Abstract
The C1Q complement protein C1QL1 is highly conserved in mammals where it is expressed in various tissues including the brain. This secreted protein interacts with Brain-specific Angiogenesis Inhibitor 3, BAI3/ADGRB3, and controls synapse formation and maintenance. C1ql1 is expressed in the inferior olivary neurons that send projections to cerebellar Purkinje cells, but its expression in the rest of the brain is less documented. To map C1ql1 expression and enable the specific targeting of C1ql1-expressing cells, we generated a knockin mouse model expressing the Cre recombinase under the control of C1ql1 regulatory sequences. We characterized the capacity for Cre-driven recombination in the brain and mapped Cre expression in various neuron types using reporter mouse lines. Using an intersectional strategy with viral particle injections, we show that this mouse line can be used to target specific afferents of Purkinje cells. As C1ql1 is also expressed in other regions of the brain, as well as in other tissues such as adrenal glands and colon, our mouse model is a useful tool to target C1ql1-expressing cells in a broad variety of tissues.
Similar content being viewed by others
Introduction
C1QL1 belongs to the subfamily of Complement component 1 Q subcomponent-Like proteins (C1QLs). This subfamily is part of the C1Q complement family characterized by a C-terminal globular gC1Q signature domain involved in the formation of hetero- or homo-trimers1,2. The N-terminal region of C1QL proteins contains two conserved cysteines allowing trimers to form higher-order oligomers3. C1QLs are absent in plants, yeasts and several invertebrates (e.g. nematodes or insects), while they are found in fish, frog and mammals3. Among mammals, the C1QL proteins are highly conserved with more than 90% amino acid identity between human and mouse orthologs3.
In rodents, C1ql1 is expressed in the brain3,4, as well as the adrenal glands, placenta, colon and testis (Mouse ENCODE transcription data: https://www.ncbi.nlm.nih.gov/gene/23829)3. In the central nervous system, its expression is mostly associated with motor-related areas. Some nuclei in the medulla, pons, midbrain, olfactory bulb, hippocampus and cortex express C1ql1 during postnatal development and adulthood3,4. In the olivo-cerebellar network, transient expression of C1ql1 is observed at postnatal day 7 (P7) in the external granular layer of the cerebellum, where the precursors of the granule cells (GCs) reside3. C1ql1 is also expressed in inferior olivary neurons (IONs) starting from birth and throughout the life of the mouse3,4. C1QL1 is a secreted synaptic protein known to be released by the IONs and to bind to its postsynaptic receptor, the Adhesion-G Protein Coupled Receptor (GPCR) Brain-specific Angiogenesis Inhibitor 3 (BAI3, ADGRB3)5, on cerebellar Purkinje cells (PCs)4,6,7. The interaction between C1QL1 and BAI3 is essential for the formation and maintenance of excitatory synapses between climbing fibers (CFs), axons from IONs, and their PC targets, as the removal of each of these two components results in the loss of a significant amount of these synapses4,6.
In order to fully understand the role of C1QL1 in different brain circuits as well as other organs, it is imperative to be able to specifically target C1ql1-expressing cells. To this aim, we developed the C1ql1cre knockin mouse model. We characterized the specificity and capacity of this mouse line as a driver using several reporter mouse lines. Finally, we show its usefulness for specific targeting of IONs in vivo using an intersectional strategy with viral vectors.
Results
Characterization of the driver capacity of the C1ql1 cre mouse line
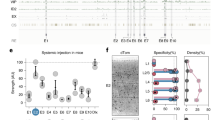
To develop a mouse line expressing the Cre recombinase under the control of the C1ql1 regulatory elements without modifying the coding sequence (CDS) of the C1ql1 gene, we targeted its last exon (in collaboration with genOway). A transgene was designed to contain two flippase recognition target (FRT) flanking a neomycin coding sequence, followed by the exon 2 of the C1ql1 gene where an ires-cre CDS cassette is inserted after the C1ql1 stop codon. This transgene was knocked in the corresponding region on chromosome 11 by homologous recombination (Fig. 1A). The neomycin sequence was removed later by breeding the transgenic mice with a flippase-expressing mouse (Fig. 1A). Identification of the C1ql1cre F0 transgenic mice containing the ires-cre CDS was subsequently performed by PCR (Fig. 1B). Mice that are homozygous for the C1ql1cre alleles are viable, fertile, normal in size and do not show any gross physical or behavioral deficits (observations on two different litters of five animals each, at birth, weaning and adult stages).
Characterization of the driver capacity of the C1ql1cre mouse line. (A) Illustration of the mouse C1ql1 locus at the 2nd exon and flanking regions. Homologous recombination was used to insert a neomycin cassette upstream of the C1ql1 exon 2 and an ires-cre cassette downstream of the stop codon in the C1ql1 exon 2. After neomycin selection and mice generation, the neomycin cassette was removed using flippase to obtain C1ql1-ires-cre knockin mice (C1ql1cre). (B) Top panel: Table of the genotyping PCR amplicon bands and their respective sizes depending on the genotype. Bottom panel: Representative cropped image of a genotyping PCR run (original gel is presented in Supp Fig. 1). (C) Schematic illustration of the breeding strategy used to generate C1ql1cre/R26cas9,-GFP double heterozygous mice. (D) Medial to lateral sagittal sections from P30 C1ql1cre/R26cas9,-GFP mouse brain were immunostained for GFP at P30. Mosaic images were acquired using a Zeiss Axiozoom V16 macroscope (a–d). EGFP was highly expressed in the cerebellum (Cb), in particular in Granule Cells (GCs), and in the Brainstem (Bs), and present in other nuclei from the Midbrain (Mb) and Olfactory Bulb (OB). Scale bars = 1000 µm. (a′–d′) High magnification of different nuclei or neurons expressing EGFP in P30 C1ql1cre/R26cas9,-GFP mice. Scale bars = 200 µm. AOB: Accessory Olfactory Bulb, AON: Anterior Olfactory Nucleus, ION: Inferior Olivary Nuclei, MM: Medial Mammillary nucleus, NLL: Nucleus of the Lateral Lemniscus, PAG: Periaqueductal Gray, PGRN: Paragigantocellular Reticular Nucleus, PRN: Pontine Reticular Nucleus, RN: Red Nucleus, SNr: Substantia Nigra pars reticulata, SUM: Supramammillary nucleus, VNC: Vestibular Nuclei, VTA: Ventral Tegmental Area.
To visualize and characterize the driver capacity of the C1ql1cre mouse line in the brain, homozygous C1ql1cre/cre mice were crossed with the Cre-dependent reporter mouse line R26cas9,-GFP/cas9,-GFP8 (Fig. 1C). In this reporter mouse, Cre-mediated recombination allows the removal of the Stop cassette before the sequence coding cas9-P2A-EGFP. Due to the ubiquitous activity of the Rosa26 locus (R26) and the use of the CAG promoter, Cas9 and soluble GFP are expressed permanently in all the cells that have expressed Cre and their progeny. This expression history was mapped in C1ql1cre/WT/R26cas9,-GFP/WT heterozygotes at P30 using immunofluorescence on brain sagittal sections with an anti-GFP antibody to increase the signal to noise ratio (Fig. 1D). The GFP-positive cells were observed in different regions of the forebrain, midbrain, brainstem and cerebellum (Fig. 1D). At low magnification, two GFP-expressing regions were identified distinctly: the brainstem and the cerebellum (Fig. 1D, a). In the brainstem, GFP expression was strong in the inferior olive (Fig. 1D, a), in concordance with previous results showing expression of C1ql1 in inferior olivary neurons (IONs) from early development until adult age3,4. In the cerebellum, many granule cells (GCs) expressed GFP in a heterogeneous manner and in an antero-posterior gradient, with highest GFP signal in the anterior lobules. Based on in situ hybridization data3, only cells in the external granular layer, not the mature GCs, express C1ql1 during the first two postnatal weeks. This suggests that Cre-dependent recombination happens at the progenitor level in the granule cell lineage. Altogether, these results show that the C1ql1cre mouse line is a powerful tool to genetically target two components of the olivo-cerebellar system, the IONs and the GCs.
At higher magnification, other GFP-expressing regions were identified in the brain of C1ql1cre/WT/R26cas9,-GFP/WT mice (Fig. 1D, a′–d′). According to the Allen Reference Brain atlas (https://mouse.brain-map.org/static/atlas), we found GFP-expressing cells in several motor-related areas in the midbrain: ventral tegmental area, periaqueductal gray (Fig. 1D, a and a′), substantia nigra pars reticulata and red nucleus (Fig. 1D, b and b′). Adjacent to these regions, several GFP-positive cells were detected in two medial nuclei of the hypothalamus: supramammillary nucleus and medial mammillary nucleus (Fig. 1D, a and a′). In the pons, two nuclei were labelled with GFP: pontine reticular nucleus (motor related-area) (Fig. 1D, b and b′) and nucleus of the lateral lemniscus (sensory-related area) (Fig. 1D, d and d′). In addition to the IONs (Fig. 1D, a and a′′), two other motor-related nuclei were expressing GFP in the medulla: the paragigantocellular reticular nucleus (Fig. 1D, b and b′′) and vestibular nuclei (Fig. 1D, c and c′′). Expression of GFP is not restricted to the midbrain or hindbrain, but can be found also in the forebrain, in the accessory olfactory bulb and anterior olfactory nucleus (Fig. 1D, c and c′). The same pattern of expression was found when C1ql1cre/cre mice were crossed with another reporter line, R26R-EYFP/WT, where the enhanced YFP (EYFP) is expressed in cells following Cre-mediated excision of the Stop cassette preceding the EYFP sequence9 (Fig. 2A and Supp Fig. 2), showing the reproducibility of neuronal targeting using the C1ql1cre mouse line. Altogether, different nuclei and regions, especially motor-related areas in the midbrain, pons and brainstem, can be targeted genetically with the C1ql1cre mouse model, in addition to the olivo-cerebellar network.
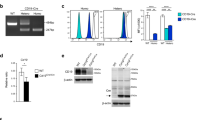
The C1ql1cre mouse line drives expression in different cell types of the olivo-cerebellar network in the brain as well as other organs. (A) Top left panel: Schematic illustration of the crosses used to generate C1ql1cre/R26R-EYFP heterozygous mice. SA, Splice Acceptor. Bottom left panel: Whole tissue protein lysates from the cerebellum, liver, adrenal gland, lung and colon of adult control (C1ql1WT/R26R-EYFP, wild-type: WT) and heterozygote (C1ql1cre/R26R-EYFP, Het) mice were subjected to denaturing electrophoresis and analyzed by immunoblot for EYFP (top blot) and β-ACTIN (bottom blot) expression. EYFP was detected in the cerebellum, adrenal gland and colon of a heterozygote C1ql1cre/R26R-EYFP mouse (original blot is presented in Supp Fig. 3). Right panel: parasagittal cerebellar sections from P28 C1ql1cre/R26R-EYFP mice were immunostained for EYFP using anti-GFP antibody. Cerebellar granule cells (GCs, green arrowheads) and axons from inferior olivary neurons (IONs, cyan arrowheads), the climbing fibers (CFs, pink arrowheads), were EYFP labelled in the cerebellum and brainstem regions, respectively. (B) Left panel: Schematic illustration of the crosses used to generate C1ql1cre/R26hM4Di,-mCitrine heterozygous mice. Right panel: parasagittal brain sections from P30 C1ql1cre/R26hM4Di,-mCitrine mice were immunostained for mCitrine using anti-GFP antibody and images were acquired using a spinning disk confocal microscope. Cerebellar granule cells (GCs, green arrowheads) and axons from inferior olivary neurons (IONs), the climbing fibers (CFs, pink arrowheads), were positive for mCitrine in the cerebellum and brainstem regions, respectively. Scale bars = 20 µm for GCs and 125 µm for IONs and CFs.
The expression of C1ql1 has been reported in several organs outside of the nervous system such as the adrenal glands and the colon using RT-PCR (Mouse ENCODE transcription data: https://www.ncbi.nlm.nih.gov/gene/23829). Using immunoblot analysis of protein extracts from different tissues of adult C1ql1cre/R26R-EYFP mice, the expression of the reporter EYFP was detected not only in the cerebellum, but also in the adrenal gland and the colon, two tissues that had C1ql1 expression reported in the Mouse ENCODE transcription database. In accordance with this database, no EYFP expression was found in extracts from the liver and the lung (Fig. 2A). These results show that the C1ql1cre mouse model is not only useful for studies of the brain, but also in any other organ containing C1ql1-expressing cells.
Normal morphology and transmission of climbing fiber/Purkinje cell synapses in C1ql1 cre/WT heterozygotes
To detect the expression of the Cre recombinase in different brain regions, RT-qPCR was performed on RNA extracts from the cerebellum, the brainstem, the hippocampus and the cortex at P14. No expression of cre was observed in extracts from control (wild-type, WT) mice, while cre was detected in all tested regions of the brain in heterozygous C1ql1cre/WT animals (Fig. 3A). As expected, the highest expression of cre was seen in the brainstem extracts where IONs are located (mean expression normalized to Rpl13a ± SEM = 6.40 ± 0.54 in the brainstem, versus 2.54 ± 0.54, 2.60 ± 0.57, and 1.53 ± 0.18 in the cerebellum, the hippocampus, and the cortex, respectively). C1QL1 loss of function leads to detectable phenotypes in climbing fiber/Purkinje cell (CF/PC) synapse formation and function in the olivo-cerebellar network when C1ql1 expression is reduced by more than 90%4,6. We assessed by RT-qPCR whether the insertion of the transgene had any impact on the expression of the endogenous C1ql1 gene. At P14 in extracts from heterozygous animals, C1ql1 mRNA levels were decreased by 50–60% in all regions tested, compared to WT animals (Fig. 3A), showing that the insertion of the ires-cre CDS cassette in the 3’UTR of C1ql1 interferes with the proper expression of the C1ql1 allele. C1ql1 expression is thus expected to be reduced drastically in C1ql1cre/cre homozygotes to levels that probably phenocopy the C1ql1 knockdown phenotype4 (data not shown). To test whether the level of decrease in C1ql1 mRNA detected in C1ql1cre/WT heterozygotes leads to functional consequences, we performed morphological and functional analysis of CF/PC synapses comparing C1ql1WT/WT and C1ql1cre/WT heterozygotes. Morphological analyses were performed in two lobules of the cerebellum: lobules VI and IX, at P30 when the olivo-cerebellar circuit is established10, using immunofluorescence against vesicular glutamate transporter 2 (VGLUT2), to label the presynaptic boutons of CFs, and against calbindin (CaBP) to label PCs (Fig. 3B, left panel). Quantification showed no major difference in the thickness of the molecular layer of the cerebellar cortex and of the extension of CF innervation on the dendritic tree of PCs (Fig. 3B, right panel). While loss of function of C1ql1 at a level > 90% leads to decreased density of VGLUT2 clusters and decreased transmission4,6, our quantification did not detect any differences in the volume and density of VGLUT2 labelled presynaptic boutons between C1ql1WT/WT and C1ql1cre/WT heterozygotes (Fig. 3B, right panel). Patch-clamp recordings of CF/PC transmission during the fourth postnatal week showed similar amplitude of EPSCs in slices from C1ql1WT/WT and C1ql1cre/WT heterozygote mice (Fig. 3C). Altogether, the expression of cre and concomitant decrease of C1ql1 expression in C1ql1cre/WT heterozygous animals do not impact morphological or electrophysiological properties of CF/PC synapses. The C1ql1cre mouse line can thus be used to manipulate the olivo-cerebellar circuit when utilized as heterozygote without detrimental effect on normal development and function of the circuit.
No effect of cre transgene knockin on the morphology and transmission of climbing fiber/Purkinje cell synapses in C1ql1cre heterozygotes. (A) The expression of cre and C1ql1 mRNAs was assessed using quantitative RT-PCR on RNA extracts from four different regions of C1ql1WT/WT (WT) or C1ql1cre/WT (C1ql1cre) mouse brains, fourteen days after birth (P14). The cre recombinase expression was detected only in C1ql1cre mice, in the cerebellum, the brainstem, the hippocampus and the cortex. The expression of C1ql1 was decreased in these regions of the brain at the same age compared to WT mice. Data are represented as mean ± SEM, unpaired student t test with Welsh’s correction, n = 5–6 animals per genotype. (B) Left panel: CF presynaptic boutons and PCs and their dendritic tree were revealed by immunostaining for VGLUT2 (green) and calbindin (CaBP; magenta) respectively, in parasagittal cerebellar sections from P30 WT and C1ql1cre mice. Images were acquired using a spinning disk confocal microscope. Scale bar = 40 µm. Right panel: analyses of the morphology of the cerebellar cortex and connectivity of CFs were performed on cerebellar lobules VI and IX. The thickness of the molecular layer, the extension of the CF synapses on the PC dendritic tree, the volume and density of CFs presynaptic boutons on PCs were not modified in lobules VI and IX of C1ql1cre heterozygous animals compared to WT. Molecular layer (ML) and CF synapse extension (CF) are indicated on the merge images. Data are represented as mean ± SEM, unpaired student t test with Welsh’s correction, n = 4 animals per genotype. Scale bars: 40 µm. (C) CF/PC transmission was assessed in WT and heterozygous C1ql1cre mice using patch-clamp recordings of the mean amplitude of the CF-evoked currents in PCs. This analysis revealed no difference between the two genotypes. Data are represented as mean ± SEM, Mann Whitney test, n = 11–12 cells per genotype.
Targeting different cell types in the olivo-cerebellar network using the C1ql1 cre mouse line
The C1ql1cre mouse line is expected to drive the expression of the Cre recombinase in one of the excitatory afferents of cerebellar PCs, the IONs, and in the precursors of the other excitatory afferent, the GCs. This targeting was indeed obtained using R26cas9,-GFP (Fig. 1) and R26R-EYFP (Fig. 2) reporter lines. The designer receptors exclusively activated by designer drugs (DREADD) are genetic tools that are widely used to probe neural circuits in the mouse brain. The hM4Di DREADD allows chemogenetic inhibition of activity in neurons that express it specifically when activated by the synthetic ligand Clozapine-N-Oxide11. In C1ql1cre/WT/R26hM4Di,-mCitrine/WT heterozygous mice, the hM4Di DREADD and soluble mCitrine are co-expressed in cells following the excision by the Cre of the Stop cassette preceding the hM4Di-P2A-mCitrine sequence12. Immunofluorescence labeling of brain sagittal sections at P30 using an anti-GFP antibody revealed expression of mCitrine in the inferior olive in the brainstem, with clear labeling of CFs in this region (Fig. 2B). mCitrine-positive GCs were also observed in the internal granular layer of the cerebellum (Fig. 2B). Of note, we observed in some instances mCitrine expression in Bergmann glia in the cerebellum when breeding a R26hM4Di,-mCitrine/WT male with C1ql1cre/cre females but not with the reverse cross (data not shown). This type of parental sex-related differences was not observed when breeding to the R26cas9,-GFP reporter line, suggesting differences linked to the reporter mouse lines.
To specifically target the IONs using C1ql1cre/WT heterozygous mice, we developed an intersectional strategy. A retrograde AAV driving the expression of a Cre-dependent VAMP2-GFP construct under the CamKII promoter was injected in the cerebellum of P0 mouse pups or P27 juvenile mice. VAMP2 is a vesicular SNARE abundantly present at presynaptic sites13. Thus, expression of VAMP2-GFP is expected to label presynaptic boutons made by IONs on their target neurons. As CaMKII is expressed in IONs but not in GCs, the conditional expression of VAMP2-GFP was specific to the inferior olive, both at P27 and P44 after P0 and P27 injections, respectively (Fig. 4A). While the breeding of C1ql1cre with, for example, the reporter mouse line R26cas9,-GFP led to labelling of the majority of ION population (Fig. 1D, a and a′′), only subpopulations of IONs are targeted using the intersectional strategy. Using immunofluorescent labeling of brain sagittal sections at P27 (Fig. 4B) or P44 (data not shown), with an anti-GFP antibody, GFP-positive CFs were observed in different lobules in the cerebellum, in particular in lobules V to X in the vermis or paravermis (Fig. 4B, top panel). At higher magnification, GFP-positive CFs were visualized along the proximal dendrites of PCs (Fig. 4B, bottom panel). No GFP-positive GCs were observed, showing the high specificity of expression in one of the two excitatory afferents of the PCs. Altogether, our results show that the C1ql1cre mouse line can be used to either target both PC excitatory inputs or only one of them, depending on the strategy that is used.
Specific targeting of inferior olivary neurons using an intersectional strategy. (A) Schematic illustration of the strategy to specifically express the mouse GFP-tagged vesicle-associated membrane protein 2 (mVAMP2) in IONs. P0 or P27 stereotaxic injections of retrograde rAAVs expressing mVAMP2-GFP, under the CamKII promoter and in a Cre-dependent manner, were performed in the cerebellum of C1ql1cre mice. Analyses were done at P27 for P0 injections or P44 for P27 injections, in parasagittal cerebellar sections. Subpopulations of IONs were labelled after anti-GFP immunostaining. Inferior Olive (IO) is indicated by dotted lines. Scale bars = 200 µm. (B) Top panel: Vermal (left) and paravermal (right) sections from C1ql1cre/WT P27 mouse injected at P0 and immunostained for GFP revealed VAMP2-GFP expressing CFs in different lobules of the cerebellum. Images were acquired using a spinning disk confocal microscope. Lobules are numbered with roman numerals. Scale bars = 500 µm. Bottom panel: CFs of transduced IONs and PCs from P27 C1ql1cre/WT mouse injected at P0 were immunostained for GFP (green) and calbindin (CaBP) (magenta), respectively. Molecular Layer (ML) and Purkinje cell layer (PCL) are indicated on the merge image. Scale bars = 20 µm.
Discussion
C1QL1 is a secreted protein involved in synapse formation and maintenance between inferior olivary neurons (IONs) and Purkinje cells (PCs) in the olivo-cerebellar circuit4,6. Since the expression of C1ql1 was identified in IONs and in the external granular layer during development, we developed a C1ql1cre mouse model by inserting an ires-cre cassette after the stop codon of C1ql1, to enable the genetic manipulation of the two sources of excitatory inputs to PCs, IONs and GCs, in vivo. Using RT-qPCR, the expression of cre is detected in the cerebellum, brainstem, hippocampus and cortex. The specificity and capacity of the C1ql1 driver was characterized by crossing the C1ql1cre mouse line with three Cre-dependent reporter mouse models: R26cas9,-GFP, R26R-EYFP and R26hM4Di,-mCitrine. Expression of the reporter proteins is observed not only in IONs and GCs, but also in some other nuclei in the brainstem, midbrain and olfactory bulb, as well as in other organs than the brain such as the adrenal gland and colon. To specifically target IONs during postnatal development and adulthood, an intersectional strategy was developed via stereotaxic injection of a retrograde rAAV2 driving expression of a Cre-dependent reporter gene in the cortex of the cerebellum at P0 or P27, resulting in the transduction of numerous IONs and CFs 2–4 weeks after the injection. In general, the C1ql1cre mouse model is a powerful genetic tool to target and manipulate C1ql1-expressing cells in the brain, in particular IONs in the brainstem, and in other organs such as the adrenal gland.
Using the Cre-dependent reporter mouse models to map the location of C1ql1-expressing cells, we show the localization of the reporter gene in several nuclei in different regions of the brain. These nuclei are involved in different functions (e.g. olfaction, arousal and consciousness), but some of them found in the brainstem, cerebellum, pons and midbrain, such as inferior olive, red nucleus, substantia nigra pars reticulata and GCs, are motor-related structures. Several studies show the expression of C1ql1 outside of the brain, in particular in adipose tissue14, the adrenal glands, colon (https://www.ncbi.nlm.nih.gov/gene/23829), and ovaries15. Our results confirm the activity of Cre recombinase in several of these organs (the adrenal gland and colon) in the C1ql1cre mouse line.
The function of C1QL1 in ovarian folliculogenesis has been investigated in mice using a full knockout model, showing its anti-apoptotic effect on granulosa cell apoptosis15. Loss of function of C1QL1 using a C1ql1 constitutive knockout mouse model results in abnormal hair cell innervation in the cochlea and loss of audition16. In addition to physiological conditions, several studies suggest that C1QL1 might be involved in some malignancies. C1QL1 is upregulated in colorectal cancer, one of the most common malignant cancers17, differentiated thyroid carcinoma18, as well as lung adenocarcinoma19. RNA-seq analyses have shown that C1ql1 expression is higher in glioblastoma compared to the normal brain17. All these studies show the diverse functions of C1QL1 in health and disease in different systems of the body. Thus, the C1ql1cre mouse is a potentially useful model to study the role of C1ql1-expressing cells in different physiological and pathological conditions.
Methods and materials
All the methods were performed in accordance with the relevant guidelines and regulations.
Animals
All mice were kept under a 12 h light: 12 h dark cycle with water and food supplied ad libitum, in the authorized animal facilities of CIRB-College de France approved by the veterinary services (D-75-05-12). All animal protocols were approved by the Comité Régional d’Ethique en Expérimentation Animale (#2001) and all the methods were performed accordingly, and in accordance with the Animals Research: Reporting of In Vivo Experiments (ARRIVE) guidelines and regulations. C1ql1cre mouse model was generated and provided by genOway. The Cre-dependent reporter mouse lines, R26cas9-GFP (B6J.129(B6N)-Gt (ROSA)26Sortm1(CAG-cas9*,-EGFP)Fezh/J, strain #026175)8, R26R-EYFP (B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J, strain #:006148)9 and R26hM4Di,-mCitrine (B6.129-Gt(ROSA)26Sortm1(CAG-CHRM4*,-mCitrine)Ute/J, strain #026219)12 were obtained from The Jackson Laboratory. All the lines are maintained on the C57BL/6J background. In all experiments, littermate mice from both sexes were used as controls for the analysis.
Viral particles and Stereotaxic injections
Injections of rAAV particles were performed in the cerebellar vermis of ice-anesthetized P0 C1ql1cre/WT heterozygous mice or isoflurane-anesthetized P27 C1ql1cre/WT heterozygous mice, at 1 mm depth from the skull and 3.2 mm relative to Bregma or at 1.5 mm depth from drilled skull and 2.7 mm from Lambda respectively, to target Purkinje cell layer. rAAV2-retrograde serotype was used to target IONs. 0.25 µl of AAV was injected per animal using pulled calibrated pipets. Retrograde rAAV-CamKII-DIO-mVAMP2/eGFP-WPRE viral particles allowing the Cre-dependent expression of mVAMP2-GFP under the control of the CamKII promoter were produced by Vector Biolabs: # AAV-275950 (RefSeq # BC055105), with a titer of 3.2 × 1013 GC/mL.
Generation of C1ql1-ires-cre (C1ql1 cre) knockin mouse
C1ql1cre mice were generated using homologous recombination (genOway). A targeting vector containing Neomycin positive-selection cassette flanked with two FRT sites, and exon 2 of C1ql1 gene where an ires-cre cassette was inserted downstream of the stop codon in this exon, was electroporated into ES cells. The homologous recombination in the targeted ES cell clones was validated by PCR and DNA sequencing. The recombined ES cells with the correct sequence were injected into blastocysts. These blastocysts were then implanted in pseudo-pregnant females. Chimerism rate was then assessed in the progeny by coat color markers comparison. Highly chimeric males (chimerism rate above 50%) were generated and crossed with C57BL/6J flippase deleter female mice to excise the Neomycin selection cassette. Sixteen heterozygous mice were screened by PCR followed by sequencing to ensure germline transmission.
PCR genotyping is carried out using two primers (forward primer: 5′-GCCCAGATGTATTCTGCCCTAGAATCC-3′; reverse primer: 5′-ATTGCACTGGCCCGCACCTAAG-3′) to detect the WT (269 bp) and the targeted knockin alleles (342 bp) (Fig. 1B). Transgenic founders were backcrossed and maintained on the C57Bl/6J background.
Characterization of C1ql1 cre knockin mouse
Gene expression analysis
The cerebellum, brainstem, hippocampus and a piece of cortex were extracted from C1ql1WT/WT or C1ql1cre/WT P14 mice and frozen immediately in liquid nitrogen, and stored at -80C°. Using the Qiagen RNeasy mini kit (Qiagen, Venlo, Netherlands, #74104), the total RNA was purified according to manufacturer’s instruction. cDNA was synthetized using 100 ng of total RNA and SuperScript™ VILO™ cDNA Synthesis Kit (Life Technologies, California, USA, #11754050) according to manufacturer’s instruction. Quantitative PCR on cDNA samples was performed using FAM-labeled C1ql1 (Taqman Gene Expression Assay, ThermoFisher, Assay ID: Mm00657289_m1) or cre (Taqman Gene Expression Assay, ThermoFisher, Assay ID: Mr00635245_cn) probes and VIC-labeled Rpl13a probe as a reference (Taqman Gene Expression Assay, ThermoFisher, Assay ID: Mm01612986-gH) and the TaqMan Universal Master Mix II, with UNG (Applied Biosystems, #4440038) according to manufacturer’s instructions. Bio-Rad CFX Manager was used for data analysis. The relative expression of C1ql1 to Rpl13a or cre to Rpl13a was calculated based on the following formula: Relative expression = (2(Cq ofC1ql1orcre – Cq ofRpl13a)) × 100.
Immunofluorescence
Mice tissues were fixed using intracardiac perfusion of 4% PFA in PBS solution. Brains were extracted, post-fixed with the same solution at 4 °C for 2–4 h, and transferred to 30% sucrose/PBS for 48 h at 4 °C for cryoprotection. 30 µm-thick sections were obtained using a freezing microtome, and kept in 0.02% NaN3 in PBS solution at 4 °C until use.
To perform immunolabeling, sections were incubated in blocking buffer (4% donkey serum and 1% Triton X-100 in PBS solution) for 30 min to 1 h at room temperature, followed by incubation with primary antibodies in 1% donkey serum and 1% Triton X-100 in PBS for overnight at 4 °C with agitation. Primary antibodies were: GFP (1:1000, chicken, ab13970, abcam or 1:2000, rabbit, ab6556, abcam), CaBP (1:2000, rabbit, CB38, Swant or 1:2000, mouse, #300, Swant) and VGLUT2 (1:7000, guinea pig, AB2251, Millipore). The slices were washed 3 times in 1% Triton X-100 in PBS for 5–10 min, followed by incubation with secondary antibodies (Alexa Fluor 488-, 568- and 647-labeled donkey/goat anti-mouse, rat, rabbit, or chicken IgGs (H + L); 1:1000, Invitrogen or Life Technologies) in 1% Triton X-100 in PBS for 1–2 h at room temperature. Then, sections were washed 3 times in 1% Triton X-100 in PBS for 5–10 min, and incubated for another 10 min at room temperature with Hoechst 33342 (0.2 mg/mL, Sigma, Gothenburg, Sweden, cat#H6024) in 0.4% Triton X-100 in PBS. Sections were mounted using ProLong Gold Antifade Reagent (Invitrogen, cat#P36930).
Image acquisition and analysis
The mosaic images for global brain morphology (Fig. 1D and Supp Fig. 2) were obtained using a Zeiss Axiozoom V16 macroscope, equipped with a digital camera (AxioCam HRm) using a 160× (pixel size: 0.4037 µm), and reconstructed using the Zeiss Zen software. Images for VGLUT2 quantifications (Fig. 3B) were acquired using a Zeiss spinning-disk confocal CSU-W1 microscope with 63× oil objective (Z-plane step size: 0.19 µm). Images for GFP analyses were acquired using the same spinning-disk confocal microscope with 63× or 25× oil objectives (single plane, Figs. 2 and 4). The mosaic images for GFP visualization were reconstructed using the stitching program from metamorph software (Fig. 4B top panel).
VGLUT2 quantification (cluster density and volume) were performed as described previously20. All the images were normalized using the quantile-based normalization plugin in Fiji. The intensity distribution of the images has been normalized using 256 quantiles for each staining. The synaptic boutons were extracted from the background using the 3D Weka Segmentation plugin (https://imagej.net/Trainable_Weka_Segmentation) after manual selection of signal and background samples. The Fiji built-in plugin 3D object counter was then used in order to count and measure every object (cluster of VGLUT2 positive signal). CF territory and ML thickness were measured manually using Fiji. All the steps were performed in blind condition.
Western blot analysis
The cerebellum, liver, adrenal gland, lung and colon were obtained from isoflurane-anesthetized adult male mice. The tissues were homogenized in lysis buffer (50mM Tris HCl (pH = 8), 1% Triton X100 (Tx; Sigma, Cat#X100) supplemented with a protease inhibitor cocktail (1:100; Sigma, Cat#P8340) and MG132 (100 µM; Sigma, Cat#C2211)). Homogenates were sonicated three times for 5 s, incubated at 4 °C for 30 min with rotation, and finally centrifuged at 14,000 r.p.m. at 4 °C for 10 min. Supernatants were denatured at 65 °C in 2X sample buffer (Sigma, Cat#S3401) prior to gel electrophoresis and western blot analysis for detection of EYFP and β-ACTIN. Immunoblotting was executed as previously described21 with the following primary antibodies: GFP/YFP (1:1000, rabbit, ab6556, abcam) and β-ACTIN conjugated to HRP (1:25,000, mouse, ab49900, abcam). An HRP-linked anti-rabbit antibody was used to target anti-GFP (1:10,000, donkey, GENA934, Sigma). HRP-conjugated antibodies were revealed using ECL Western Blotting substrate (Thermo Fisher Scientific, Cat#32209) chemiluminescent solutions and images acquired on a Fusion FX7 system (Vilber Lourmat, Île-de-France, France).
Electrophysiology
Acute parasagittal cerebellar slices were obtained from C1ql1cre mice from P25 to P30. 200 µm-thick slices were cut at room temperature with a Campden Ci 7000 smz microtome in (in mM): Sucrose 120, NaCl 60, KCl 2.5, D(+)Glucose 25, NaHCO3 25, NaH2PO4 1.25, CaCl2 0.1, MgCl2 3, ascorbic acid 0.4, myo-inositol 3, NaPyruvate 2, pH = 7.3–7.4. Slices were then transferred and allowed to recover for one hour at room temperature in the following solution (in mM): NaCl 125, KCl 2.5, D(+)Glucose 25, NaHCO3 25, NaH2PO4 1.25, CaCl2 2, MgCl2 1, ascorbic acid 0.4, myo-inositol 3, NaPyruvate 2, pH = 7.3–7.4, oxygenated. This solution was used to fill the stimulation pipette and, complemented with picrotoxin (100 µM), was used as external solution. Borosilicate glass pipettes with 2–5 MΩ resistance were used for recordings and filled with the following internal solution (in mM): CsCl2 155, Hepes 10, EDTA 5, QX314 5, pH = 7.35 adjusted with CsOH. Responses to CF stimulation were recorded at a holding membrane potential of − 10 mV in Purkinje cells of lobule VI using a MultiClamp 700B amplifier (Molecular Devices, CA) and acquired using the freeware WinWCP written by John Dempster (https://pureportal.strath.ac.uk/en/datasets/strathclyde-electrophysiology-software-winwcp-winedr). Series resistance was compensated by 80–90% and cells were discarded if significant changes were detected. CF-mediated responses were identified by the typical all-or-none response and strong depression displayed by the second response elicited during paired-pulse stimulations (20 Hz). Electrophysiological data were analyzed using the software Clampfit 10.7 (Molecular Devices).
Statistical analysis
All the data were analyzed and represented using GraphPad Prism. They are represented as mean ± SEM. The student’s t-test was used to measure the differences between two groups when they had normal distribution. The non-parametric Mann–Whitney test was used when the two groups didn’t have normal distribution.
Data availability
All data are available in the main text.
References
Kishore, U. et al. C1q and tumor necrosis factor superfamily: Modularity and versatility. Trends Immunol. 25(10), 551–561 (2004).
Ghai, R. et al. C1q and its growing family. Immunobiology 212(4), 253–266 (2007).
Iijima, T., Miura, E., Watanabe, M. & Yuzaki, M. Distinct expression of C1q-like family mRNAs in mouse brain and biochemical characterization of their encoded proteins. Eur. J. Neurosci. 31(9), 1606–1615 (2010).
Sigoillot, S. M. et al. The secreted protein C1QL1 and its receptor BAI3 control the synaptic connectivity of excitatory inputs converging on cerebellar Purkinje cells. Cell Rep. 10(5), 820–832 (2015).
Marc F. B., David C. M., Thomas C. S. The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins. PNAS https://doi.org/10.1073/pnas.1019577108 (2011).
Kakegawa, W. et al. Anterograde C1ql1 signaling is required in order to determine and maintain a single-winner climbing fiber in the mouse cerebellum. Neuron 85(2), 316–329 (2015).
Lanoue, V. et al. The adhesion-GPCR BAI3, a gene linked to psychiatric disorders, regulates dendrite morphogenesis in neurons. Mol. Psychiatry. 18(8), 943–950 (2013).
Platt, R. J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159(2), 440–455 (2014).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).
Watanabe, M. & Kano, M. Climbing fiber synapse elimination in cerebellar Purkinje cells. Eur. J. Neurosci. 34(10), 1697–1710 (2011).
Armbruster, B. N., Li, X., Pausch, M. H., Herlitze, S. & Roth, B. L. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. 104(12), 5163–5168 (2007).
Zhu, H. et al. Cre dependent DREADD (Designer Receptors Exclusively Activated by Designer Drugs) mice. Genesis N Y N 54(8), 439–46 (2016).
Schoch, S. et al. SNARE function analyzed in synaptobrevin/VAMP knockout mice. Science 294(5544), 1117–1122 (2001).
Guan, H., Shi, T., Liu, M., Wang, X. & Guo, F. C1QL1/CTRP14 is largely dispensable for atherosclerosis formation in apolipoprotein-E-deficient mice. J. Cardiovasc. Dev. Dis. 9(10), 341 (2022).
Lu, X. et al. Deficiency of C1QL1 reduced murine ovarian follicle reserve through intraovarian and endocrine control. Endocrinology 163(6), bqac048 (2022).
Qi, Y. et al. Deletion of C1ql1 Causes hearing loss and abnormal auditory nerve fibers in the mouse cochlea. Front. Cell. Neurosci. 15, 713651. https://doi.org/10.3389/fncel.2021.713651 (2021).
Qiu, X. et al. Profiles of differentially expressed genes and overexpression of NEBL indicates a positive prognosis in patients with colorectal cancer. Mol. Med. Rep. 17(2), 3028–3034 (2018).
Celestino, R. et al. CRABP1, C1QL1 and LCN2 are biomarkers of differentiated thyroid carcinoma, and predict extrathyroidal extension. BMC Cancer 18, 68 (2018).
Gao, Y. J., Chen, F. & Zhang, L. J. C1q-like 1 is frequently up-regulated in lung adenocarcinoma and contributes to the proliferation and invasion of tumor cells. J. Chemother. 33(7), 476–485 (2021).
Veleanu, M. et al. Modified climbing fiber/Purkinje cell synaptic connectivity in the cerebellum of the neonatal phencyclidine model of schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 119(21), e2122544119 (2022).
González-Calvo, I. et al. Sushi domain-containing protein 4 controls synaptic plasticity and motor learning. eLife 10, e65712 (2021).
Acknowledgements
We would like to thank Yves Dupraz for the development of tools for stereotaxic injections in neonates, the Venance team for sharing the R26R-EYFP mouse line, and the personnel from the CIRB animal and imaging facilities.
Funding
This work was supported by funding from: European Research Council ERC consolidator grant SynID 724601 (FS), Q-life ANR-17-CONV-0005 (FS), ANR-10-LABX-54 MEMO LIFE (FS), INCA PEDIAHR21-014 (FS), Sorbonne Université ED158 (MAP), Collège de France (MAP), Ecole des neurosciences de Paris Ile-de-France (SM), La Ligue Nationale Contre le Cancer (SM).
Author information
Authors and Affiliations
Contributions
Conceptualization: F.S. Methodology: F.S., S.M., S.M.S., M.A.P. Investigation: S.M., S.M.S., F.S., M.A.P. Visualization: S.M., S.M.S., F.S. Funding acquisition: F.S. Project administration: F.S. Supervision: F.S. and S.M.S. Writing—original draft: S.M. and S.M.S. Writing—review and editing: S.M.S., S.M., F.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Moghimyfiroozabad, S., Paul, M.A., Sigoillot, S.M. et al. Mapping and targeting of C1ql1-expressing cells in the mouse. Sci Rep 13, 17563 (2023). https://doi.org/10.1038/s41598-023-42924-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-42924-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.