Abstract
Obesity has become a common global problem. Some obese people can be metabolically healthy. Gene-environment interaction can be important in this context. This study aimed to assess the interaction between dietary fat quality indices and the Melanocortin 4 receptor (MC4R) gene in metabolically healthy and unhealthy overweight and obese women. This cross-sectional study was conducted on 279 women with overweight and obesity. The definition of metabolically healthy and unhealthy phenotypes was done according to Karelis criteria. Dietary assessment was done using a 147-item validated semi-quantitative food frequency questionnaire and dietary fat quality was assessed by cholesterol-saturated fat index (CSI) and the ratio of omega-6/omega-3 (N6/N3) essential fatty acids. MC4R was genotyped by polymerase chain reaction-restriction fragment length polymorphism technique. A generalized linear model was used to evaluate the interaction between dietary fat quality indices and the MC4R gene in both crude and adjusted models. Study subjects with higher ratio of N6/N3 had higher homeostatic model assessment for insulin resistance (HOMA IR) index (P = 0.03) and other variables showed no difference according to the tertile of CSI and N6/N3. Participants with the C allele of MC4R rs17782313 had lower height (P < 0.001) and higher HOMA index (P = 0.01). We found that the CC genotype of MC4R interacts with the N6/N3 ratio on the metabolically unhealthy phenotype in the crude model (β = 9.94, CI 2.49–17.39, P = 0.009) and even after adjustment for all confounders (β = 9.002, CI 1.15–16.85, P = 0.02, β = − 12.12, CI 2.79–21.46, P = 0.01). The data of this study can justify one inconsistency observed in society, regarding dietary recommendations about metabolic health status. Those with CC genotype, are more likely to have an unhealthy phenotype with an increase in N6/N3 as one fat quality indices than those who do not have CC genotype. We found the interaction of dietary fat quality indices such as N6/N3 and the MC4R gene in metabolically unhealthy overweight and obese women.
Similar content being viewed by others
Introduction
Obesity has become a very important health issue worldwide and its prevalence has increased dramatically, becoming an uncontrollable epidemic1,2. Complications of obesity, such as cardiovascular diseases, diabetes, and cancer can cause serious health problems for people and high costs to the health care system3,4. Obese people have different phenotypes with different metabolic risks5. A group of obese people who do not have metabolic complications is considered metabolically healthy obese (MHO) people6,7. Meanwhile, obese individuals with obesity-related metabolic complications are known as metabolically unhealthy obese (MUO)8. Obesity is a multifactorial phenomenon in which genetic and environmental factors (such as diet) and their interaction can affect its occurrence1. So far, many genes have been associated with the risk of obesity such as Iroquois homeobox protein 3 (IRX3) and retinitis pigmentosa GTPase regulator-interacting protein 1 like (RPGRIP1L)9. Therefore, gene-environment interaction can be considered one of the most important determinants of obesity risk10,11.
The existence of diverse findings can probably be attributed to changes in the genetic background of individuals and gene-diet interactions12,13. Variation in the melanocortin-4 receptor (MC4R) gene is known as the most common genetic cause of obesity14. Variants of MC4R-rs17782313 can affect food intake, total energy intake, fat intake, appetite, and consequently the occurrence of obesity15,16. On the other hand, protective effects on obesity have also been seen in some variants in the MC4R gene17. MC4R rs17782313 has three types of genotypes: CC, TC, and TT. The relationship between these genotypes and obesity and related factors has been investigated in some studies. Findings of a cross-sectional study showed interactions between CC genotype and high stress, high appetite, and high energy and fat intake are likely associated with higher BMI. As a result, it seems that people with C allele of MC4R rs17782313 are more susceptible to overweight or obesity18. A study conducted in Asia showed that the minor allele C can be associated with a 1.55 times increase in the risk of obesity, while the homozygous CC genotype can have a stronger effect and is associated with a 2.43 times increase in the risk of obesity in women19.
Results of a cross-sectional study showed that participants with low-frequency alleles of MC4R rs17782313 had a greater risk of metabolically unhealthy obesity20. In addition, the result of a previous study showed that the probability of metabolically healthy obesity was higher in patients with the T/C genotype of MC4R rs1778231321. Therefore, genetic variants in MC4R rs17782313 are considered as an important factor to understand the cause and type of obesity phenotypes22.
Studies have shown that total dietary fat intake is related to the risk of obesity23,24. However, today there is more focus on the quality of consumed fat than its quantity25. To investigate the effect of type of fat intake, previous studies have suggested dietary fat quality indices, such as cholesterol-saturated fat index (CSI) and the ratio of omega-6 to omega-3 (N6/N3). CSI was introduced in 1986 by Connor et al.26 and N6/N3 was proposed by Simopoulos et al.27. Findings regarding the relationship between the type of dietary fat and the risk of obesity are inconsistent. Several studies have shown a positive association between saturated fat intake and obesity28,29, while the opposite finding has been observed in studies30,31. Also, regarding the consumption of polyunsaturated fatty acid (PUFA), positive23, negative32, and null33 associations with obesity have been observed. In a cross-sectional study, an inverse relationship between N6/N3 ratio and general and abdominal obesity was seen34. Also, results of a study showed that subjects with metabolically unhealthy obesity had a higher intake of n-6/n-3 PUFA ratio than metabolically healthy obese person35. In addition, the results of a study showed a positive association between CSI and one of the risk markers of cardiovascular diseases in overweight and obese people36.
Despite the importance of investigating the interaction of genes and diet, which can help make dietary recommendations more specific to people, human studies in this field are limited and inconclusive. In this cross-sectional study, we aimed to assess the interaction between dietary fat quality indices and the MC4R gene in metabolically healthy and unhealthy overweight and obese women.
Materials and methods
Study population
The participants of this cross-sectional study were overweight and obese women. 279 women from the health centers of Tehran University of Medical Sciences were included according to the inclusion criteria. Body mass index (BMI) of 25–40 kg/m2 and age range of 18–68 were the inclusion criteria. We excluded participants who had the following conditions: history of malignancies, acute or chronic diseases, cardiovascular disease, all types of diabetes, thyroid disease, renal or hepatic disease, taking drugs to lower blood pressure, sugar and blood lipids, taking weight loss supplements or using a specific diet in the past year, smoking and presence of pregnancy, lactation or menopause. All methods were carried out in accordance with relevant guidelines and regulations. All participants completed written informed consent. Also, the ethics committee of the TUMS approved the study protocol. (Ethics number: IR.TUMS.MEDICINE.REC.1401.498).
Anthropometric and blood pressure measurements
Weight, body mass index, fat-free mass (FFM), and body fat (BF) percentage were measured by a bioelectrical impedance analyzer (BIA) (InBody 770 scanner from InBody Co. (Seoul, Korea)) according to the manufacturer’s protocols37. Participants had to take off their shoes, sweaters, and coats and not have metal objects such as earrings, rings, and watches with them. Height was measured in a standing position without shoes using a non-elastic tape with an accuracy of 0.5 cm. Regarding the waist circumference (WC) and hip circumference (HR) of participants, the narrowest part of the waist (after expiration) and the largest part of the hip were measured respectively using an elastic tape with an accuracy of 0.5 cm. Then waist to hip ratio (WHR) was calculated by the formula.
Definition of metabolically healthy and unhealthy phenotypes
Karelis criteria38 were used to classify participants in terms of metabolic health. In this way, 5 items were examined: (1) Triglycerides ⩽ 1.7 mmol/L, (2) Low-density lipoprotein (LDL) ⩽ 2.6 mmol/L and no treatment, (3) High-density lipoprotein (HDL) ⩾ 1.3 mmol/L and no treatment, (4) C-reactive protein (CRP) ⩽ 3.0 mg/L and 5) homeostatic model assessment for insulin resistance (HOMA-IR) ⩽ 2.7. If ⩾ 4 items were present, the individual was considered metabolically healthy.
Physical activity assessment
The physical activity (PA) of the participants in the last week was assessed by the reliable and validated International Physical Activity Questionnaire-short form (IPAQ) and measured in terms of metabolic equivalent hours per week (METs-h/week)39.
Biochemical and hormonal determination
Biochemical evaluations were carried out in the Nutrition and Biochemistry Laboratory of the School of Nutritional and Dietetics, TUMS. After 10–12 h of fasting, serum samples were collected. First samples were centrifuged, stored at − 80 °C, and analyzed using a single assay technique. Triglyceride (TG) was assayed using glycerol-3-phosphate oxidase–phenol 4-amino antipyrine peroxidase (GPOPAP) enzymatic endpoint40. Also, high-density lipoprotein (HDL) and low-density lipoprotein (LDL) cholesterol were evaluated by direct enzymatic clearance assay41. High sensitive C reactive protein (hs-CRP) level was assessed by immunoturbidimetric assay. Randox Laboratories (Hitachi 902) kits were used for all assessments. The level of insulin was measured and HOMA-IR was calculated according to this formula: HOMA-IR = insulin (Mu/mL) × fasting glucose (mmol/L)/22.542.
Dietary intake assessment
Assessment of the participants’ dietary intake in the last year was done using a 147-item validated semi-quantitative standard food frequency questionnaire (FFQ)43. Individuals were asked to report the consumption frequency of each food item on a daily, weekly, monthly, or yearly basis and the questionnaire was completed by an expert dietician. The intake of macronutrients, micronutrients, and total energy were analyzed by the NUTRITIONIST 4 (First Data Bank, San Bruno, CA) food analyzer44.
Dietary fat quality indices
Dietary fat quality was assessed by two indices: (1) cholesterol-saturated fat index (CSI) and (2) the ratio of N6/N3 essential fatty acids. Food intakes extracted from FFQ were used to calculate both of these indices. CSI was calculated according to this formula CSI = (1.01 × g saturated fat) + (0.05 × mg cholesterol)26. N6/N3 was calculated by dividing N6/N3 contents of foods27.
The Genotype determination
The polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP) technique was used for genotyping MC4R rs17782313 and rs1333048 single-nucleotide polymorphisms (SNPs) (genotypes C&T).
To extract MC4R Genomic deoxyribonucleic acid (DNA), we used 200 mL of whole blood and the GeneAll Mini Columns Type kit (GeneAll, South Korea). Extracted DNA, was used to evaluation of 2 SNPs reported near the MC4R gene, rs17782313, and rs17700633 SNP, which was performed. Using a specific primer (89 forward primer 5AAGTTCTACCTACCATGTTCTTGG-3 and reverse primer 5-90TTCCCCCTGAAGCTTTTCTTGTCATTTTGAT-3), polymerase chain reaction (PCR) was performed on these SNPs. PCR was performed using 50 ng/ml DNA, 10 mmol of each primer, and 1 M dimethyl sulfide (total volume equal to 20 μl). The amplification steps included the following items, respectively: primary denaturation at 94 °C (5 min), 35 cycles of denaturation at 60 °C (1 min), annealing at 94 °C (45 s), extension at 72 °C (1 min), and final extension at 72 °C (10 min). The SNPs rs7041 (Thr, 420, Lys) and rs4588 (Asp, 416, Glu) were auscultated by StyI and HaeIII enzymes according to the following procedure: The StyI enzymes (1μL) and HaeIII enzymes (1μL) each one separately added to the PCR product (5μL), distilled water (D.W.) (8μL) and Buffer Y. Tango (10 × 1μL). Products obtained from the digestion process were stained (with ethidium bromide) on a 2% agarose gel and imaging was performed. To confirm PCRFLP results, 10% of the sample were sequenced directly. Sequencing was performed using an ABI PRISM 3730 automated sequencer (Applied Biosystems, Foster City, CA, USA)45, and as a result, fragments containing 3 genotypes, CC, CT, and TT, were distinguished.
Statistical analyses
The normality distribution of data was checked by Kolmogorov–Smirnov test. Demographic characteristics of individuals were presented as mean ± standard deviation, minimum and maximum. One-way analysis of variance (ANOVA) was performed to compare anthropometric indices, lipid profile, hs-CRP level, insulin and HOMA-IR between individuals. In order to eliminate the effect of confounding factors, analysis of covariance (ANCOVA) was performed. Post-hoc multiple comparison analysis (Bonferroni corrected) was employed to look into the mean differences between the groups. A generalized linear model (GLM) was used to evaluate the interaction between dietary fat quality indices and MC4R gene in both crude and adjusted models. The results of the analyzes were adjusted for BMI, age, physical activity and energy intake. SPSS version 23.0 (SPSS, Chicago, IL, USA) was used for data analyzed. P-value < 0.05 was considered statistically significant and interaction P-value < 0.1 was determined as mariginally significant.
Ethics approval and consent to participate
This study was supported by grants from the Tehran University of Medical Sciences (TUMS), Tehran, Iran. Each individual was informed completely regarding the study protocol and provided a written and informed consent form before taking part in the study.
Results
Descriptive characteristics of the study sample
The current study included 279 women who were either overweight or obese. Individuals' age, weight, BMI, CSI, and N6 per N3 mean and standard deviation (SD) were 36.84 ± 8.45 years, 79.99 ± 10.88 kg, 30.73 ± 3.72 kg/m2, 12.65 ± 5.29, and 12.65 ± 0.10 respectively. Among the genotypes of 279 obese women with the MC4R gene, 40.9% of participants had TT, 26.2% TC and 33% had CC genotypes.
General characteristics of study population according to tertiles of CSI and N6/N3 in obese and overweight women
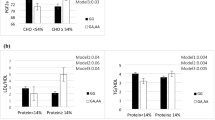
Table 1 shows the key characteristics of the study population concerning the tertile categories of CSI and N6/N3 in obese and overweight women. Before adjusting for confounders, the results displayed a significant difference across the CSI category for age (P = 0.02) which disappeared after adjustment (P = 0.27). Other variables showed no association with the tertiles of CSI before and after adjustment. No variables had a significant association with N6/N3 tertiles in the crude model, but after controlling for confounding factors, a higher HOMA index was associated with a higher N6/N3 (P = 0.03) (Table 1). Figure 1 shows the estimated marginal means for HOMA index to identify between which tertiles of N6/N3 the differences could be found.
General characteristics of study population according to MC4R rs17782313 in obese and overweight women
There were no significant differences in anthropometric measurements and biochemical variables in crude models, but the C allele carrier of MC4R showed lower height (P = < 0.001) and higher HOMA index (P = 0.01) after adjustment (Table 2). Figures 2 and 3 shows the estimated marginal means for height and HOMA index, respectively to identify between which tertiles of MC4R the differences could be found.
Dietary intake of study population according to tertiles of CSI and N6/N3 in obese and overweight women
Higher CSI was linked to more intake of refined grains, vegetables, fish, poultry, egg, red meat, protein, carbohydrate, total fat, cholesterol, PUFA, saturated fatty acid (SFA), linolenic acid, Eicosapentaenoic acid (EPA), and docosahexaenoic acid (DHA). Across the higher tertile of N6/N3, we found lower consumption of carbohydrates, monounsaturated fatty acid (MUFA), PUFA, oleic acid, and linoleic acid (Table 3).
The interaction between MC4R rs17782313 and CSI and N6/N3 on metabolically unhealthy phenotype
Using the GLM, we found no association between MC4R rs17782313 polymorphism and CSI on MUH phenotype in a multivariate-adjusted model controlling for the covariates. But the CC genotype of MC4R rs17782313 interacts with the N6/N3 ratio on the metabolically unhealthy phenotype. We found that interaction between CC genotype and N6/N3 on metabolically unhealthy phenotype in the crude model (β = 9.94, CI 2.49–17.39, P = 0.009) and even after adjustment for all confounders (β = 9.002, CI 1.15–16.85, P = 0.02, β = − 12.12, CI 2.79–21.46, P = 0.01), This means that those who homozygously have the risk allele as CC genotype, are more likely to have an unhealthy phenotype with an increase in N6/N3 than those who do not have CC genotype (Table 4).
Discussion
This study showed those who have the CC genotype are more likely to have an unhealthy obesity phenotype with higher N6/N3 ratio than those who have the CC genotype with lower intake of N6/N3 ratio.
This cross-sectional study reported that according to tertiles of fat quality indices, a higher HOMA IR index was accompanied by higher ratio of N6/N3. Among the genotypes of 279 obese women with the MC4R gene, higher CSI was linked with more refined grain, vegetables, fish, poultry, egg, red meat, protein, carbohydrate, total fat, cholesterol, PUFA, SFA, linolenic acid, EPA, and DHA. Across the higher tertile of N6/N3, we found lower consumption of carbohydrates, MUFA, PUFA, oleic acid, and linoleic acid.
A multivariate-adjusted model controlling for covariates found no interaction between the MC4R rs17782313 polymorphism and CSI on the MUH phenotype. But the CC genotype of MC4R rs17782313 interacts with the N6/N3 ratio on the metabolically unhealthy phenotype. Most previous studies support our result. For example, C allele carriers significantly had higher total cholesterol and TG levels in comparison to the TT genotype of MC4R in Brazilian obese children and adolescents, which can cause dyslipidemia46. It was additionally established that the risk of diabetes in carriers of the C allele increased by 14% regardless of the BMI. While another study showed that total cholesterol and LDL were not associated with different genotypes of MC4R47,48. Some other studies explain the interaction of the CC genotype of MC4R with the N6/N3 ratio on the metabolically unhealthy phenotype as follows: one paper expressed that body weight and BMI were higher in the CC and CT groups compared with individuals in the TT group, which can cause an unhealthy phenotype49; another reported the presence of the C allele in Mexican adults can have a possible influence on increasing fasting glucose levels48,50; but the CC genotype in European, is just associated with higher BMI and waist, but not with other variables of metabolic disorder51; another one a found higher prevalence of hyperglycemia and diabetes in women with the CT/CC genotype and explained this association by the higher obesity prevalence in this group52. Aside from fat tissue size, the distribution of body fat accumulation, whether subcutaneous Aside from fat tissue size, the distribution of body fat accumulation, whether subcutaneous or viscerally, is more important46 because visceral adipocytes are more metabolically active and can lead to the development of insulin resistance (IR) and all-cause mortality.us or viscerally, is more important46 because visceral adipocytes are more metabolically active and can lead to the development of insulin resistance (IR) and all-cause mortality53,54,55. Moreover, higher visceral adipose tissue (VAT)/subcutaneous adipose tissue (SAT) ratios may be associated with increased metabolic and cardiovascular risk, independently of BMI and VAT content56. We found in our study that CC genotype is associated with metabolically unhealthy situations regardless of BMI or visceral fat level, and this may happen because of lower height or higher HOMA index in C allele carriers.
In our study, a higher intake of the ratio of N6/N3 was associated with a higher HOMA IR index. As a result, two studies expressed that a higher N6/N3 ratio has a worse effect on HOMA-IR and quantitative insulin-sensitivity check index (QUICKI) indices57,58. In addition, an article reported that type 1 diabetes (DT1) was positively correlated with foods rich in PUFAs28 But in contrast to our result, another study found no effect of flax seed on HOMA-IR, although the N6/N3 ratio was lower in this group59. Some other studies examined the benefits of lower intake of N6/N3 ratio, failed to show a significant effect on insulin sensitivity60 the result we found is because of the effect of higher N6/N3 ratio on the expression of inflammatory markers57 which can induce HOMA-insulin resistance61.
Participants with the C allele had lower height and higher HOMA index. Our finding in this regard was in line with previous observational studies48 another study found that males with CC genotype had higher serum glucose levels compared with the other genotypes (TT and TC), the study expressed that C-allele carriers in the rs17782313 have an increased susceptibility to MUHO compared to the T-allele carriers20. T Schritter et al. demonstrated that in subjects carrying the C allele homozygous or the heterozygous form, the insulin response reduced as compared to TT genotype21. But no statistically significant difference in HOMA index and height status across MC4R rs17782313 genotypes among women and men was seen in some studies62.
Higher CSI was linked with more consumption of refined grain, vegetable, fish, poultry, egg, red meat, protein, carbohydrate, total fat, cholesterol, PUFA, SFA, linolenic acid, EPA, and DHA, and higher tertile of N6/N3 associate with lower intake of carbohydrate, MUFA, PUFA, oleic acid, and linoleic acid. According to previous studies higher intake of protein especially animal protein associated with higher cholesterol and SFA intake which can cause higher CSI index63,64 We know that consuming foods that are rich in omega 6 and have higher ratio of N6/N3, accompanied with lower intake of MUFA, omega 3 and oleic acid, and this can induce inflammation65. It is important to mention the strengths of the present study, first, was its novelty, to our knowledge it is the first study that investigated the interaction of dietary fat quality indices and the MC4R gene in metabolically healthy and unhealthy overweight and obese women. Other strengths are that we used validated FFQ questionnaires based on the Iranian population. As our strengths, there are some limitations to our study, first the cross-sectional design of our study, ruling out any causal relationship. The second limitation is the reliance on self-reported dietary data, which can cause information bias. Since our study only included overweight and obese women, we cannot generalize the results of this study to all women in the population.
Conclusion
The present study explains the interaction between genetics and the environment. Higher ratio intake of N6/N3 in CC genotype associates with unhealthy phenotype. In addition, C allele carrier of MC4R showed lower height and higher HOMA index. These results can help us to have better dietary recommendations about metabolic health status.
Data availability
The datasets generated and/or analyzed during the current study are not publicly available due to the restrictions and the research rules of the Tehran University of Medical Sciences (TUMS) but are available from the corresponding author on reasonable request.
Abbreviations
- ANCOVA:
-
Analysis of covariance
- ANOVA:
-
Analysis of variance
- BF:
-
Body fat
- BIA:
-
Bioelectrical impedance analyser
- BMI:
-
Body mass index
- CRP:
-
C-reactive protein
- CSI:
-
Cholesterol-saturated fat index
- DHA:
-
Docosahexaenoic acid
- DNA:
-
Deoxyribonucleic acid
- EPA:
-
Eicosapentaenoic acid
- FFM:
-
Fat free mass
- FFQ:
-
Food frequency questionnaire
- GLM:
-
Generalized linear model
- GPOPAP:
-
Glycerol-3-phosphate oxidase–phenol 4-amino antipyrine peroxidase
- HDL:
-
High-density lipoprotein
- HOMA-IR:
-
Homeostatic model assessment for insulin resistance
- HR:
-
Hip circumference
- HS-CRP:
-
High sensitive C-reactive protein
- IPAQ:
-
International physical activity questionnaire
- IRX3:
-
Iroquois homeobox protein 3
- LDL:
-
Low-density lipoprotein
- MET:
-
Metabolic equivalent
- MC4R:
-
Melanocortin 4 receptor
- MHO:
-
Metabolically healthy obese
- MUFA:
-
Monounsaturated fatty acid
- MUO:
-
Metabolically unhealthy obese
- N6/N3:
-
Omega-6/omega-3
- PA:
-
Physical activity
- PCR-RFLP:
-
Polymerase chain reaction-restriction fragment length polymorphism
- PUFA:
-
Polyunsaturated fatty acid
- QUICKI:
-
Quantitative insulin-sensitivity check index
- RPGRIP1L:
-
Retinitis pigmentosa GTPase regulator-interacting protein 1 like
- SD:
-
Standard deviation
- SFA:
-
Saturated fatty acid
- SAT:
-
Subcutaneous adipose tissue
- SNPs:
-
Single-nucleotide polymorphisms
- TG:
-
Triglyceride
- VAT:
-
Visceral adipose tissue
- WC:
-
Waist circumference
- WHR:
-
Waist to hip ratio
References
Castro, I. RRd. Obesity Prevention and Control: The Urgent Need for Effective Public Policies (SciELO Brasil, 2017).
Golden, A. & Kessler, C. Obesity and genetics. J. Am. Assoc. Nurse Pract. 32(7), 493–496 (2020).
Alizadeh, S. et al. Metabolic phenotypes of obese, overweight, and normal weight individuals and risk of chronic kidney disease: A systematic review and meta-analysis. Arch. Endocrinol. Metab. 63, 427–437 (2019).
Tremmel, M., Gerdtham, U.-G., Nilsson, P. M. & Saha, S. Economic burden of obesity: A systematic literature review. Int. J. Environ. Res. Public Health 14(4), 435 (2017).
Vecchié, A. et al. Obesity phenotypes and their paradoxical association with cardiovascular diseases. Eur. J. Intern. Med. 48, 6–17 (2018).
Brochu, M. et al. What are the physical characteristics associated with a normal metabolic profile despite a high level of obesity in postmenopausal women?. J. Clin. Endocrinol. Metab. 86(3), 1020–1025 (2001).
Karelis, A. D. et al. The metabolically healthy but obese individual presents a favorable inflammation profile. J. Clin. Endocrinol. Metab. 90(7), 4145–4150 (2005).
Wildman, R. P. et al. The obese without cardiometabolic risk factor clustering and the normal weight with cardiometabolic risk factor clustering: Prevalence and correlates of 2 phenotypes among the US population (NHANES 1999–2004). Arch. Intern. Med. 168(15), 1617–1624 (2008).
Cheng, M. et al. Computational analyses of obesity associated loci generated by genome-wide association studies. PLoS ONE 13(7), e0199987 (2018).
Lavebratt, C., Almgren, M. & Ekström, T. Epigenetic regulation in obesity. Int. J. Obes. 36(6), 757–765 (2012).
Qi, L. Gene–diet interaction and weight loss. Curr. Opin. Lipidol. 25(1), 27 (2014).
Daya, M. et al. Obesity risk and preference for high dietary fat intake are determined by FTO rs9939609 gene polymorphism in selected Indonesian adults. Asia Pac. J. Clin. Nutr. 28(1), 183–191 (2019).
Kim, J. et al. Intraocular pressure, glaucoma, and dietary caffeine consumption: A gene–diet interaction study from the UK Biobank. Ophthalmology 128(6), 866–876 (2021).
Yu, K., Li, L., Zhang, L., Guo, L. & Wang, C. Association between MC4R rs17782313 genotype and obesity: A meta-analysis. Gene 733, 144372 (2020).
Khalilitehrani, A., Qorbani, M., Hosseini, S. & Pishva, H. The association of MC4R rs17782313 polymorphism with dietary intake in Iranian adults. Gene 563(2), 125–129 (2015).
Magno, F. C. C. M. et al. Association of the MC4R rs17782313 polymorphism with plasma ghrelin, leptin, IL6 and TNFα concentrations, food intake and eating behaviors in morbidly obese women. Eat. Weight Disorders-Stud. Anorex. Bulim. Obes. 26, 1079–1087 (2021).
Zujko, M. E. et al. Dietary Total Antioxidant Capacity and Dietary Polyphenol Intake and Prevalence of Metabolic Syndrome in Polish Adults: A Nationwide Study. Oxid. Med. Cell Longev. 2018, 7487816. https://doi.org/10.1155/2018/7487816 (2018).
Rahati, S., Qorbani, M., Naghavi, A. & Pishva, H. Association and interaction of the MC4R rs17782313 polymorphism with plasma ghrelin, GLP-1, cortisol, food intake and eating behaviors in overweight/obese Iranian adults. BMC Endocr. Disord. 22(1), 234 (2022).
Rana, S., Rahmani, S. & Mirza, S. MC4R variant rs17782313 and manifestation of obese phenotype in Pakistani females. RSC Adv. 8(30), 16957–16972 (2018).
Gao, L. et al. MC4R Single Nucleotide Polymorphisms Were Associated with Metabolically Healthy and Unhealthy Obesity in Chinese Northern Han Populations. Int. J. Endocrinol. 2019, 4328909. https://doi.org/10.1155/2019/4328909 (2019).
ElhamKia, M. et al. The interaction between dietary total antioxidant capacity and MC4R gene and HOMA-IR in metabolically healthy and unhealthy overweight and obese women. Nutr. Metab. Insights. 15, 11786388221105984 (2022).
Mozafarizadeh, M. et al. Evaluation of FTO rs9939609 and MC4R rs17782313 polymorphisms as prognostic biomarkers of obesity: A population-based cross-sectional study. Oman Med. J. 34(1), 56 (2019).
Raatz, S. K., Conrad, Z., Johnson, L. K., Picklo, M. J. & Jahns, L. Relationship of the reported intakes of fat and fatty acids to body weight in US adults. Nutrients 9(5), 438 (2017).
Wang, L., Wang, H., Zhang, B., Popkin, B. M. & Du, S. Elevated fat intake increases body weight and the risk of overweight and obesity among Chinese adults: 1991–2015 trends. Nutrients 12(11), 3272 (2020).
Mozaffarian, D. & Ludwig, D. S. The 2015 US dietary guidelines: Lifting the ban on total dietary fat. JAMA 313(24), 2421–2422 (2015).
Connor, S. L. et al. The cholesterol/saturated-fat index: An indication of the hypercholesterolaemic and atherogenic potential of food. Lancet (Lond. Engl.). 1(8492), 1229–1232 (1986).
Simopoulos, A. P. The importance of the ratio of omega-6/omega-3 essential fatty acids. Biomed. Pharmacother. Biomed. Pharmacother. 56(8), 365–379 (2002).
Brayner, B. et al. Dietary patterns characterized by fat type in association with obesity and type 2 diabetes: A longitudinal study of UK Biobank participants. J. Nutr. 151(11), 3570–3578 (2021).
Lopez-Lopez, D. E., Saavedra-Roman, I. K., Calizaya-Milla, Y. E. & Saintila, J. Food Addiction, Saturated Fat Intake, and Body Mass Index in Peruvian Adults: A Cross-Sectional Survey. J. Nutr. Metab. 2021, 9964143. https://doi.org/10.1155/2021/9964143 (2021).
Forouhi, N. G. et al. Dietary fat intake and subsequent weight change in adults: Results from the European prospective investigation into cancer and nutrition cohorts. Am. J. Clin. Nutr. 90(6), 1632–1641 (2009).
Wu, F. et al. Individual SFA intake and risk of overweight/obesity: Findings from a population-based nationwide cohort study. Br. J. Nutr. 128(1), 75–83 (2022).
Cardel, M., Lemas, D. J., Jackson, K. H., Friedman, J. E. & Fernández, J. R. Higher intake of PUFAs is associated with lower total and visceral adiposity and higher lean mass in a racially diverse sample of children. J. Nutr. 145(9), 2146–2152 (2015).
Nimptsch, K., Berg-Beckhoff, G. & Linseisen, J. Effect of dietary fatty acid intake on prospective weight change in the Heidelberg cohort of the European prospective investigation into cancer and nutrition. Public Health Nutr. 13(10), 1636–1646 (2010).
Suara, S. B. et al. Dietary fat quantity and quality in relation to general and abdominal obesity in women: A cross-sectional study from Ghana. Lipids Health Dis. 19(1), 1–13 (2020).
Arsic, A. et al. Metabolically healthy obesity: Is there a link with polyunsaturated fatty acid intake and status?. Can. J. Physiol. Pharmacol. 99(1), 64–71 (2021).
MoussaviJavardi, M. S., Madani, Z., Movahedi, A., Karandish, M. & Abbasi, B. The correlation between dietary fat quality indices and lipid profile with Atherogenic index of plasma in obese and non-obese volunteers: a cross-sectional descriptive-analytic case-control study. Lipids Health Dis. 19, 1–9 (2020).
A T. Body composition analyzer; BC-418. Instruction Manual (2015).
Karelis, A. D., Brochu, M. & Rabasa-Lhoret, R. Can we identify metabolically healthy but obese individuals (MHO)?. Diabetes Metab. 30(6), 569–572 (2004).
Aadahl, M. J. T. Validation of a new self-report instrument for measuring physical activity. Med. Sci. Sports Exerc. 35(7), 1196–1202 (2003).
Purcell, G. V., Behenna, D. B. & Walsh, P. R. Evaluation of the BMC glucose oxidase/peroxidase-4-aminophenazone-phenol procedure for glucose as adapted to the technicon SMAC. Clin. Chem. 25(10), 1844–1846 (1979).
Miller, W. G. et al. Seven direct methods for measuring HDL and LDL cholesterol compared with ultracentrifugation reference measurement procedures. Clin. Chem. 56(6), 977–986 (2010).
Basurto, L. et al. Differences between metabolically healthy and unhealthy obesity in PAI-1 level: Fibrinolysis, body size phenotypes and metabolism. Thromb. Res. 180, 110–114 (2019).
Mirmiran, P. E. F., Mehrabi, Y., Hedayati, M. & Azizi, F. Reliability and relative validity of an FFQ for nutrients in the Tehran lipid and glucose study. Public Health Nutr. 13(5), 654–662 (2010).
Ghaffarpour, M., Houshiar-Rad, A. & Kianfar, H. J. The manual for household measures, cooking yields factors and edible portion of foods. Tehran Nashre Olume Keshavarzy. 7(213), 42–58 (1999).
Pooyan, S. et al. A high-protein/low-fat diet may interact with vitamin D-binding protein gene variants to moderate the risk of depression in apparently healthy adults. Lifestyle Genom. 11(1), 64–72 (2018).
Fernandes, A. E. et al. Associations between a common variant near the MC4R gene and serum triglyceride levels in an obese pediatric cohort. Endocrine 49, 653–658 (2015).
Huang, W., Sun, Y. & Sun, J. Combined effects of FTO rs9939609 and MC4R rs17782313 on obesity and BMI in Chinese Han populations. Endocrine 39, 69–74 (2011).
Qi, L., Kraft, P., Hunter, D. J. & Hu, F. B. The common obesity variant near MC4R gene is associated with higher intakes of total energy and dietary fat, weight change and diabetes risk in women. Hum. Mol. Genet. 17(22), 3502–3508 (2008).
Resende, C. M. M. et al. The polymorphism rs17782313 near MC4R gene is related with anthropometric changes in women submitted to bariatric surgery over 60 months. Clin. Nutr. 37(4), 1286–1292 (2018).
Mejía-Benítez, A. et al. Analysis of the contribution of FTO, NPC1, ENPP1, NEGR1, GNPDA2 and MC4R genes to obesity in Mexican children. BMC Med. Genet. 14, 1–6 (2013).
Zobel, D. P. et al. Variants near MC4R are associated with obesity and influence obesity-related quantitative traits in a population of middle-aged people: Studies of 14,940 Danes. Diabetes 58(3), 757–764 (2009).
Raskiliene, A., Smalinskiene, A., Kriaucioniene, V., Lesauskaite, V. & Petkeviciene, J. Associations of MC4R, LEP, and LEPR polymorphisms with obesity-related parameters in childhood and adulthood. Genes 12(6), 949 (2021).
Ibrahim, M. M. Subcutaneous and visceral adipose tissue: Structural and functional differences. Obes. Rev. 11(1), 11–18 (2010).
Katzmarzyk, P., Mire, E. & Bouchard, C. Abdominal obesity and mortality: The Pennington Center longitudinal study. Nutr. Diabetes 2(8), e42-e (2012).
Brodowski, J. et al. Searching for the relationship between the parameters of metabolic syndrome and the rs17782313 (T>C) polymorphism of the MC4R gene in postmenopausal women. Clin. Interv. Aging. 12, 549–555. https://doi.org/10.2147/CIA.S129874 (2017).
Kaess, B. et al. The ratio of visceral to subcutaneous fat, a metric of body fat distribution, is a unique correlate of cardiometabolic risk. Diabetologia 55, 2622–2630 (2012).
Roškarić, P., Šperanda, M., Mašek, T., Verbanac, D. & Starčević, K. Low dietary n6/n3 ratio attenuates changes in the NRF 2 gene expression, lipid peroxidation, and inflammatory markers induced by fructose overconsumption in the rat abdominal adipose tissue. Antioxidants. 10(12), 2005 (2021).
Ma, F., Zhu, Z., Ma, X. & Li, S. N-3 polyunsaturated fatty acids (PUFAS) dietary improves the metabolic syndrome induce by high fat food. Int. J. Clin. Exp. Med. 12(1), 1261–1268 (2019).
Taylor, C. G., Noto, A. D., Stringer, D. M., Froese, S. & Malcolmson, L. Dietary milled flaxseed and flaxseed oil improve N-3 fatty acid status and do not affect glycemic control in individuals with well-controlled type 2 diabetes. J. Am. Coll. Nutr. 29(1), 72–80 (2010).
Griffin, M. D. et al. Effects of altering the ratio of dietary n-6 to n-3 fatty acids on insulin sensitivity, lipoprotein size, and postprandial lipemia in men and postmenopausal women aged 45–70 y: The OPTILIP Study. Am. J. Clin. Nutr. 84(6), 1290–1298 (2006).
Park, K., Steffes, M., Lee, D.-H., Himes, J. H. & Jacobs, D. Association of inflammation with worsening HOMA-insulin resistance. Diabetologia 52(11), 2337–2344 (2009).
Khodarahmi, M., Kahroba, H., Jafarabadi, M. A., Mesgari-Abbasi, M. & Farhangi, M. A. Dietary quality indices modifies the effects of melanocortin-4 receptor (MC4R) rs17782313 polymorphism on cardio-metabolic risk factors and hypothalamic hormones in obese adults. BMC Cardiovasc. Disord. 20(1), 1–10 (2020).
Sauvaget, C., Nagano, J., Hayashi, M. & Yamada, M. Animal protein, animal fat, and cholesterol intakes and risk of cerebral infarction mortality in the adult health study. Stroke 35(7), 1531–1537 (2004).
Delimaris I. Adverse Effects Associated with Protein Intake above the Recommended Dietary Allowance for Adults. ISRN Nutr. 2013, 126929. https://doi.org/10.5402/2013/126929 (2013).
Simopoulos, A. P. Importance of the ratio of omega-6/omega-3 essential fatty acids: evolutionary aspects. Omega-6/omega-3 essential fatty acid ratio. Sci. Evid. 92, 1–22 (2003).
Acknowledgements
The authors thank the laboratory of Nutrition Sciences and Dietetics at Tehran University of Medical Sciences (TUMS). We are grateful to all the participants for their contributions to this research. This study was approved by the research ethics committee of the Tehran University of Medical Sciences (TUMS), Tehran, Iran with ethics number IR.TUMS.MEDICINE.REC.1401.498. All participants signed a written informed consent that was approved by this committee before enrollment in the study.
Funding
This study was supported by the Tehran University of Medical Sciences (TUMS), Tehran, Iran (Granted Number: 1401-2-212-59190).
Author information
Authors and Affiliations
Contributions
K.M. and N.R. designed the search and conducted sampling, M.N., M.F., and R.G.-E. wrote the paper; N.R. and F.G. performed statistical analysis. All authors read and approved the final manuscript. All authors approved the final manuscript and gave consent for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rasaei, N., Fallah, M., Nemati, M. et al. Investigation the interaction of dietary fat quality indices and the MC4R gene in metabolically healthy and unhealthy overweight and obese women. Sci Rep 13, 12183 (2023). https://doi.org/10.1038/s41598-023-38988-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-38988-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.