c.93G>A, MSH2 c.211+9C>G, MSH3 c.3133G>A, EXO1 c.1765G>A, TP53 c.215G>C, CASP3 c.-1191A>G and c.-182-247G>T, FAS c.-1378G>A and c.-671A>G and FASL c.-844C>T SNVs in outcome of 109 patients treated with CDDP chemoradiation. Genotypes were identified in genomic DNA by PCR-based methods. Conventional criteria and tests analyzed response and survival. Patients with XPC c.2815AC or CC had 3.43 times more chances of presenting partial response or stable disease. Patients with FAS c.-671GG, GSTM1 present plus XPC c.2815AA, or plus XPD c.934GG, or plus XPD c.2251AA, or plus TP53 c.215GC or CC, and XPD c.2251AA plus XPF c.2505TT had up to 2.70 and 2.37 times more chances of presenting tumor progression and evolving to death, respectively. Our data indicate, for the first time, preliminary evidence that combined SNVs of CDDP metabolism act as independent prognostic factors and can be used to select patients for distinct treatments.
Similar content being viewed by others
Introduction
Cisplatin (CDDP)-based chemotherapy administered with radiotherapy (RT) remains the standard treatment for patients with locally advanced head and neck (HN) squamous cell carcinoma (SCC)1,2.
CDDP interacts mainly with DNA to form inter and intra-strand crosslinks3. The DNA lesions induced by CDDP activate different pathways, including intracellular detoxification4,5, DNA repair5,6,7, and apoptosis5,8.
Glutathione S-transferase proteins, encoded by mu 1 (GSTM1), theta 1 (GSTT1), and pi 1 (GSTP1) genes, conjugate CDDP to glutathione and contribute to its detoxification4,5. Proteins encoded by xeroderma pigmentosum C (XPC), D (XPD), F (XPF), excision repair cross-complementation group 1 (ERCC1), mutL homolog 1 (MLH1), mutS homolog 2 (MSH2), 3 (MSH3) and exonuclease 1 (EXO1) genes act on the nucleotide excision repair (NER) and the mismatch repair (MMR) pathways and remove DNA lesions of CDDP in cells5,6,7. When lesions in DNA overwhelm repair capacity, the biological effect favors apoptosis; the intrinsic and extrinsic apoptosis pathways are activated by modulation of enzymes encoded by tumor protein p53 (TP53), caspase 3 (CASP3), Fas cell surface death receptor (FAS) and Fas ligand (FASL) genes5,8 inducing damaged cells to death.
CDDP chemoradiation effects in HNSCC patients are variable, and this variability has been associated with single nucleotide variants (SNVs) on genes that act on CDDP metabolism pathways9,10,11,12,13,14,15,16,17,18. The functional roles of main SNVs19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34 are presented in Table 1.
The GSTM111, XPD c.934G>A9,10,11, XPD c.2251A>C9,10,12, FAS c.-671A>G13 and FASL c.-844C>T13 SNVs altered relapse-free (RFS), disease-free (DFS), event-free survival (EFS) and/or overall survival (OS) of HNSCC and oropharynx SCC patients treated with CDDP and/or RT, respectively.
Previous analyses of this prospective study conducted by our group demonstrated that GSTP1 c.313A>G17, XPD c.934G>A15, EXO1 c.1762G>A15,16, FAS c.-671A>G18 SNVs of single pathways influenced response rate (RR) and/or PFS, RFS, EFS, and OS of HNSCC patients treated with CDDP chemoradiation. As the final effects of treatment with CDDP chemoradiation possibly depend on the balance of actions of all CDDP metabolic pathways and not in single ways, the present analysis aimed to verify whether the association of defects involving intracellular detoxification, DNA repair, and apoptosis altered even more HNSCC patients´ outcome in almost the same cohort of patients.
Materials and methods
Patients and clinicopathological aspects
Previously published data on outcomes of HNSCC patients diagnosed at the Clinical Oncology Service of the General Hospital of the University of Campinas between June 2011 and February 2014, which focused on the roles of SNVs on genes of intracellular detoxification (n = 90)17, NER (n = 90)15, and MMR (n = 90)16, and intrinsic and extrinsic apoptosis (n = 109)18 pathways, were compiled and seen together in the current analysis of this prospective study. All patients were selected for CDDP chemoradiation as definitive treatment due to unresectable tumors, refusal of surgery related to expected sequels or an organ preservation protocol. Patients not candidates for CDDP treatment or under induction, adjuvant, or palliative therapy were excluded. Creatinine clearance greater than 45 ml/min was required.
The data related to age at diagnosis, gender, tobacco and alcohol consumption, histological grade, stage, human papillomavirus (HPV) type 16 status, and time to treatment delivery were obtained from the patient charts. As previously reported, patients were classified as smokers or non-smokers and drinkers or abstainers35. The tumor was diagnosed according to World Health Organization criteria36 and staged by the American Joint Committee of Cancer37. HPV was tested by P16 immunohistochemistry, as previously described38,39. The interval between the date of diagnosis and the date of treatment initiation was considered the time of treatment delivery.
The study was conducted according to the Declaration of Helsinki and approved by the University of Campinas Ethics Committees (no 274/2011; CAAE: 0218.0.146.000-11). Informed consent was obtained from all subjects and/or their legal guardians before the beginning of the study.
Treatment, response rate, and survival
The single daily fractionated RT (70 Gy at 2 Gy/day) with concurrent bolus CDDP (80–100 mg/m2), given on days 1, 22, and 43, were administered to HNSCC patients; those with consistent side effects during treatment received CDDP at a lower dose (50–75 mg/m2)16. Patients who failed to respond to their initial treatment regimen or relapsed received intravenous methotrexate as palliative chemotherapy40. RR to CDDP chemoradiation was assessed as complete response (CR), partial response (PR,) or stable disease (SD), using Response Evaluation Criteria in Solid Tumors (RECIST) guidelines version 1.141.
EFS was defined from the date of diagnosis and the date of progression, relapse, or death by disease. OS was determined from the date of diagnosis and death by any causes or last follow-up. Patients were followed at 3-month intervals, and the end of the follow-up period considered for the present study was June 2022.
Genotyping
Genotyping was performed in DNA from peripheral blood samples of patients by multiplex polymerase chain reaction (PCR) for GSTM1 and GSTT142, PCR plus enzymatic digestion for GSTP1 c.313A>G (rs1695)43, XPC c.2815A>C (rs2228001)44, XPD c.934G>A (rs1799793) and c.2251A>C (rs13181)23, XPF c.2505T>C (rs1799801)45, ERCC1 c.354C>T (rs11615)46, MLH1 c.93G>A (rs1800734)47, MSH2 c.211+9C>G (rs2303426)48, EXO1 c.1765G>A (rs1047840)49, TP53 c.215G>C (rs1042522)50, CASP3 c.-1191A>G (rs12108497)30 and c.-182-247G>T (rs4647601)30, FAS c.-1378G>A (rs2234767)51 and c.-671A>G (rs1800682)52, FASL c.-844C>T (rs763110)51 and real-time PCR for MSH3 c.3133G>A (rs26279)16. PCR conditions and primers used are shown in Supplementary Table S1. Positive and negative controls were used in reactions. The 10% of genotype determinations were replicated in independent experiments with total concordance.
Statistical analysis
Single SNVs and combinations of two SNVs with biological significance were analyzed in the study. To analyze the roles of deleterious isolated and combined genotypes related to normal or greater detoxification of CDDP, reduced apoptosis of cells damaged by CDDP, and regular or greater repair of lesions induced by CDDP based on information presented in Tables 1 and S2, in the outcome of HNSCC patients was the focus of the study. Differences between clinicopathological aspects and genotypes of SNVs in RR were analyzed by Fisher’s exact test in univariate Cox analysis. Only variables with more than ten individuals in each group and presenting P-values ≤ 0.20 were included in the multivariate Cox analysis, and the logistic regression model assessed associations of variables and RR, with odds ratios (OR) values and 95% confidence intervals (95% CI). Significant results were internally validated using a bootstrap resampling study to investigate the stability of risk estimates (1,000 replications). Kaplan–Meier method, log-rank test, and univariate and multivariate Cox analyses identified variables predicting EFS and OS, with hazard ratios (HR) values and 95% CI. All variables with P-value ≤ 0.20 in univariate Cox regression analysis were included in multivariate analysis.
All statistical tests were done using the SPSS 15.0 software (SPSS Incorporation, Chicago, IL, USA), and significance was achieved when P-values were ≤ 0.05.
Results
Clinicopathological aspects of patients
The median age at diagnosis of 109 HNSCC patients enrolled in the study was 56 years. Most of the patients were male, smokers, and drinkers, and had tumors located in the larynx and pharynx, moderately differentiated tumors, and tumors at advanced stages. Median BMI was within the ordinarily acceptable range, and HPV type 16 was negative in all analyzed cases (Table 2).
Response rate
The CR, PR, and SD were seen in 23.9%, 70.4%, and 5.7% of 88 available HNSCC patients, respectively.
All clinicopathological aspects and single and combined SNVs with biological significance in response to CDDP chemoradiation in 88 patients evaluated are presented in Supplementary Tables S3 and S4, respectively.
Factors with significant associations with the response rates to CDDP and RT are presented in Table 3. In univariate analysis, patients with T3 or T4 tumors, N2 or N3 nodal status, and FAS c.-1378GG genotype had 2.83, 4.77, and 2.67 times more chances of presenting PR or SD than those with T1 or T2 tumors, N1 or N2 nodal status and FAS c.-1378GA or AA genotype, respectively. In multivariate analysis, patients with T3 or T4 tumors, N2 or N3 nodal status, XPC c.2815AC or CC genotype had 3.05, 4.32, and 3.43 times more chances of presenting PR and SD than patients with the remaining aspects, respectively. Combined genotypes of analyzed SNVs did not consistently alter the response to CDDP chemoradiation.
Survival
The impact of all clinicopathological aspects and isolated and combined genotypes of detoxification, DNA repair, and apoptosis-related SNVs in the survival of 109 patients are presented in Supplementary Table S5.
Factors with significant associations with patients’ survival are presented in Table 4. The median follow-up time of HNSCC patients was 22 months (range: 3–126).
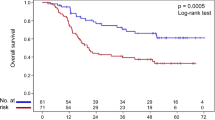
At 24 months of follow-up, EFS was lower in patients with specific tumor aspects and genotypes (Kaplan–Meier estimates). In univariate Cox analysis, patients with T3 or T4 tumors, tumors at III or IV stage, ERCC1 c.354CC or CT, GSTM1 present plus XPC c.2815AA, GSTM1 present plus XPD c.934GG, GSTM1 present plus XPD c.2251AA, GSTM1 present plus XPF c.2505TT, GSTM1 present plus CASP3 c.-1191GG or AG, XPC c.2815AA plus XPD c.2251AA, XPD c.934GG plus XPF c.2505TT, XPD c.2251AA plus XPF c.2505TT, and ERCC1 c.354TT plus FASL c.-844CC had up to 4.50 times more chances of presenting progression, relapse, or death by disease effects than others. In multivariate analysis, patients with T3 or T4 tumor, FAS c.-671GG, GSTM1 present plus XPC c.2815AA, GSTM1 present plus XPD c.934GG, GSTM1 present plus XPD c.2251AA, GSTM1 present plus XPF c.2505TT, GSTM1 present plus TP53 c.215CC or GC, GSTM1 present plus CASP3 c.-1191GG or AG, XPD c.2251AA plus XPF c.2505TT, and ERCC1 c.354TT plus FASL c.-844CC had up to 2.69 times more chances of presenting progression, relapse, or death by disease effects than others.
OS was lower in patients with specific tumor aspects and genotypes at 24 months of follow-up (Kaplan–Meier estimates). Lower OS was also observed in patients with specific tumor aspects and genotypes at 24 months of follow-up (Kaplan–Meier estimates). In univariate Cox analysis, patients with T3 or T4 tumors, tumors at III or IV stage, ERCC1 c.354CC or CT, GSTM1 present plus XPC c.2815AA, GSTM1 present plus XPD c.934GG, GSTM1 present plus XPD c.2251AA, GSTM1 present plus CASP3 c.-1191GG or AG, XPD 934GG plus XPF c.2505TT, XPD c.2251AA plus XPF c.2505TT, and ERCC1 c.354TT plus MSH3 c.3133AA had up to 5.12 times more chances of evolving to death by any cause than remaining patients. In multivariate analysis, patients with T3 or T4 tumor, FAS c.-671GG, FAS c.-671GG or AG, GSTM1 present plus XPC c.2815AA, GSTM1 present plus XPD c.934GG, GSTM1 present plus XPD c.2251AA, GSTM1 present plus TP53 c.215CC or GC, XPD c.934GG plus XPF c.2505TT, and XPD c.2251AA plus XPF c.2505TT had up to 2.37 times more chances of evolving to death by any cause than the remaining patients.
EFS and OS of HNSCC patients with GSTM1 plus XPC c.2815A>C, GSTM1 plus XPD c.934G>A, GSTM1 plus XPD c.225A>C, and GSTM1 plus TP53 c.215G>C are presented in Fig. 1.
Discussion
The effects of CDDP and RT have been associated with genetic variability in distinct metabolic pathways9,10,11,12,13,14,15,16,17,18. Since patients may inherit defects in more than one pathway, we investigated in the current analysis of this prospective study the roles of eighteen SNVs involved in intracellular detoxification, DNA repair, and apoptosis pathways in the outcome of HNSCC patients treated with CDDP chemoradiation. We analyzed only combinations of two SNVs to obtain more consistent results.
We found that patients with large local tumors (T3 or T4) or tumors with large extensions to lymph nodes (N2 or N3) had 3.05 and 4.32 more chances of presenting PR or SD than others in multivariate Cox analysis. The worse response to therapy in these patients was expected in the study because when the tumor size advances, cells change the epithelial-mesenchymal transition53,54, having chemotherapy resistance due to abnormalities of cellular drug accumulation, DNA repair, and cytosolic drug inactivation as consequences55.
In the present multivariate Cox analysis, we observed that patients with XPC c.2815AC or CC had 3.43 times more chances of achieving PR or SD than others. We found an association between XPD c.934G>A and haplotype of XPD c.934G>A and c.2251A>C SNVs15 and EXO1 c.1765G>A and haplotype of EXO1 c.1765G>A and c.2270C>T SNVs17, but not of XPC c.2815A>C15, with response to CDDP chemoradiation in part of this cohort of patients (n = 88) in a previous analysis of NER or MMR pathways, respectively. It is possible the inclusion of new HNSCC patients (n = 21) in the current study may shed light on the real roles of the SNVs in response to CDDP chemoradiation. The association of XPC c.2815AC or CC with worse response to CDDP and RT was not expected in the study because the C allele of XPC c.2815A>C SNV was previously associated with lower DNA repair22, which seems to induce a better response to therapy. Nevertheless, Khan (2000)56 did not demonstrate a clear difference in the rate of nucleotide excision repair in the evaluation of the A and C alleles. Thus, further functional studies are needed to define the fundamental role of XPC c.2815A>C SNV in the DNA repair of CDDP lesions.
We found in multivariate Cox analysis that patients with advanced tumor size, T3 or T4, had 2.33 more chances of presenting tumor progression, relapse of tumor or death by effects of the disease, and 1.94 more chances of evolving to death than those with localized tumors in multivariate Cox analysis. These associations were also seen in other studies57,58,59, and again, these results may be attributed to changes in morphology and behavior of tumor cells during tumor growth53,54, favoring the dissemination of tumors60,61 and short survival55.
We observed in multivariate Cox analysis that patients with the FAS c.-671GG genotype had lower EFS and OS than those with the remaining genotypes and nearly two times more chances of presenting tumor progression, relapse of the tumor, or death than others. Our group previously published this result in the analysis of SNVs on genes of apoptosis pathways in the same cohort of patients18. The allele G was previously associated with reduced apoptosis of colorectal cancer cells because it affects the coupled binding of transcription factors SP1 and STAT1 to chromatin, altering complex recruitment for transcriptional activation33. It is also biologically plausible that the allele G attenuates transcriptional activation mediated by the SP1/STAT1 FAS complex in HNSCC, which in turn dampens the apoptotic pathway of FAS due to its dysregulated expression32,62, favoring the survival of tumor cells, tumor progression or relapse of tumor and death in patients with FAS c.-671GG genotype. Combinations of FAS c.-671A>G genotypes with other SNVs did not alter the EFS of HNSCC patients in the current analysis. The number of patients stratified by combined genotype may not have been sufficient to identify associations with patient survival.
Patients with GSTM1 present plus XPC c.2815AA, GSTM1 present plus XPD c.934GG (HR: 2.45 for EFS, HR: 2.37 for OS), GSTM1 present plus XPD c.2251AA (HR: 1.93 for EFS, HR: 1.90 for OS), GSTM1 present plus TP53 c.215GC or CC (HR: 1.98 for EFS, HR: 1.93 for OS), and XPD c.2251AA plus XPF c.2505TT (HR: 2.0 for EFS, HR: 1.87 for OS) had more chances of tumor progression, relapse of tumor or death than others, but changes in survival of patients with distinct genotypes of isolated GSTM1, XPD c.934G>A, XPD c.2251A>C, TP53 c.215G>C, and XPF c.2505T>C SNVs were not found in the current multivariate Cox analysis.
The presence of GSTM19, XPD c.934GG11,12, and XPD c.2251AA12 genotypes were associated with shorter EFS in previous studies conducted by other groups. XPD c.934AA genotype was associated with lower EFS (HR: 2.12) and OS (HR: 2.04), but XPF c.2505T>C did not alter the survival of 90 HNSCC patients treated with CDDP and/or RT in a previous analysis of this prospective study, which focused on SNVs on genes of NER pathway15. It is worth commenting that previous and current analyses were based on a small number of patients with XPD c.934AA genotype (n = 10) and were adjusted by different variables. Isolated TP53 c.215G>C SNV did not alter the survival of 109 HNSCC enrolled in previous18 and current analyses of this prospective study.
As far as our knowledge goes, there are no studies about associations of genotypes of different pathways of CDDP metabolism with the survival of HNSCC patients treated by CDDP chemoradiation. Associations of the above-mentioned combined genotypes with short survival were expected in the study. GSTM1 present enhances CDDP detoxification of cells4 and the alleles A of XPC c.2815A>C22, G of XPD c.934G>A23, A of XPD c.2251A>C23, T of XPF c.2505T>C24, and C of TP53 c.215G>C29 SNVs induce greater DNA repair and less apoptosis of damaged cells, respectively, favoring higher survival of tumor cells and lower survival of HNSCC patients. It is possible that the sum or synergism of functional abnormalities, such as detoxification and apoptosis, as seen in GSTM1 present plus TP53 c.215GC or CC combined genotype, and repair double defect, as seen in XPD c.2251AA plus XPF c.2505TT is necessary to alter the survival of HNSCC cells and HNSCC patients´ survival in the current analysis, and this the most plausible explanation for the association of combined genotypes but not of isolated genotypes with the survival of patients enrolled in the recent analysis of this prospective study.
It is worth commenting that patients’ survival was not substantially altered by isolated or combined genotypes of GSTP1 c.313A>G, EXO1 c.1765G>A, and MSH3 c.3133G>A SNVs in the current analysis of this study, but GSTP1 c.313GG genotype was associated with lower EFS in a previous analysis of the same cohort of patients (n = 90)14 and EXO1 c.1765GG and MSH3 c.3133GG genotypes were associated with lower EFS and OS, respectively, in a large sample of HNSCC (n = 397) analyzed previously by our group16. The number of patients and statistical adjustments in previous and current analyses may explain differences in results found by our group.
In summary, our data present isolated XPC c.2815A>C and FAS c.-671A>G SNVs, and for the first time, associations of GSTM1 with XPC c.2815A>C, XPD c.934G>A, XPD c.2251A>C and TP53 c.215G>C, and XPD c.2251A>C with XPF c.2505T>C SNVs, as independent factors for the outcome of HNSCC patients treated with CDD chemoradiation. We are aware that although a considerable number of patients were included in this complex and prospective pharmacogenetic study, we believe that a larger cohort of patients and additional functional analyses of XPC c.934G>A SNV in DNA repair may shed light on the roles of the SNVs in response and survival of HNSCC patients treated with CDDP chemoradiation. Thus, we believe that if our data is validated in further studies, specific SNVs on genes of CDDP metabolism can be used to select HNSCC with a high risk for unfavorable outcomes for a differentiated treatment.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Vermorken, J. B. et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N. Engl. J. Med. 359, 1116–1127 (2008).
Siu, L. L. et al. Effect of standard radiotherapy with cisplatin vs accelerated radiotherapy with panitumumab in locoregionally advanced squamous cell head and neck carcinoma: A randomized clinical trial. JAMA Oncol. 3, 220–226 (2017).
Deavall, D. G., Martin, E. A., Horner, J. M. & Roberts, R. Drug-induced oxidative stress and toxicity. J. Toxicol. 8, 645460 (2012).
Hayes, J. D. & Strange, R. C. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology 61, 154–166 (2000).
Siddik, Z. H. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene 22, 7265–7279 (2003).
Friedberg, E. C. How nucleotide excision repair protects against cancer. Nat. Rev. Cancer. 1, 22–33 (2001).
Kunkel, T. A. & Erie, D. A. DNA mismatch repair. Annu. Rev. Biochem. 74, 681–710 (2005).
Thorburn, A. Death receptor-induced cell killing. Cell Sign. 16, 139–144 (2004).
Quintela-Fandino, M. et al. DNA-repair gene polymorphisms predict favorable clinical outcome among patients with advanced squamous cell carcinoma of the head and neck treated with cisplatin-based induction chemotherapy. J. Clin. Oncol. 24(26), 4333–4339 (2006).
Zhong, S. et al. Effects of ERCC2 Lys751Gln (A35931C) and CCND1 (G870A) polymorphism on outcome of advanced-stage squamous cell carcinoma of the head and neck are treatment dependent. Cancer Epidemiol. Biomark. Prev. 20(11), 2429–2437 (2011).
Mahimkar, M. B. et al. Polymorphisms in GSTM1 and XPD genes predict clinical outcome in advanced oral cancer patients treated with postoperative radiotherapy. Mol. Carcinog. 51, 94–103 (2012).
Song, X. et al. Variants in nucleotide excision repair core genes and susceptibility to recurrence of squamous cell carcinoma of the oropharynx. Int. J. Cancer. 133(3), 695–704 (2013).
Nogueira, G. A. et al. Association between genetic polymorphisms in DNA mismatch repair-related genes with risk and prognosis of head and neck squamous cell carcinoma. Int. J. Cancer 137(4), 810–818 (2015).
Zhang, F. et al. Apoptotic variants as predictors of risk of oropharyngeal cancer recurrence after definitive radiotherapy. Int. J. Cancer 137(10), 2454–2461 (2015).
Lopes-Aguiar, L. et al. XPD c.934G>A polymorphism of nucleotide excision repair pathway in outcome of head and neck squamous cell carcinoma patients treated with cisplatin chemoradiation. Oncotarget 8(10), 16190–16201 (2017).
Nogueira, G. A. S. et al. Polymorphisms in DNA mismatch repair pathway genes predict toxicity and response to cisplatin chemoradiation in head and neck squamous cell carcinoma patients. Oncotarget 9(51), 29538–29547 (2018).
Pincinato, E. C. et al. GSTM1, GSTT1 and GSTP1 Ile105Val polymorphisms in outcomes of head and neck squamous cell carcinoma patients treated with cisplatin chemoradiation. Sci. Rep. 27(9), 9312 (2019).
Costa, E. F. D. et al. FAS and FASL variations in outcomes of tobacco- and alcohol-related head and neck squamous cell carcinoma patients. Tumour Biol. 42(7), 1010428320938494 (2020).
Mcilwain, C. C., Townsend, D. M. & Tew, K. D. Glutathione S-transferase polymorphisms: Cancer incidence and therapy. Oncogene 25(11), 1639–1648 (2006).
Moyer, A. M. et al. Glutathione S-transferase T1 and M1: Gene sequence variation and functional genomics. Clin. Cancer Res. 13(23), 7207–7216 (2007).
Watson, M. A., Stewart, R. K., Smith, G. B., Massey, T. E. & Bell, D. A. Human glutathione S-transferase P1 polymorphisms: Relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis 19(2), 275–280 (1998).
Zhu, Y. et al. Modulation of DNA damage/DNA repair capacity by XPC polymorphisms. DNA Repair. 7(2), 141–148 (2008).
Spitz, M. R. et al. Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res. 61(4), 1354–1357 (2001).
Vaezi, A. et al. XPF expression correlates with clinical outcome in squamous cell carcinoma of the head and neck. Clin. Cancer Res. 17(16), 5513–5522 (2011).
Yu, J. et al. Comparison of two human ovarian carcinoma cell lines (A2780/CP70 and MCAS) that are equally resistant to platinum but differ at codon 118 of the ERCC1 gene. Int. J. Oncol. 16(3), 55–60 (2000).
Perera, S., Mrkonjic, M., Rawson, J. B. & Bapat, B. Functional effects of the MLH1-93G>A polymorphism on MLH1/EPM2AIP1 promoter activity. Oncol. Rep. 25(3), 809–815 (2011).
Marra, G. et al. Tolerance of human MSH2+/- lymphoblastoid cells to the methylating agent temozolomide. Proc. Natl. Acad. Sci. 98(13), 7164–7169 (2001).
Wei, K. et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 17(5), 603–614 (2003).
Dumont, P., Leu, J. I., Della Pietra, A. C., George, D. L. & Murphy, M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potentials. Nat. Genet. 33(3), 357–365 (2003).
Jang, J. S. et al. Identification of polymorphisms in the Caspase-3 gene and their association with lung cancer risk. Mol. Carcinog. 47(5), 383–390 (2008).
Chen, K. et al. CASP3 polymorphisms and risk of squamous cell carcinoma of the head and neck. Clin. Cancer Res. 14(19), 6343–6349 (2008).
Sibley, K. et al. Functional FAS promoter polymorphisms are associated with increased risk of acute myeloid leukemia. Cancer Res. 63(15), 4327–4340 (2003).
Wang, S. et al. FAS rs2234767 and rs1800682 polymorphisms jointly contributed to risk of colorectal cancer by affecting SP1/STAT1 complex recruitment to chromatin. Sci Rep. 6, 19229 (2016).
Wu, J. et al. A novel polymorphic CAAT/enhancer-binding protein beta element in the FasL gene promoter alters Fas ligand expression: a candidate background gene in African American systemic lupus erythematosus patients. J. Immunol. 170(1), 132–138 (2003).
Hayes, R. B. Tobacco and alcohol use and oral cancer in Puerto Rico. Cancer Causes Control. 10(1), 27–33 (1999).
El-Naggar, A. K., Chan, J. K. C., Grandis, J. R., Takata, T. & Slootweg, P. J. WHO classification of head and neck Tumours. Lyon. 4, 347 (2017).
Edge, S. B. et al. American Joint Committee on Cancer: Cancer Staging Manual Vol. 7, 21–97 (Springer, 2010).
Begum, S., Gillison, M. L., Ansari-Lari, M. A., Shah, K. & Westra, W. H. Detection of human papillomavirus in cervical lymph nodes: A highly effective strategy for localizing the site of tumor origin. Clin. Cancer Res. 9(17), 6469–6475 (2003).
Singhi, A. D. & Westra, W. H. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer 116(9), 2166–2173 (2010).
Forastiere, A. A. et al. Randomized comparison of cisplatin plus fluorouracil and carboplatin plus fluorouracil versus methotrexate in advanced squamous-cell carcinoma of the head and neck: A Southwest oncology group study. J. Clin. Oncol. 10(8), 1245–1251 (1992).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumors: revised recist guideline. Eur. J. Cancer. 45(2), 228–247 (2009).
Arruda, V. R. et al. Increased risk for acute myeloid leukemia in individuals with glutathione S-transferase mu 1 (GSTM1) and theta 1 (GSTT1) gene defects. Eur. J. Haematol. 66(6), 383–388 (2001).
Hohaus, S. et al. Glutathione S-transferase P1 genotype, and prognosis in Hodgkin’s lymphoma. Clin. Cancer Res. 11(6), 2175–2179 (2005).
Liang, J. et al. XPC gene polymorphisms and risk of idiopathic azoospermia or oligozoospermia in a Chinese population. Int. J. Androl. 32(3), 235–241 (2009).
Povey, J. E. et al. DNA repair gene polymorphisms and genetic predisposition to cutaneous melanoma. Carcinogenesis 28(5), 1087–1093 (2007).
Ma, B. B. et al. Multicenter phase II study of gemcitabine and oxaliplatin in advanced nasopharyngeal carcinoma: Correlation with excision repair cross-complementing-1 polymorphisms. Ann. Oncol. 20(11), 1854–1859 (2009).
Park, S. H. et al. 93G>A polymorphism of hMLH1 and risk of primary lung cancer. Int. J. Cancer. 112(4), 678–682 (2004).
Jung, C. Y. et al. Polymorphisms in the hMSH2 gene and the risk of primary lung cancer. Cancer Epidemiol. Biomark. Prev. 15(4), 762–768 (2006).
Tsai, M. H. et al. Interaction of Exo1 genotypes and smoking habit in oral cancer in Taiwan. Oral Oncol. 45(9), 90–94 (2009).
Honma, H. N. et al. Influence of p53 codon 72 exon 4, GSTM1, GSTT1 and GSTP1*B polymorphisms in lung cancer risk in a Brazilian population. Lung Cancer 61(2), 152–162 (2008).
Zhang, X. et al. Functional polymorphisms in cell death pathway genes FAS and FASL contribute to the risk of lung cancer. J. Med. Genet. 42(6), 479–484 (2005).
Huang, Q. R., Danis, V., Lassere, M., Edmonds, J. & Manolios, N. Evaluation of a new Apo-1/Fas promoter polymorphism in rheumatoid arthritis and systemic lupus erythematosus patients. Rheumatology 38(7), 645–651 (1999).
Kalluri, R. & Weinberg, R. A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119(6), 1420–1428 (2009).
Thiery, J. P., Acloque, H., Huang, R. Y. & Nieto, M. A. Epithelial-mesenchymal transitions in development and disease. Cell 139(5), 871–890 (2009).
Amable, L. Cisplatin resistance and opportunities for precision medicine. Pharmacol. Res. 106, 27–36 (2016).
Khan, S. G. A new xeroderma pigmentosum group C poly (AT) insertion/deletion polymorphism. Carcinogenesis 21(10), 1821–1825 (2000).
Adelstein, D. J. et al. An intergroup phase III comparison of standard radiation therapy and two schedules of concurrent chemoradiotherapy in patients with unresectable squamous cell head and neck cancer. J Clin Oncol. 21(1), 92–98 (2003).
Martins, R. G. et al. Cisplatin and radiotherapy with or without erlotinib in locally advanced squamous cell carcinoma of the head and neck: a randomized phase II trial. J. Clin. Oncol. 31(11), 1415–1421 (2013).
López, R. V. M. et al. Human papillomavirus (HPV) 16 and the prognosis of head and neck cancer in a geographical region with a low prevalence of HPV infection. Cancer Causes Control 25(4), 461–471 (2014).
Keshamouni, V. G. & Schiemann, W. P. Epithelial–mesenchymal transition in tumor metastasis: A method to the madness. Fut. Oncol. 5(8), 1109–1111 (2009).
López-Novoa, J. M. & Nieto, M. A. Inflammation and EMT: An alliance towards organ fibrosis and cancer progression. EMBO Mol. Med. 1, 303–314 (2009).
Look, D. C., Pelletier, M. R., Tidwell, R. M., Roswit, W. T. & Holtzman, M. J. Stat1 depends on transcriptional synergy with Sp1 (∗). J. Biol. Chem. 270(51), 30264–30267 (1995).
Acknowledgements
We would like to thank all patients who accepted to participate in this study. This work was supported by the São Paulo Research Foundation (FAPESP) (Grant Number 2012/01807-2).
Author information
Authors and Affiliations
Contributions
A.M.C.F. participated in statistical analysis, manuscript writing, and revision. J.M.C.A. participated in analyses of radiological exams, and the revision of the manuscript. L.T.M. and G.J.L. participated in statistical analysis, and manuscript revision. C.S.P.L. participated in research planning, study design, manuscript writing, and revision. All authors have read and agreed with the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ferreira, A.M.C., Altemani, J.M.C., Macedo, L.T. et al. Genetic variability in cisplatin metabolic pathways and outcome of locally advanced head and neck squamous cell carcinoma patients. Sci Rep 13, 16762 (2023). https://doi.org/10.1038/s41598-023-44040-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-44040-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.