Abstract
Colon adenocarcinoma (COAD) is a common malignant tumor, and the role of the protein PFKFB4 in glycolysis and pentose phosphate pathways is crucial. Researchers investigated the clinical significance of PFKFB4 in COAD by studying its expression in 79 tissue samples using immunohistochemistry. We found that PFKFB4 expression was significantly higher in COAD patients, particularly in the sigmoid colon. Interestingly, high PFKFB4 expression was associated with both improved overall survival (OS) and post-progression survival (PPS) in COAD patients. Further analysis revealed that genes associated with PFKFB4 were linked to various metabolic pathways, including amino acid biosynthesis, glycolysis, gluconeogenesis, glucose metabolism, and inflammatory response. PFKFB4 expression also showed correlations with the infiltration of different immune cell types in COAD patients, such as CD8+ T cells, CD4+ T cells, regulatory T cells (Tregs), macrophages, neutrophils, dendritic cells, active mast cells, and resting NK cells. Overall, the relationship between PFKFB4 expression and the prognosis of COAD is complex and diverse, possibly playing different roles at different stages of the disease. Moreover, its mechanism might involve interactions with various metabolic pathways and immune infiltration in the tumor microenvironment. These findings provide valuable insights into the potential role of PFKFB4 as a biomarker or therapeutic target in COAD.
Similar content being viewed by others
Introduction
Glycolysis is a crucial metabolic process that takes place in human cells when glucose is broken down. It produces substrates essential for various metabolic pathways, including fatty acid and cholesterol synthesis, the pentose phosphate pathway (PPP), and the tricarboxylic acid cycle (TCA)1. The Warburg effect, also known as aerobic glycolysis, is a phenomenon in which tumor cells, even in the presence of oxygen, have a preference for metabolizing glucose through glycolysis instead of mitochondrial oxidative phosphorylation (OXPHOS), leading to significant lactic acid production2,3.
Phosphofructokinase 2 (PFK2) is a bifunctional enzyme with both kinase and phosphatase activities. In humans, four PFK2 isozymes exist: PFKFB1, PFKFB2, PFKFB3, and PFKFB44. Among these isozymes, phosphoric acid fructose kinase 1, phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (PFKFB4) plays a vital role in regulating the flux through the glycolytic and pentose phosphate pathways and ATP synthesis. It accomplishes this by controlling the levels of fructose 2,6-bisphosphate, the most potent allosteric effector. These pathways and ATP synthesis significantly impact tumor development5,6.
Studies have demonstrated that PFKFB4 is highly expressed in various types of human cancers, such as bladder cancer7, prostatic small cell neuroendocrine carcinoma (SCNC)8, pancreatic cancer, gastric cancer9, glioblastoma10, and acute myeloid leukemia (AML)11. Colon adenocarcinoma (COAD) is a prevalent form of cancer worldwide. Recent epidemiological data ranks it as the fourth most common cancer in China, with the fifth and fourth highest mortality rates for males and females, respectively12. However, the role of PFKFB4 in COAD remains unclear. Therefore, this study aimed to investigate the association between PFKFB4 expression levels and the clinicopathological characteristics and prognosis of COAD patients. Additionally, we analyzed co-expression gene signatures and immune infiltration features related to PFKFB4 expression using public databases to explore the potential function of PFKFB4 in COAD.
Results
Clinicopathological characteristics of patients with COAD
A total of 79 COAD patients were included in the study, comprising 43 women and 36 men, with a median age of 62 years (range, 27–81 years). Among them, 45 cases involved the colon as the tumor site, while 34 cases were associated with the sigmoid colon. The mean tumor diameter was 4.50 ± 0.2 cm, ranging from 1 to 10 cm. Serosa involvement was absent in 13 cases, and 27 cases exhibited lymph node metastases. Tumor differentiation was categorized as well, moderate, or poor in 1, 40, and 27 cases, respectively. Distant metastases were observed in 3 out of the 79 cases. Dukes' stage classification included 11 cases at stage A, 38 cases at stage B, 26 cases at stage C, and 3 cases at stage D. Detailed clinicopathological profiles of the patients are summarized in Table 1. Based on the above data, the relationship between PFKFB4 expression and various clinicopathological features can be obtained. PFKFB4 expression significantly differed between colon and sigmoid colon cases (p = 0.0210). However, age, gender, tumor size, differentiation, pT stage, pN stage, and pM stage did not show significant associations with PFKFB4 expression. Dukes’ stage had a trend toward an association with PFKFB4 expression but did not reach statistical significance (p = 0.1183).
Higher PFKFB4 expression in COAD tissues than in adjacent normal tissues
IHC analysis was performed to evaluate PFKFB4 expression levels in COAD tissues. Positive PFKFB4 expression was observed in nearly all tumor and adjacent normal tissues. Figure 1 shows representative IHC staining of PFKFB4 in colon cancer tissues. PFKFB4 was mainly expressed in the cytoplasm and nucleus, and it was diffusely distributed in the tumor tissues. The expression range and intensity of PFKFB4 were stronger in the nucleus than in the cytoplasm. Semi-quantification of PFKFB4 expression in COAD and adjacent normal tissues revealed significantly higher expression in tumor tissues compared to adjacent normal tissues (Fig. 2a). Moreover, PFKFB4 expression was significantly higher in COAD tissues than in matched adjacent normal tissues in the 15 pairs of matched samples analyzed (Fig. 2b).
Representative immunohistochemical images of PFKFB4 expression in COAD tissues. This figure presents representative immunohistochemical images depicting the expression of PFKFB4 in COAD tissues. The images demonstrate the staining intensity at different magnifications: Negative staining (corresponding normal (control) tissue) at magnifications (a) × 100 and (e) × 400. Weak staining at magnifications (b) × 100 and (f) × 400. Moderate staining at magnifications (c) × 100 and (g) × 400. Strong positive staining at magnifications (d) × 100 and (h) × 400.
Elevated PFKFB4 expression in cancerous tissues compared with adjacent normal tissues. (a) The figure presents a box plot displaying the distribution of PFKFB4 staining scores in adjacent normal tissues (n = 15) and COAD tissues (n = 79). The box plot shows the median with the interquartile range. Statistical analysis was performed using the unpaired Student’s t-test to calculate the p-value. (b) A comparison of PFKFB4 expression between 15 pairs of matched neighboring normal tissues and COAD tissues. The analysis utilized the paired Student’s t-test to determine the p-value.
Differential expression analysis of PFKFB4 in COAD patients
A pan-cancer analysis of PFKFB4 expression using the TIMER2.0 database demonstrated increased PFKFB4 mRNA levels in multiple malignancies, including COAD (Fig. 3).
PFKFB4 expression in various cancer types. This figure depicts the expression levels of PFKFB4 in different types of cancer. The data for PFKFB4 expression were analyzed using the TIMER2.0 database, encompassing multiple cancer types. The significance levels for the analysis are represented as follows: *p < 0.05, **p < 0.01, and ***p < 0.001.
Prognostic analysis of PFKFB4 in COAD patients
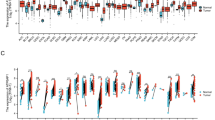
By utilizing the online Kaplan–Meier plotter database, we investigated whether there existed a difference in survival time between COAD patients with high and low PFKFB4 expression. This comprehensive analysis aimed to provide deeper insights into the clinical significance of PFKFB4 in COAD patients. As shown in Fig. 4, the group with low PFKFB4 expression exhibited median survival times of 32 months for RFS, 72 months for OS, and 53 months for PPS. In contrast, the group with high PFKFB4 expression demonstrated median survival times of 42 months for RFS, 130 months for OS, and 25 months for PPS.
Relationship between high expression of PFKFB4 and RFS, OS, and PPS in COAD patients. This figure illustrates the association between high expression of PFKFB4 and three clinical outcomes in COAD patients: Relapse-Free Survival (RFS), Overall Survival (OS), and Post-Progression Survival (PPS). The analysis utilized Kaplan–Meier survival analysis on data obtained from the Kaplan–Meier plotter database. A subsequent log-rank test was performed to assess the statistical significance of the results.
Functional analysis of co-expressed genes of PFKFB4 in COAD patients
Co-expression analysis using the CAMOIP database identified 10,189 PFKFB4-related genes in COAD. GO enrichment analysis of these genes revealed 271 associated functional annotations, comprising 190 for biological processes, 52 for cellular components, and 29 for molecular functions (Fig. 5a). KEGG pathway enrichment analysis showed 44 biological pathways, including 1 related to cellular processes, 8 to processing environmental information, 6 to human diseases, 19 to metabolism, and 10 to organic systems (Fig. 5b). Additionally, 26 Reactome functional annotations were obtained (Fig. 5c). The top 25 biological processes (BP), the top 15 cellular components (CC), and the top 10 molecular functions (MF) functional annotations were listed based on the Count values of the three classification functional annotations in GO, respectively. Enrichment analysis and display of these functional annotations were performed. The top 20 metabolic pathways with a count value in KEGG and the top 20 functional annotations with count value in Reactome were listed for enrichment analysis, and the bar chart was drawn. Correspondingly, bubble charts were created to show the top 20 functional annotations or pathways from GO, KEGG, and Reactome, respectively (Fig. 6).
Histogram of functional analysis of co-expressed genes of PFKFB4 in COAD. In this figure, we present the functional analysis of co-expressed genes associated with PFKFB4 in COAD. A total of 10,189 PFKFB4-related genes in COAD were identified through co-expression analysis. These co-expressed genes were further subjected to classification and functional analyses using the following enrichment databases: (a) Gene Ontology (GO) enrichment analysis; (b) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis; (c) Reactome enrichment analysis. The CAMOIP database was used to perform these enrichment analyses.
Bubble diagrams of functional analysis of co-expressed genes of PFKFB4 in COAD. This figure presents the functional analysis of co-expressed genes associated with PFKFB4 in COAD. A total of 10,189 PFKFB4-related genes in COAD were identified through co-expression analysis. The top 20 enriched pathways from the co-expressed genes of PFKFB4 were analyzed using the following enrichment databases: (a) Gene Ontology (GO) enrichment analysis; (b) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis; (c) Reactome enrichment analysis. The enrichment analyses were performed using the CAMOIP database.
The top five GO functional annotations related to PFKFB4 in COAD are neutrophil activation, granulocyte activation, myeloid leukocyte activation, leukocyte degranulation, and leukocyte activation involved in immune response. The top five pathways on the PFKFB4-related KEGG pathways list are fructose and mannose metabolism, biosynthesis of amino acids, herpes simplex virus 1 infection, mucin type O-glycan biosynthesis, and osteoclast differentiation. The top five Reactome functional annotations associated with PFKFB4 are neutrophil degranulation, formation of the cornified envelope, innate immune system, keratinization, and antimicrobial peptides.
The PFKFB4 expression and immune infiltration level in COAD patients
Using the TIMER2.0 database, we investigated whether PFKFB4 expression contributes to the development of COAD by affecting immune cell infiltration. Our analysis showed that PFKFB4 gene expression in CRC was significantly correlated with the infiltration of CD8+ T cells, CD4+ T cells, regulatory T cells (Tregs), macrophages, neutrophils, dendritic cells, and natural killer (NK) cells (Supplementary Table S1).
Furthermore, we analyzed TCGA-COAD data using the CAMOIP database to explore the impact of PFKFB4 gene levels on various immune cells. Our results indicated a positive correlation between PFKFB4 expression and the infiltration of CD4+ T cells, gamma delta T cells, macrophages, myeloid dendritic cells, activated mast cells, and resting NK cells in COAD (Fig. 7).
Correlation of PFKFB4 expression with immune infiltrates in COAD. This figure demonstrates the correlation between PFKFB4 expression and immune infiltrates in COAD. The immune cell scores estimated by different methods were compared between different groups using the Mann–Whitney U-test based on the CAMOIP database. Comparison of immune cell scores estimated by (a) CIBERSORT, (b) EPIC, (c) IPS, (d) MCPcounter, and (e) quanTIseq, respectively. The significance levels for the comparisons are indicated as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Discussion
Most cancer cells generate energy through a process known as “aerobic glycolysis,” characterized by high glucose uptake and glycolysis, followed by lactic acid fermentation in the cytosol, rather than the oxidative phosphorylation and citric acid cycle observed in normal cells13. Numerous human cancers, including glioblastoma, gastric cancer, pancreatic cancer, bladder cancer, and SCNC, exhibit elevated expression levels of PFKFB4, one of the four isoenzymes of phosphofructokinase14. PFKFB4 regulates glycolysis in cells by generating the glycolytic signaling molecule F-2,6-BP and subsequently hydrolyzing it, thus limiting tumor development15. However, an increasing number of studies have uncovered a connection between the occurrence, growth, and prognosis of specific malignancies and the overexpression of PFKFB4 in different tumors16. For instance, patients with low PFKFB4 expression in gastric cancer had significantly longer overall survival (OS), initial progression survival, and post-progression survival durations compared to those with high PFKFB4 expression17.
PFKFB4 plays a pivotal role in controlling the metabolic fluxes of the glycolytic and pentose phosphate pathways (PPP), which are the primary mechanisms tumor cells utilize to metabolize glucose. Studies have shown that PFKFB4 controls the formation of reactive oxygen species (ROS) by directing glucose metabolic intermediates to the PPP in various cancer cells16,18,19. Moreover, PFKFB4 by reducing prostate cancer cells fructose-2,6-bisphosphate level, make the glucose-6-phosphate guide the PPP, reduces the glycolytic pathway activity, make the NADPH and glutathione levels, eventually reduce the oxidative stress and death of tumor cells, prevent prostate cancer cells apoptosis, promote the growth of tumor20.
In addition to its role in glycolysis, glycolytic enzymes are now recognized to participate in various physiological and pathological processes21. Notably, PFKFB4 expression was significantly higher in solid breast cancers compared to healthy tissues. Researchers have found that the protein kinase PFKFB4 phosphorylates the oncogenic steroid receptor coactivator-3 (SRC-3), enhancing its transcriptional activity and promoting the development of breast cancer22. Furthermore, PFKFB4 is involved in regulating the cell cycle, autophagy, and tumor metastasis23. Nevertheless, few studies have investigated the relationship between PFKFB4 and COAD.
This study has identified a potential correlation between COAD and PFKFB4 expression in clinical samples. The results indicate that COAD tissues exhibit significantly higher levels of PFKFB4 expression compared to adjacent normal tissues. Furthermore, patients with COAD in the sigmoid colon had considerably greater levels of PFKFB4 expression than those with COAD in other parts of the colon. This may be linked to the fact that sigmoid colon is the most common site of COAD, which is located in the right colon and tends to be more malignant24. However, the investigation did not find any direct relationship between PFKFB4 expression and patient age, gender, tumor differentiation, size, depth of invasion, lymph node metastasis, or distant metastasis. This could be due to the inadequate samples.
The clinical samples collected in this study demonstrated a high expression of PFKFB4 in COAD patients. Regarding the prognosis of COAD patients, the corresponding data revealed that high expression of PFKFB4 was associated with improved OS. This finding suggests that COAD patients with high PFKFB4 expression may have better survival rates after treatment, indicating a positive correlation with prognosis before disease progression.
However, intriguingly, the data also showed that PPS was worse in COAD patients with high PFKFB4 expression. This suggests that after disease progression in COAD patients, high expression of PFKFB4 may be linked to shorter survival and an unfavorable prognosis. This finding implies that high expression of PFKFB4 might play a role in promoting the late progression of COAD.
To delve deeper into this seemingly contradictory result, we first conducted gene enrichment analysis of the co-expressed genes associated with PFKFB4 using the CAMOIP database, followed by functional annotation and enrichment analysis. This analysis revealed several PFKFB4-related pathways in COAD, including up-regulation of granulocyte activation, myeloid leukocyte activation, inflammatory response, biosynthesis of amino acids, fructose and mannose metabolism, glycolysis, gluconeogenesis, and glucose metabolism. These findings suggest that COAD patients with high PFKFB4 expression may have a higher tendency to utilize the glycolytic pathway for metabolic biosynthesis, which could be associated with enhanced immunological responses and increased immune infiltration.
Proverbially, the tumor microenvironment (TME) influences the development and recurrence of tumors, and immune cells in TME have been found to either promote or prevent tumor growth25,26. Moreover, immune infiltration, a key element of the TME, has been shown to impact tumor development and response to therapy27. Using the TIMER2.0 and CAMOIP databases, we observed a robust correlation between PFKFB4 expression and the levels of infiltration of various immune cells, including CD8+ T cells, CD4+ T cells, regulatory T cells (Tregs), macrophages, neutrophils, dendritic cells, activated mast cells, and resting NK cells. This suggests that PFKFB4 may also be a marker for immune status and provide information for immunotherapies.
Accordingly, we speculate that the relationship between high expression of PFKFB4 and prognosis of COAD patients is complex and diverse, and it may play different roles in different stages of COAD, leading to the seemingly contradictory results between OS and PPS. This seemingly paradoxical result may be due to the complex biological function of PFKFB4 in COAD. High expression of PFKFB4 may be beneficial to inhibit the proliferation of tumor cells in the initial stage, thereby associated with better OS. However, when the disease progresses to an advanced stage, PFKFB4 may begin to play a role in promoting tumor progression, resulting in a lower prognostic value for post-progression survival. These results may involve a variety of metabolic pathways such as biosynthesis of amino acids, glycolysis, gluconeogenesis, glucose metabolism, and inflammatory response, and a variety of infiltration of immune cells in the body, such as CD8+ T cells, CD4+ T cells, Tregs, macrophages, neutrophils, dendritic cells, active mast cells, and resting NK cells. They work together to produce a complex diversity of the relationship between PFKFB4 expression and the prognosis of COAD patients.
5-FU is a chemotherapeutic agent used in the treatment of colon cancer. However, it can also indirectly affect glycolysis by disrupting the DNA repair process, leading to cell death. In addition, several studies have shown that alterations in glycolytic metabolism may affect the sensitivity of cancer cells to chemotherapeutic agents such as 5-FU. In some cases, increased glycolysis was associated with increased 5-FU sensitivity28. The effect of 5-FU on DNA can indirectly affect the glycolysis pathway in cancer cells. PFKFB4, as a glycolytic regulator, may play a role in determining the response of tumor cells to 5-FU treatment. The balance between glycolysis and response to chemotherapy is an ongoing area of research in oncology29. Clearly, this study may provide relevant ideas and evidence for the role of PFKFB4 in the anti-colon cancer mechanism of 5-FU.
Unfortunately, our study has some limitations. Firstly, our clinical data might be affected by sampling errors due to the limited sample size. Secondly, although we used the TCGA database for additional analysis to address the issue of inadequate sample size, the amount of PFKFB4 gene samples in COAD remains insufficient. Furthermore, based on informatics analysis for clinical databases, it is reasonable to draw this conclusion. Admittedly, this is also the deficiency of our study, and we will conduct experimental analysis at the cellular level in the follow-up research plan in order to further verify our conclusion. Therefore, more research is needed to validate our findings and explore further the molecular linkages, clinical applications, and mechanisms of PFKFB4 in COAD.
Conclusion
The expression of PFKFB4 is significantly higher in COAD tissue, with particularly elevated levels observed in the sigmoid colon compared to other regions of the colon. Moreover, the relationship between high PFKFB4 expression and the prognosis of COAD patients is intricate and diverse, as it may assume distinct roles at different stages of COAD. The underlying mechanism may involve multiple metabolic pathways, including biosynthesis of amino acids, glycolysis, gluconeogenesis, glucose metabolism, and inflammatory response. Future studies are essential to gain a comprehensive understanding of the function and potential therapeutic significance of PFKFB4 in COAD. These investigations will provide valuable insights and contribute to a better prognostic evaluation of COAD patients.
Materials and methods
Patients and specimens
Tumor tissue samples were collected from 79 patients diagnosed with colon COAD who were admitted to Ningxia Medical University General Hospital (Yinchuan, China) between September 2016 and December 2017. Among these patients, only 15 tumor tissues had matched adjacent normal tissues (located at least 2 cm from the tumor margin) due to the tissue section generation process. The samples were fixed with 4% polyformaldehyde at room temperature for 24 h and subsequently embedded in paraffin wax. The tissues were then sectioned into 5-µm-thick slices for immunohistochemical (IHC) analysis. Pathological diagnosis was performed by two senior pathologists. Clinicopathological data of the patients are presented in Table 1, although 2 cases lacked information on tumor differentiation, and 1 case lacked information on tumor size, depth of invasion, lymph node metastasis, and Dukes’ stage. The experiments involving human subjects were carefully examined and authorized by the Medical Research Ethics Review Committee of the General Hospital of Ningxia Medical University (Yinchuan, China). The participants/patients provided written informed consent prior to their participation in the investigation. All research was carried out in accordance with relevant guidelines and regulations.
Immunohistochemical staining
The expression levels of PFKFB4 in COAD tissues were determined using immunohistochemical (IHC) analysis. Formalin-fixed and paraffin-embedded tissue sections were heated at 65 °C for 1 h, followed by deparaffinization in xylene and rehydration in a series of ethanol solutions. To block endogenous peroxidase activity, the sections were treated with 3% hydrogen peroxide for 10 min at room temperature after three washes with PBS. Antigen retrieval was performed using EDTA buffer (pH 8.0). Subsequently, the sections were blocked with 10% goat serum (Cat. No. ZLI-9022; OriGene Technologies) for 10 min at 37 °C. Next, the sections were incubated with anti-PFKFB4 antibody (1:100; Cat. No. ab137785; Abcam) at 37 °C for 1 h, followed by overnight incubation in a humid environment at 4 °C. The next day, the sections were washed three times with PBS and then incubated with HRP-conjugated peroxidase working solution (Cat. No. P0448; Dako; Agilent Technologies, Inc.) for 30 min at 37 °C. After treating the sections with the chromogen substrate 3,3′-diaminobenzidine for 10 min at room temperature, counterstaining with hematoxylin was performed for 1 min at room temperature. The slides were then dehydrated in progressively higher concentrations of alcohol and xylene before being sealed with neutral balsam and glass coverslips.
To assess PFKFB4 expression levels, a semi-quantitative method was used, where a light microscope was employed to view and capture images of the sections17,30. Two experienced pathologists graded the intensity of staining as follows: 0 for negative, 1 for light yellow, 2 for tan, and 3 for brown. Additionally, the total number of stained cells was scored based on the proportion of positively stained cells, which were categorized into four groups: 0 for no positively stained cells, 1+ for ≤ 10% positive cells, 2+ for 11–50% positive cells, 3+ for 51–75% positive cells, and 4+ for > 75% positively stained cells. The final score was determined by multiplying the staining intensity score and the proportion of positively stained cells score. To distinguish samples with high and low expression, a cutoff value based on the median IHC score was used.
Expression analysis
To address statistical bias caused by the limited sample size in the analysis of PFKFB4 expression in COAD, we employed the TIMER2.0 online platform (http://timer.cistrome.org/) to access the PFKFB4 expression data across various tumors and evaluate the expression in adjacent non-cancerous tissues31.
Survival analysis
We conducted survival analysis for COAD using the Kaplan–Meier plotter database (https://kmplot.com/analysis) and performed a log-rank test32. The survival analysis focused on Affymetrix ID 206246_at, which corresponds to PFKFB4. The assessment included a total of 1302 COAD patients for relapse-free survival (RFS), 550 patients for overall survival (OS), and 145 patients for post-progression survival (PPS).
Functional enrichment analysis
We utilized the CAMOIP online platform to identify genes associated with PFKFB4 in COAD based on Cancer Genome Atlas (TCGA)-COAD data [16]. Our screening criteria for these genes were set at p-values < 0.05 and FDR (q-value) < 0.25. Detailed annotation information about Kyoto Encyclopedia of Genes and Genomes (KEGG), Reactome, and Gene Ontology biological process (GO) was obtained for these selected genes. Visualization of the associated functional annotations or pathways was achieved using the resources provided by the web platform, in the form of a histogram (available at https://www.omicsstudio.cn) and a bubble chart (available at https://cloud.oebiotech.cn), respectively.
Analysis of immune cell infiltration
To investigate the association between PFKFB4 expression and immune cell infiltration in COAD, we utilized the TIMER2.0 database and the CIBERSORT algorithm31. Tumor purity-adjusted partial Spearman’s correlation was used to evaluate the relationship between PFKFB4 mRNA expression levels and immune infiltration levels. Statistical significance was considered at a p-value threshold of < 0.05.
Additionally, we employed the CAMOIP database and the CIBERSORT algorithm to assess the immune cells associated with PFKFB4 expression in COAD33. The CIBERSORT algorithm allowed us to evaluate the following immune cell types: B cells naive, B cells memory, plasma cells, T cells CD8+, T cells CD4+ naive, T cells CD4+ memory resting, T cells CD4 memory activated, T cells follicular helper, T cells regulatory (Tregs), T cells gamma delta, NK cells resting, Monocytes, Macrophages M0, M1, and M2, Mast cells resting, Mast cells active, Dendritic cells resting, Dendritic resting cells, Dendritic activated cells, Neutrophils, and Eosinophils34. Moreover, we used the EPIC algorithm to score B cells, cancer-associated fibroblasts (CAFs), CD4+ T cells, CD8+ T cells, endothelial cells, macrophages, NK cells, and other cells. The IPS algorithm allowed us to rate MHC molecules, effector cells, suppressor cells, and checkpoint molecules35. Finally, we employed the MCPcounter method to evaluate T cells, CD8+ T cells, cytotoxic lymphocytes, B lineage, NK cells, monocytic lineage, myeloid dendritic cells, neutrophils, endothelial cells, and fibroblasts36. Additionally, we used the quanTIseq method to appraise B cells, macrophages M1 and M2, monocytes, neutrophils, NK cells, T cells CD4+ and CD8+ , Tregs, dendritic cells, and other immune cells37.
Statistical analysis
Statistical analysis and graphical representations were performed using GraphPad Prism 9.4 (La Jolla, CA, USA). The relationship between PFKFB4 expression and clinicopathological features was assessed using either a χ2 test or Fisher’s exact test. We employed a two-tailed Student’s t-test to compare COAD tissues and adjacent normal tissues, while the Mann–Whitney U test was used to compare immune cells between the PFKFB4 high expression group and low expression group. Statistical significance was defined as p < 0.05.
Data availability
The authors of this study will provide the clinical data sets used or analyzed in the investigation upon a reasonable request. The study utilized publicly accessible datasets, which can be found on the websites of TIMER2.0 (http://timer.cistrome.org/), Kaplan–Meier plotter (https://kmplot.com/analysis), and CAMOIP (http://www.camoip.net/). All necessary information can be accessed through these websites.
Change history
09 December 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41598-023-49212-z
References
Koppenol, W. H., Bounds, P. L. & Dang, C. V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337. https://doi.org/10.1038/nrc3038 (2011).
Warburg, O. On the origin of cancer cells. Science 123, 309–314. https://doi.org/10.1126/science.123.3191.309 (1956).
Warburg, O. On respiratory impairment in cancer cells. Science 124, 269–270 (1956).
Rider, M. H. et al. 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: Head-to-head with a bifunctional enzyme that controls glycolysis. Biochem. J. 381, 561–579. https://doi.org/10.1042/BJ20040752 (2004).
Chesney, J. et al. Fructose-2,6-bisphosphate synthesis by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 (PFKFB4) is required for the glycolytic response to hypoxia and tumor growth. Oncotarget 5, 6670–6686. https://doi.org/10.18632/oncotarget.2213 (2014).
Gao, R. et al. CD44ICD promotes breast cancer stemness via PFKFB4-mediated glucose metabolism. Theranostics 8, 6248–6262. https://doi.org/10.7150/thno.28721 (2018).
Yun, S. J. et al. PFKFB4 as a prognostic marker in non-muscle-invasive bladder cancer. Urol. Oncol. 30, 893–899. https://doi.org/10.1016/j.urolonc.2010.08.018 (2012).
Li, W. et al. The role of CD44 in glucose metabolism in prostatic small cell neuroendocrine carcinoma. Mol. Cancer Res. 14, 344–353. https://doi.org/10.1158/1541-7786.MCR-15-0466 (2016).
Bobarykina, A. Y. et al. Hypoxic regulation of PFKFB-3 and PFKFB-4 gene expression in gastric and pancreatic cancer cell lines and expression of PFKFB genes in gastric cancers. Acta Biochim. Pol. 53, 789–799 (2006).
Goidts, V. et al. RNAi screening in glioma stem-like cells identifies PFKFB4 as a key molecule important for cancer cell survival. Oncogene 31, 3235–3243. https://doi.org/10.1038/onc.2011.490 (2012).
Wang, G., Li, S., Xue, K. & Dong, S. PFKFB4 is critical for the survival of acute monocytic leukemia cells. Biochem. Biophys. Res. Commun. 526, 978–985. https://doi.org/10.1016/j.bbrc.2020.03.174 (2020).
Zheng, R. et al. Cancer incidence and mortality in China, 2016. J. Natl. Cancer Center 2, 1–9. https://doi.org/10.1016/j.jncc.2022.02.002 (2022).
Ward, P. S. & Thompson, C. B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 21, 297–308. https://doi.org/10.1016/j.ccr.2012.02.014 (2012).
Warmoes, M. O. & Locasale, J. W. Heterogeneity of glycolysis in cancers and therapeutic opportunities. Biochem. Pharmacol. 92, 12–21. https://doi.org/10.1016/j.bcp.2014.07.019 (2014).
Guo, Y. et al. Systematic review with meta-analysis: HIF-1alpha attenuates liver ischemia-reperfusion injury. Transplant Rev. (Orlando) 29, 127–134. https://doi.org/10.1016/j.trre.2015.05.001 (2015).
Yi, M. et al. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 and 4: A pair of valves for fine-tuning of glucose metabolism in human cancer. Mol. Metab. 20, 1–13. https://doi.org/10.1016/j.molmet.2018.11.013 (2019).
Wang, F. et al. PFKFB4 as a promising biomarker to predict a poor prognosis in patients with gastric cancer. Oncol. Lett. 21, 296. https://doi.org/10.3892/ol.2021.12557 (2021).
Dasgupta, S. et al. Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer. Nature 556, 249–254. https://doi.org/10.1038/s41586-018-0018-1 (2018).
Strohecker, A. M. et al. Identification of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase as a novel autophagy regulator by high content shRNA screening. Oncogene 34, 5662–5676. https://doi.org/10.1038/onc.2015.23 (2015).
Ros, S. et al. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 is essential for p53-null cancer cells. Oncogene 36, 3287–3299. https://doi.org/10.1038/onc.2016.477 (2017).
Lu, Z. & Hunter, T. Metabolic kinases moonlighting as protein kinases. Trends Biochem. Sci. 43, 301–310. https://doi.org/10.1016/j.tibs.2018.01.006 (2018).
Truong, T. H. et al. PELP1/SRC-3-dependent regulation of metabolic PFKFB kinases drives therapy resistant ER(+) breast cancer. Oncogene 40, 4384–4397. https://doi.org/10.1038/s41388-021-01871-w (2021).
Kotowski, K. et al. Role of PFKFB3 and PFKFB4 in cancer: Genetic basis, impact on disease development/progression, and potential as therapeutic targets. Cancers (Basel) 2021, 13. https://doi.org/10.3390/cancers13040909 (2021).
Yagi, Y. et al. Sigmoid colon cancer arising in a diverticulum of the colon with involvement of the urinary bladder: A case report and review of the literature. BMC Gastroenterol. 14, 90. https://doi.org/10.1186/1471-230X-14-90 (2014).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437. https://doi.org/10.1038/nm.3394 (2013).
Eissmann, M. F., Buchert, M. & Ernst, M. IL33 and mast cells-the key regulators of immune responses in gastrointestinal cancers?. Front. Immunol. 11, 1389. https://doi.org/10.3389/fimmu.2020.01389 (2020).
Liu, K. et al. Reprogramming the tumor microenvironment by genome editing for precision cancer therapy. Mol. Cancer 21, 98. https://doi.org/10.1186/s12943-022-01561-5 (2022).
Ghanbari Movahed, Z., Rastegari-Pouyani, M., Mohammadi, M. H. & Mansouri, K. Cancer cells change their glucose metabolism to overcome increased ROS: One step from cancer cell to cancer stem cell?. Biomed. Pharmacother. 112, 108690. https://doi.org/10.1016/j.biopha.2019.108690 (2019).
Zhang, Y. et al. Targeting glucose metabolism enzymes in cancer treatment: Current and emerging strategies. Cancers (Basel) 2022, 14. https://doi.org/10.3390/cancers14194568 (2022).
Guo, Z. et al. TELO2 induced progression of colorectal cancer by binding with RICTOR through mTORC2. Oncol. Rep. 45, 523–534. https://doi.org/10.3892/or.2020.7890 (2021).
Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–W514. https://doi.org/10.1093/nar/gkaa407 (2020).
Lanczky, A. & Gyorffy, B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 23, e27633. https://doi.org/10.2196/27633 (2021).
Lin, A. et al. CAMOIP: A web server for comprehensive analysis on multi-omics of immunotherapy in pan-cancer. Brief. Bioinform. 2022, 23. https://doi.org/10.1093/bib/bbac129 (2022).
Zhou, R. et al. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I-III colon cancer. Cancer Immunol. Immunother. 68, 433–442. https://doi.org/10.1007/s00262-018-2289-7 (2019).
Peng, L. et al. Immune infiltration and clinical outcome of super-enhancer-associated lncRNAs in stomach adenocarcinoma. Front. Oncol. 12, 780493. https://doi.org/10.3389/fonc.2022.780493 (2022).
Song, S. & Shu, P. Expression of ferroptosis-related gene correlates with immune microenvironment and predicts prognosis in gastric cancer. Sci. Rep. 12, 8785. https://doi.org/10.1038/s41598-022-12800-6 (2022).
Plattner, C., Finotello, F. & Rieder, D. Deconvoluting tumor-infiltrating immune cells from RNA-seq data using quanTIseq. Methods Enzymol. 636, 261–285. https://doi.org/10.1016/bs.mie.2019.05.056 (2020).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (grant no. 81860442), and the Ningxia Natural Science Foundation (grant nos. 2022AAC02027, 2022AAC03475), and the Scientific Research Project of Ningxia Medical University (XZ2020006).
Author information
Authors and Affiliations
Contributions
X.G., X.D., and Y.H. were responsible for the study's conception, data analysis, and manuscript writing. FW and CG examined and corrected the manuscript. The data were gathered by Y.Z. L.D. was in charge of managing and supervising the project. The article's submission was reviewed and approved by all of the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained errors in the Abstract and in Figure 4. Full information regarding the corrections made can be found in the correction for this Article.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gu, X., Dai, X., Huang, Y. et al. Differential roles of highly expressed PFKFB4 in colon adenocarcinoma patients. Sci Rep 13, 16284 (2023). https://doi.org/10.1038/s41598-023-43619-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-43619-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.