Abstract
Bacterial colonization is mediated by fimbriae, which are thin hair-like appendages dispersed from the bacterial surface. The aggregative adherence fimbriae from enteroaggregative E. coli are secreted through the outer membrane and consist of polymerized minor and major pilin subunits. Currently, the understanding of the structural morphology and the role of the minor pilin subunit in the polymerized fimbriae are limited. In this study we use small-angle X-ray scattering to reveal the structural morphology of purified fimbriae in solution. We show that the aggregative fimbriae are compact arrangements of subunit proteins Agg5A + Agg3B which are assembled pairwise on a flexible string rather than extended in relatively straight filaments. Absence of the minor subunit leads to less compact fimbriae, but did not affect the length. The study provides novel insights into the structural morphology and assembly of the aggregative adherence fimbriae. Our study suggests that the minor subunit is not located at the tip of the fimbriae as previously speculated but has a higher importance for the assembled fimbriae by affecting the global structure.
Similar content being viewed by others
Introduction
The initial phase of bacterial colonization depends on the ability to attach to a surface. The attachment of the bacteria is mediated by the fimbriae, which are located on the surface of the bacteria. Enteroaggregative E. coli (EAEC) is an emerging pathogen, which has been recognized as a major cause of diarrhea worldwide1,2,3,4. The pathogenesis of EAEC is determined by its ability to colonize the intestinal tract mediated by the aggregative adherence fimbriae (AAFs)5,6,7,8,9. Aggregative adherence fimbriae belong to the chaperone-usher (CU) family of adhesins, a highly divergent pathway common to many gram-negative bacteria. The CU pathway is used for assembly and secretion of the fimbriae, and all members harbor a periplasmic chaperone, an outer membrane usher protein, and at least one fimbrial subunit10,11,12,13. Currently, five different AAFs variants have been genetically characterized, in which all fimbrial clusters encodes 4 genes that confer a “stacked brick” adhesion pattern between the bacterial cells and the eukaryotic cells6,7,9,14,15. Previous transmission electron microscopy (TEM) pictures of AAF/I, AAF/II and AAF/III show that the AAFs are long, flexible filaments with estimated diameters of 3–5 nm6,14.
The AAFs are composed of two pilin subunits, for AAF/V the major subunit is Agg5A and the minor subunit Agg3B9. The two pilin subunits harbor an immunoglobulin‐like fold which lacks the final antiparallel G‐strand, resulting in an unstructured N‐terminal hydrophobic groove16,17. When secreted into the periplasm, the pilin domains form a complex with the chaperone (donor strand complementation), thereby stabilizing the subunits and preventing them from auto‐assembly. At the usher, each pilin subunit is inserted into a neighboring pilin subunit, replacing the G-strand from the chaperone with G-strands from neighboring pilin subunit (donor strand exchange), resulting in a polymerized fimbriae which is secreted through the outer membrane10,11,12,17. A study done by Andrea Berry et al., suggests the minor subunit forms the tip of the fimbriae, by accepting the donor strand (DS) from the terminal major pilin subunit17. Deletion of the minor pilin subunit has shown not to have an effect on adhesion but instead, the minor pilin subunit has been shown to be the contributor to the release of proinflammatory cytokines from mammalian cells18,19,20.
The monomeric structure of Agg5A and other AAF variants have been resolved by NMR, but these are monomers rather than long polymerized fimbriae16,17. In order to understand the interactions between the EAEC AAF fimbriae and the host receptors, it is crucial to unravel the structural architecture of the fimbriae and the role of the minor pilin subunit in the fimbrial polymerization.
In this study we use small-angle X-ray scattering (SAXS) to characterize the polymerized fimbriae in solution from a strain harboring the full fimbriae cluster expressed from a constitutive promoter as well as a fimbriae mutant (ΔAgg3B) where the Agg3B subunit has been deleted. ΔAgg3B was still able to adhere to plastic surface and to form biofilms, suggesting that ΔAgg3B fimbriae are functional. The SAXS data suggest that the AAF/V are flexible sticky chains which fold together in solution and that the presence of Agg3B affects the global conformations of the fimbriae. Studies of long polymerized proteins in solution are limited and SAXS based findings contributes to new knowledge on the structural assembly of the AAFs.
Results
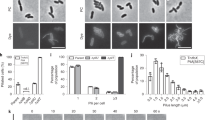
The full fimbrial cluster (AAF/V) was cloned as previously described into the plasmid vector pACYC184 and expressed in the non-fimbriated E. coli strain HB1019. To test the involvement of the minor pilin subunit in the pilus polymerization, the minor pilin subunit was deleted from the fimbrial cluster (ΔAgg3B) and tested for its functionality by biofilm formation in the microtiter plate assay21. The bacteria were incubated in microtiter plates for 24 h at 37 °C and then the biofilm was visualized by crystal-violet staining. The results showed that there was no statistical difference between the full fimbriae cluster and ΔAgg3B (P = 0.12), suggesting that the minor pilin subunit of AAF/V is not attributable for the adhesive phenotype of AAF/V and that ΔAgg3B expresses functional fimbriae (Fig. 1).
Deletion of minor pilin subunit (ΔAgg3B) does not affect adhesion and biofilm formation. The full fimbriae cluster and ΔAgg3B was inserted into E. coli HB101 and adhesion to plastic surface was performed. The results are represented as the means ± standard errors of the means for eight replicates and represent one of three independent experiments performed with similar results. ****P < 0.0001; ***P < 0.001.
Purification of whole fimbriae, dsc-Agg5A and dsc-Agg5AB
Whole polymerized fimbriae from the wildtype and ΔAgg3B were purified as large soluble aggregates that eluted in a broad peak during size exclusion chromatography. Since monomeric subunits lack the antiparallel G-strand which makes them uncapable of folding without a neighboring subunit, donor strand complementation (dsc) of the fimbrial subunits was necessary17. To elucidate the role of the minor pilin subunit (Agg3B), a chimeric construct containing both the major pilin subunit (Agg5A) and minor pilin subunit (Agg3B) was created. Mature Agg5B was fused to the N-terminal of the mature Agg5A with an intervening 10-residue linker (DNKQNATAVA), resulting in the fused construct dsc-Agg5AB.The N-terminal donor strand of Agg5A was fused to the C-terminus of Agg5A with a 4-residue linker (DNKQ) (Fig. 2).
Both proteins were expressed in the E. coli cytoplasm and refolded as previously described16,17. During purification of the subunits, two bands eluted during gel filtration, corresponding to a monomer and dimer. The monomer fractions were used for the SAXS.
Structural characterization of fimbriae
Two fimbriae types were subjected to SAXS analysis, together with engineered subunit samples (Table 1)17. Scattering curves obtained from both the constructs and fimbriae samples are plotted and the individual dilution series are superimposed (Fig. 3).
The radius of gyration (Rg) of a particle is a well-defined measure of the overall size of a single homogeneous particle of any shape or form. Guinier analysis (Supplementary data) gave estimates of the gyration radii for all four sample types (Table 1).
At low q, the curve is leveled off, for the subunit samples, allowing us to determine an estimation of the molecular weight and aggregation number of these two samples (Supplementary material). Though, molecular weight determination using Guinier approximation of SAXS data should allow for a 10% inaccuracy, it clearly illustrates how dsc-Agg5A dimerizes as the dominant species in solution, while the dsc-Agg5AB aggregates into a larger structure with an average size close to 7 subunits. As the dilution curves overlay superimpose perfectly, the assembly is not concentration dependent, indicating a specific or irreversible interaction between dsc-agg5AB. The dimerization of dsc-Agg5A is further corroborated by fitting of the SAXS curves to other filament model molecules (Supplementary Fig. S2, Supplementary Table S1), as well as the NMR structure of dsc-Agg5A (Supplementary Fig. S1), leading up to an ab initio simulation (Supplementary Fig. S3A). When modeling the dimeric structure of dsc-Agg5B, we relied on ab initio techniques guided by the monomer NMR structure of dsc-Agg5A. Though the resolved structures for dsc-Agg5AB do change from prediction to prediction, they are consistently large and elongated (Supplementary Fig. S3B).
The purified fimbriae species aggregate in a slightly different manner, contrary to the subunit’s aggregation pattern. Radii of gyration are larger for the fimbriae Agg5A compared to the fimbriae Agg5A + Agg3B. The latter is shifted down relative the Agg5A fimbriae at the log–log plot (Supplementary Fig. S4). This drop in scattering intensity would normally indicate that the average molar mass of fimbriae Agg5A + Agg3B is lower than the Agg5A fimbriae. This can in principle be a result of structural differences, which then would be expected to disappear at high q, resulting in overlapping curve patterns. However, the fact that the curves align parallel at high q, indicate an error in the measured concentration, and indeed the nominal concentration in Table 1 is unusually high.
By using SAXS to analyze it, it is possible to characterize the arrangement of the subunits in the filament. However, this does rely on validated models which can be made using idealized structures such as rods, spheres, disks, or random coils, and the power of Monte Carlo simulations. SAXS data can be directly compared to idealized arrangements such as rods, discs, spheres, or random coils, which give rise to characteristic slopes in plots of log(I) vs log(q). Fimbriae might be expected to form extended rod-like structures in solution, which would give a slope of − 1 in such a plot, however Supplementary Fig. S4 clearly shows that this is not the case. Instead, the overall slope is close to − 2, which would correspond to an ideal random coil22. We conclude that purified fimbriae in solution do not extend like rods, but form more compact arrangements.
Supplementary Figure S4 shows that the ideal coil model is not a perfect fit, as the curves show features other than the overall slope. Also, that model is not physically relevant, as different parts of the coil would be free to overlap. Therefore, an explicit model of chains of proteins was constructed, where linear chains of subunits are treated as hard spheres which interact with an attractive potential (Supplementary Fig. S5, Supplementary Table S2). The model can be sampled by Monte Carlo simulation (Fig. 4) and can be compared with experimental SAXS data as described in the Supporting Information.
Model fits to Agg5A and Agg5A + Agg3B fimbriae data. The insets show the aligned trajectories (blue dots) and arbitrarily chosen structures (orange chains). Molecular representations are made using VMD version 1.9.3.23.
The fitting parameters (Table 2), which also show the dimeric subunit structure used in the modelling, results in a rather flexible chain with ӨMax = π/2, and thus higher flexibilities were not considered. Table 2 also lists c/cnom, the ratio of the fitted concentration to the nominal value listed in Table 1.
Both the Agg5A and Agg5A + Agg3B fimbriae samples give very similar model parameters and arrange in linear assemblies of approximately 40 dimeric subunits, spaced 50 and 55 Å apart, respectively. The major difference between these fimbriae is the degree of stickiness, where the Agg5A + Agg3B fimbriae have a higher tendency to fold together onto themselves, expressed as a (β) higher sticking energy in the model (Supplementary Fig. S6). This is what gives both the clearer plateau at intermediate q and the smaller radius of gyration. The higher stickiness of fimbriae containing subunit Agg3B is consistent with the aggregation of the Agg5A/Agg3B complex discussed above. Similarly, to stickiness we can model stiffness (Θmax) on chain structure and predicted scattering. As the chain is made stiffer, its scattering pattern transitions from that of the flexible and self-avoiding coil to that of an ideal rod, with a slope of − 1 in the log–log plot, as expected (Supplementary Fig. S7).
Discussion
The AAFs are a divergent group which has been implicated in multiple phenotypes related to pathogenesis. AAFs have been linked to adherence to a variety of cell lines, ability to form biofilm on biotic and abiotic surfaces, adherence to erythrocytes and stimulation of the proinflammatory cytokine (IL-8)5,9,18,19,24,25,26. Currently, no specific receptor has been identified for AAF/V but a recent study suggests that AAF/II binds to heparan sulfate proteoglycans on human colonoids5.
In this study, we investigated the structural characteristics of AAF/V by SAXS. Adherence fimbriae are known to form bundles, and the fact that they fold together into sticky balls in solution is perhaps to be expected6,7. Fimbriae naturally sit packed and aligned together on the bacterial surface. It is likely that a propensity for these chains to associate is a necessity to be able to form the bundles, which are often seen in microscopic images. However, the results demonstrated that the different fimbriae types had the ability to assemble into well-defined flexible strings. Furthermore, as concentration variations did not affect the arrangement, unspecific aggregation resulting in miss folding could be excluded. Thus, the results are in support of a biological relevant arrangement of the subunits into longer flexible chains. The results corroborated previous TEM of the AAFs which has demonstrated presence of long flexible fibers dispersed from the bacterial surface. AAF/I and AAF/II have shown to be long and bundled (2–5 nm), whereas AAF/III was most commonly observed in individual filaments6,7.
Interpretation of the SAXS data requires use of models, which were generated using simple physical assumptions. The models rest on the key assumption that the known structures of the monomer subunits are intact and do not undergo unfolding as part of the dimerization step. This assumption is supported by structural characterization of other fimbriae types17.
We also sought to elucidate the role of the minor pilin subunit by constructing a ΔAgg3B mutant and investigate how this affected the polymerized fimbriae. Previous studies have shown that the minor pilin subunit from AAF/II is not attributable for the adhesive phenotype of AAF/II but instead related to induction of the proinflammatory cytokine IL-818,19. No difference was observed between the wildtype and ΔAgg3B in biofilm formation, confirming that ΔAgg3B mutant is not involved in the adhesion and confirming that the ΔAgg3B fimbriae are functional. The fact that the presence of the Agg3B subunit affects the global structure of the fimbriae suggests that it is present to a significant degree in the chains. This observation does not match the hypothesis that Agg3B is only present in one copy at the very tip of the fimbriae to terminate DSC17. Such a small presence could not alter the propensity of the entire chain to stick together.
Materials and methods
Dsc-Agg5A purification
The dsc-Agg5A was purified as previously described16. Briefly, the cells were grown in LB until OD600 reached 0.6, following induction by 1 mM IPTG, which was followed by overnight incubation at 37 °C before the cells were harvested by centrifugation. The cells were lyzed by sonication under denaturing conditions before being purified with Ni–NTA (Qiagen). The eluate was first dialyzed against 50 mM sodium acetate pH 5, 50 mM NaCl, 1 M urea, which was followed by a second dialysis against the same buffer but without urea. Agg5a was further purified by gel filtration using a Superdex 75 gel-filtration column (GE Healthcare).
Dsc-chimeric Agg5AB
A donor strand complemented construct of dsc-Agg5AB was ordered from Genscript. Agg3B was fused to the N-terminal of the mature Agg5A with an intervening 10-residue linker (DNKQNATAVA) (Fig. 2).
The N-terminal donor strand of Agg5A was fused to the C-terminus of Agg5A with a 4-residue linker (DNKQ). Dsc-Agg5AB was cloned into a pQE‐30 plasmid (Qiagen) containing a vector encoded N‐terminal His6 tag and purified according to the same method as described for dsc-Agg5A.
Construction of Agg5ACD clone
Whole purified fimbriae were obtained from HB101(pDKAAF5) expressing the whole AAF/V cluster as previously described9. To obtain a construct without the minor pilin subunit, an Agg5ACD mutant was generated for this study. Briefly, a reverse PCR was carried out on pDKAAF5 with the primers GCGCGCCTCGAGGTATAGTTTTGGGAAGATAACAGTAT and GCGCGCCTCGAGAGATTCCTTCTGCTATATGCATA to delete the B subunit. Amplified DNA was digested with the restriction enzyme xhoI and ligated. The correct deletion was verified by sanger sequencing with the primers GCTATAGATAACCCACTGTACAAG and GGTGTATCTGAGTG GATTGTCAGA.
Microtiter dish biofilm formation assay
Biofilm formation has been shown to correlate with AAF adhesion9,21. To test if ΔAgg3B was able to express functional fimbriae, the microtiter dish biofilm formation assay was performed as described previously9,21.
Purification of polymerized fimbriae
Purification of whole polymerized fimbriae were purified by the method by Izquierdo et al.26. The presence of AAF/V fimbriae was confirmed by SDS-page.
Small angle X-ray scattering
The SAXS experiments were carried out on the bioSAXS BM29 beamline at the European Synchrotron Radiation Facility (ESRF) in Grenoble, France, using a X-ray wave length of λ = 0.9919 Å and a sample-detector distance of d = 287.2 mm, providing a q-range of 0.00423–0.49 Å−1.
Scattered X-ray intensity is a function of the angle between the incoming beam and the scattering direction (2θ). This angle is most conveniently expressed in terms of the magnitude of the wave momentum transfer (q), which in terms of the wavelength (λ) and the half-angle (θ) is q = (4π/λ) × sin(θ). The quantity q carries units of inverse length, and is therefore directly comparable to the length scales present in the sample. All scattering data in this report are presented as angle-dependent intensities, I(q).
Measurements were performed at 10 °C and data were collected in 20 individual one-second exposures. The individual data frames were merged, scaled to absolute values using water as a reference and background subtracted using the ATSAS software27. Four samples were analyzed at different concentrations. Arbitrary constants of 6.2 × 10–2 and 2.5 × 10–2 cm2/g have been respectively subtracted from the A and A + B data to account for inaccuracies in the buffer subtraction. Dsc-Agg5A and dscAgg5AB, were analyzed individually, as well complete fimbriae assembled in the producer strains, either as a fimbria composed of purely subunit Agg5AA or a fimbriae species composed of a mixture of subunit Agg5A and Agg3B (Table 1).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files and the following https://doi.org/10.5281/zenodo.7404371.
References
Hebbelstrup Jensen, B., Olsen, K. E. P., Struve, C., Krogfelt, K. A. & Petersen, A. M. Epidemiology and clinical manifestations of enteroaggregative Escherichia coli. Clin. Microbiol. Rev. 27, 614–630 (2014).
Jenkins, C. Enteroaggregative Escherichia coli. 27–50 (2018). https://doi.org/10.1007/82_2018_105.
Estrada-Garcia, T., Perez-Martinez, I., Bernal-Reynaga, R. & Zaidi, M. B. Enteroaggregative Escherichia coli: A pathogen bridging the North and South. Curr. Trop. Med. Rep. https://doi.org/10.1007/s40475-014-0018-7 (2014).
Navarro-Garcia, F. & Elias, W. P. Autotransporters and virulence of enteroaggregative E. coli. Gut Microbes 2, 13–24 (2011).
Rajan, A. et al. Enteroaggregative E. coli adherence to human heparan sulfate proteoglycans drives segment and host specific responses to infection. PLoS Pathog. 16, e1008851 (2020).
Czeczulin, J. R. et al. Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect. Immun. 65, 4135–4145 (1997).
Nataro, J. P. et al. Aggregative adherence fimbria I expression in enteroaggregative Escherichia coli requires two unlinked plasmid regions. Infect. Immun. 61, 1126–1131 (1993).
Savarino, S. J., Fox, P., Deng, Y. & Nataro, J. P. Identification and characterization of a gene cluster mediating enteroaggregative Escherichia coli aggregative adherence fimbria I biogenesis. J. Bacteriol. 176, 4949–4957 (1994).
Jønsson, R. et al. Novel aggregative adherence fimbria variant of enteroaggregative Escherichia coli. Infect. Immun. 83, 1396–1405 (2015).
Busch, A. & Waksman, G. Chaperone–usher pathways: Diversity and pilus assembly mechanism. Philos. Trans. R. Soc. B Biol. Sci. 367, 1112–1122 (2012).
Nuccio, S.-P. & Bäumler, A. J. Evolution of the chaperone/usher assembly pathway: Fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71, 551–575 (2007).
Werneburg, G. T. & Thanassi, D. G. Pili assembled by the chaperone/usher pathway in Escherichia coli and Salmonella. EcoSal Plus 8, 10–1128 (2018).
Geibel, S. & Waksman, G. The molecular dissection of the chaperone–usher pathway. Biochim. Biophys. Acta BBA Mol. Cell Res. 1843, 1559–1567 (2014).
Bernier, C., Gounon, P. & le Bouguénec, C. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect. Immun. 70, 4302–4311 (2002).
Boisen, N., Struve, C., Scheutz, F., Krogfelt, K. A. & Nataro, J. P. New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect. Immun. 76, 3281–3292 (2008).
Jønsson, R. et al. Structural and functional studies of Escherichia coli aggregative adherence fimbriae (AAF/V) reveal a deficiency in extracellular matrix binding. Biochim. Biophys. Acta BBA Proteins Proteom. 1865, 304–311 (2017).
Berry, A. A. et al. Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli. PLoS Pathog. 10, e1004404 (2014).
Farfan, M. J., Inman, K. G. & Nataro, J. P. The major pilin subunit of the AAF/II fimbriae from enteroaggregative Escherichia coli mediates binding to extracellular matrix proteins. Infect. Immun. 76, 4378–4384 (2008).
Harrington, S. M., Strauman, M. C., Abe, C. M. & Nataro, J. P. Aggregative adherence fimbriae contribute to the inflammatory response of epithelial cells infected with enteroaggregative Escherichia coli. Cell Microbiol. 7, 1565–1578 (2005).
Elias, W. P., Czeczulin, J. R., Henderson, I. R., Trabulsi, L. R. & Nataro, J. P. Organization of biogenesis genes for aggregative adherence fimbria II defines a virulence gene cluster in enteroaggregative Escherichia coli. J. Bacteriol. 181, 1779–1785 (1999).
Boll, E. J. et al. Role of enteroaggregative Escherichia coli virulence factors in uropathogenesis. Infect. Immun. 81, 1164–1171 (2013).
Warren, B. E. X-ray Diffraction (Dover Publications, Inc., 1990).
Humphrey, W., Dalke, A. & Schulten, K. VMD—Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Boll, E. J. et al. The fimbriae of enteroaggregative Escherichia coli induce epithelial inflammation in vitro and in a human intestinal xenograft model. J. Infect. Dis. 206, 714–722 (2012).
Boll, E. J. et al. Enteroaggregative Escherichia coli adherence fimbriae drive inflammatory cell recruitment via interactions with epithelial MUC1. MBio 8, 10–1128 (2017).
Izquierdo, M. et al. Identification of cell surface-exposed proteins involved in the fimbria-mediated adherence of enteroaggregative Escherichia coli to intestinal cells. Infect. Immun. 82, 1719–1724 (2014).
Manalastas-Cantos, K. et al. ATSAS 3.0: Expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 54, 343–355 (2021).
Acknowledgements
We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities and we would like to thank the staff of beamline BM29 for providing assistance. The project was a part of NXUS, funded equally by the Capital region of Denmark and University of Copenhagen.
Author information
Authors and Affiliations
Contributions
R.J., A.B., S.R.M., G.V.J., N.S.G. contributed to the experimental data and analyses, L.A., S.M., K.A.K., and H.J. contributed in the study design, formulating the scientific questions as well as securing funding. All authors have read and agreed to the contented of the work presented.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jønsson, R., Björling, A., Midtgaard, S.R. et al. Aggregative adherence fimbriae form compact structures as seen by SAXS. Sci Rep 13, 16516 (2023). https://doi.org/10.1038/s41598-023-42079-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-42079-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.