Abstract
Despite the abnormal expression of 18S rRNA m6A methyltransferase METTL5 being reported in some types of human malignancies, but its effect on hepatocellular carcinoma (HCC) remains to be unclear. This study aims to elucidate the influences of METTL5 on the carcinogenesis and progression of HCC. Expressions of METTL5 gene, transcript, protein, and promoter methylation in HCC were examined through multiple databases, c-BioPortal was used to confirm the genomic alterations of METTL5, the biological functions, target networks of kinases and microRNAs of METTL5, and its interactive differential genes were investigated through LinkedOmics. The possible correlation of METTL5 with the tumor-related infiltration of immune cells for HCC were explored comprehensively by using the online tools of TIMER and TISIDB. Expressions of METTL5 gene, mRNA, and protein were considerably overexpressed in HCC samples in comparison with healthy samples. The high methylation of the METTL5 promoter was observed in HCC tissues. Elevated METTL5 expression exhibited unfavorable survival outcomes in HCC patients. METTL5 expression were enriched in the signaling pathways of ribosome and oxidative phosphorylation, mismatch repair, and spliceosome through the involvement of several cancer-related kinases and miRNAs. The METTL5 expression has a positive correlation with the infiltration degree of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in HCC. Marker genes of tumor immune-infiltrated cells have strong connection with METTL5. Furthermore, the upregulation of METTL5 was strongly correlated with the immune regulation of immunomodulators, chemokines, and chemokine receptors in the immune microenvironment. The oncogenesis and development of HCC are closely related to METTL5 expression, and the overexpression of METTL5 resulted in the poor survival outcome of HCC patients by regulating tumor immune microenvironment.
Similar content being viewed by others
Introduction
Liver cancer remains a global threat to the physical and mental health of humans and is characterized by high mortality and great incidence1. Hepatocellular carcinoma (HCC or LIHC) is among the most prominent subtypes of all primary intrahepatic malignancies, encompassing approximately 90% of all liver cancer cases. HCC was the sixth most prevalent malignancy in terms of morbidity and is considered the third most frequent cause in terms of malignancy related mortality all over the world2. The diagnoses of HCC were identified on the basis of radiologic image, serologic molecular markers, or pathologic findings. However, a large proportion of HCC cases from the initial diagnosis involved terminal stage cases accompanied by high mortality. Despite the rapid advances in the effective treatment strategies of hepatectomy, chemotherapy, radiotherapy, immunotherapy, and targeted therapy, the survival outcome of HCC patients remains unsatisfactory because of its highly invasive nature3. The five-year survival rate of a small percentage of HCC-diagnosed population was expected to exceed 15% because of its easy recurrence and metastasis4. Thus, the potential diagnostic and prognostic indexes of HCC are urgently needed to provide new therapeutic HCC targets.
Ribosomal RNA (rRNA) comprised approximately 80% of the total cellular RNA in eukaryotes. rRNA modification emerged as a momentous post-transcriptional gene regulation process. The N6 methyladenosine (m6A) modification that occurred on rRNA influenced the modulation of ribosome structure and function5,6. Methyltransferase N6-adenosine (METTL5) served as a rRNA m6A and was recently identified to specifically catalyze human 18S rRNA N6 methylation at adenosine 1832 site7, which possessed vital importance in regulating the function and development of ribosomes7,8.
Abnormal METTL5 expression was observed in numerous human malignancy types. METTL5 expression was highly upregulated in lung adenocarcinoma and was bound up with short survival time7,9. METTL5 overexpression promoted translation initiation and cell growth in breast cancer7,8. The effects of METTL5 on the tumorigenesis and the development of HCC remains to be rarely investigated.
The expression characteristics of METTL5 in HCC samples were further explained in this research. We aim to evaluate the underlying molecular mechanisms responsible for HCC onset and the potential role of METTL5 expression on HCC prognosis.
Results
Expression levels of METTL5 gene in HCC
We calculated the differential expression of the METTL5 gene in various types of cancers and matched normal tissues by means of online searching at the GEPIA database (Fig. 1A). The METTL5 gene was markably elevated among HCC specimens in comparison with surrounding healthy tissues (Fig. 2A). Further analyses were adopted for assessing METTL5 gene expression in relation to tumor stage, and the METTL5 gene expression gradually increased among the advent of HCC progression via GEPIA (Fig. 2B). Such finding was consistent with the outcomes obtained from the Timer database (Fig. 1B). The aforementioned data implied that METTL5 gene overexpression may play a significant role in the tumorigenesis for HCC.
Expression levels of METTL5 gene in Pan-cancer. (A) Expression profiles of the METTL5 gene in different cancer types and paired normal tissue samples from the GEPIA database. (B) Expression profiles of the METTL5 gene in different cancer types and paired normal tissue samples from the TIMER database.
Expression profiles of the METTL5 gene in hepatocellular carcinoma (LIHC). (A) Expression profiles of the METTL5 gene in LIHC and paired of normal tissues from the GEPIA database. (B) Correlation between expression of METTL5 gene and Cancer stage of LIHC through the GEPIA database. (C) Promoter methylation levels of METTL5 in LIHC were evaluated by box plot using the UALCAN database. (D) METTL5 alterations in LIHC through the cBioPortal database.
Potential prognostic effects of METTL5 gene expression on HCC
We evaluated the prognostic significance of METTL5 expression on HCC patients using GEPIA. METTL5 gene overexpression is disadvantageous in terms of the overall survival time (OS) (Fig. 3A) and disease-free survival (DFS) (Fig. 3B) of HCC patients.
Comparisons of the effects of high and low expression levels of METTL5 gene on survival time of hepatocellular carcinoma (LIHC) patients using GEPIA database. (A) and (B) Revealed that elevated expression levels of METTL5 gene were associated with worse overall survival and disease free survival for LIHC patients.
Expression profiles of METTL5 transcript in HCC
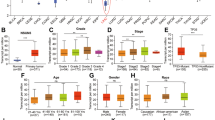
The available expression data of the METTL5 transcript in multiple HCC tissues were evaluated online from the UALCAN and HPA databases. Findings from both databases revealed that the intensified mRNA expression of METTL5 was observed in HCC specimens in comparison with adjacent healthy tissues via UALCAN (Fig. 4A) which is consistent with the outcomes of HPA database(Fig. 4B). Further analyses were used to assess the METTL5 transcript expression in relation to various clinical characteristics of HCC by using UALCAN. The results proved that METTL5 mRNA in HCC specimens was up-regulated in terms of age from 21 to 40, N1, stage 4, grade 4, Asian race, TP53 mutation, and male patients (Fig. 5).
Expression profiles of the METTL5 transcript in hepatocellular carcinoma (LIHC): (A) expression profiles of the METTL5 transcript in different cancer types and paired normal tissue samples from the UALCAN database. (B) Expression profiles of the METTL5 gene in different cancer types from the HPA database.
Expression of METTL5 transcription in different clinical characteristics of patients with liver hepatocellular carcinoma (LIHC) via UALCAN. (A) Correlation of METTL5 transcription with different patients age. (B) Correlation of METTL5 transcription with different patients sex. (C) Correlation of METTL5 transcription with different patients race. (D) Correlation of METTL5 transcription with different nodal metastasis status. (E) Correlation of METTL5 transcription with different cancer grade. (F) Correlation of METTL5 transcription with different cancer stage. (G) Correlation of METTL5 transcription with different TP53 mutation Status. (I) Correlation of METTL5 transcription with different histological subtypes.
Potential prognostic effect of METTL5 transcript expression on HCC
We further evaluated the impact of METTL5 mRNA expression on the survival outcomes of HCC patients using the KM database. The results showed that METTL5 mRNA overexpression had a close association with poor OS and PFS in HCC patients (Fig. 6A and B). We also confirmed that increased METTL5 mRNA expression was strongly linked with unfavorable OS and PFS in stage 3, stage 3 + 4, AJCC T3, drinking alcohol hobby, no infection with Hepatitis virus diseases, and male HCC patients (Table 1). The substantially increased expression of METTL5 transcript had link with disadvantageous survival via UALCAN (Fig. 6C). These data implied that the overexpression of METTL5 mRNA in HCC predicted poor prognostic outcomes.
Comparisons of the effects of high and low expression levels of METTL5 transcript on survival time of hepatocellular carcinoma (LIHC) patients. (A) and (B) Elevated expression levels of METTL5 transcript were associated with worse overall survival and progression free survival for LIHC patients using K-M plotter. (C) Effect of METTL5 transcript expression on LIHC patient survival via UALCAN.
Expression profiles of METTL5 protein in HCC patients
METTL5 protein expression was substantially elevated in liver cancer when compared with paired normal tissues through the UALCAN database (Fig. 7A and B). The upregulation of METTL5 protein was commonly detected in 21–40 year-olds and female patients via UALCAN (Fig. 7C and D).
METTL5 transcription in subgroups of patients with hepatocellular carcinoma via UALCAN. (A) Boxplot showing relative expression of METTL5 transcription in pancancer samples. (B) Boxplot showing relative expression of METTL5 transcription in normal and LIHC samples. (C) Boxplot showing relative expression of METTL5 transcription in normal individuals with different age in LIHC patients. (D) Boxplot showing relative expression of METTL5 transcription in normal individuals of gender in LIHC patients.
Potential prognostic effects of METTL5 protein expression on HCC patients
We further evaluate the impact of METTL5 protein expression on the survival outcomes of HCC patients using the HPA database. The high expression of METTL5 protein generally indicated the poor survival outcomes of HCC patients (Fig. 8A).
The prognostic value and biological function of METTL5 protein in LIHC samples. (A) Comparisons of the effects of high and low expression levels of METTL5 protein on survival time of hepatocellular carcinoma(LIHC) patients using HPA database. (B) Protein–protein interaction network of METTL5 was constructed by GeneMANIA.
Genetic alteration of METTL5 in HCC patients
Hypermethylation of METTL5 gene was displayed in HCC in comparison with paired healthy specimens (Fig. 2C), inferring that the upregulation of METTL5 methylation contributed to METTL5 overexpression in HCC. Genetic alterations of METTL5 in HCC patients were analyzed using the cBioPortal tools. Our findings revealed that the genomic alterations of METTL5 occurred in 0.7% of HCC patients (Fig. 2D). All these results revealed that METTL5 genomic alteration indeed occur in HCC patients.
Gene enrichment analysis of METTL5 in HCC patients
A total of 8600 genes correlated with METTL5 were shown in the volcano plot via LinkedOmics (Fig. 9A), which comprised 3568 positively correlative genes and 5032 adversely correlative genes. The top 50 most prominent genes with positive and negative connections with METTL5 were displayed (Fig. 9B and C). The METTL5-related enriched GO term and KEGG pathway were analyzed. Significant biological processes (BPs) indicated that METTL5-related genes were intimately engaged in translational initiation, protein localization to endoplasmic reticulum, mitochondrial gene expression, ribonucleoprotein complex biogenesis, and ncRNA processing (Fig. 10A). These interactive genes served as cellular components of ribosome, mitochondrial protein complex, cytosolic part, mitochondrial inner membrane, and respiratory chain (Fig. 10B). The molecular function enrichment of METTL5-related genes is mainly projected in ribosome, rRNA binding, electron transfer activity, unfolded protein binding, and oxidoreductase activity. Acting on a Heme group of donors with Heme-copper terminal oxidase activity (Fig. 10C), METTL5 were proven to largely participate in the signaling pathways of ribosome, oxidative phosphorylation, spliceosome, Parkinson disease, proteasome, and Huntington disease (Fig. 10D).
Genes differentially expressed in correlation with METTL5 in hepatocellular carcinoma via LinkedOmics. (A) A Pearson test was used to analyze correlations between METTL5 and genes differentially expressed in LIHC. (B) Heat maps showing the top 50 genes positively correlated with METTL5 in LIHC. (C) Heat maps showing the top 50 genes negatively correlated with METTL5 in LIHC.
Co-expressed protein correlated with METTL5
We further evaluated the major protein that interacted with METTL5 through the Metascape database. The results showed 20 types of METTL5 interactive proteins.
PPI network of METTL5
The PPI network was used to visualize the interaction between METTL5 and its relevant protein through the use of GeneMANIA tools. METTL5 was mainly enriched in the regulation of S-adenosylmethionine-dependent methyltransferase activity, methyltransferase activity, transferase activity, transferring one-carbon groups, methylation, RNA modification, and N-methyltransferase activity (Fig. 8B).
METTL5 networks of kinase and microRNA (miRNA) in HCC
The top 5 most remarkable kinase-targets of METTL5 were primarily involved in the myosin light chain kinase, myosin light chain kinase3, myosin light chain kinase family member 4, aurora kinase B, and ribosomal protein S6 kinase A4 (Fig. 11B). The miRNA-target network was positively relevant to MIR-127, MIR-423, and MIR-510 (Fig. 11A).
Correlation between METTL5 expression and immune biomarker in HCC
Immune infiltration had a significant impact on tumor progression. No apparent change of immune cell infiltration under different copy numbers of METTL5 existed in HCC specimens (Fig. 12B). The TIMER database was used to comprehensively evaluate the correlation of METTL5 expression to immune cell infiltration in HCC. METTL5 expression showed positive correlation with B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cell infiltration levels in HCC (Fig. 12A). The cox regression analysis was established to estimate the effects of METTL5 and immune cell infiltration on the survival outcomes of HCC patients. The results indicated that the infiltrating degree of B cells (coef = − 8.430, p = 0.017) and CD8+ T cells (coef = − 6.787, p = 0.008) had a positive correlation with the survivorship risk of HCC patients. Dendritic cells (coef = 5.563, p = 0.002) and METTL5 (coef = 0.562, p = 0.001) were available as independent risk factors of survival in HCC cases, whereas CD4+ Tcell (p = 0.159), macrophage (p = 0.106), and neutrophils (p = 0.804) had no impact on the survival of HCC patients. METTL5 expression also had a positive link with PD1 (PDCD1) and CTLA4 in HCC based on the TIMER and GEPIA databases (Fig. 12C and D). The expression levels of METTL5 had a strong connection with 16 gene markers of immune cell in HCC samples (Table 2).
Correlation of METTL5 expression with immune infiltration in hepatocellular carcinoma(LIHC). (A) Correlation between the expression of METTL5 and the abundance of immune infiltration in LIHC at Timer database. (B) The infiltration level of various immune cells under different copy numbers of METTL5 in LIHC. (C) Correlation of METTL5 expression with immune checkpoint inhibitors.
METTL5 expression associated with immunomodulators in HCC
TISIDB database was used to explore the possible correlation between METTL5 and various immune signatures. METTL5 expression had a positive correlation with the several immunostimulators of NT5E, PVR, TNFRSF4, TNFRSF14, and TNFRSF18 (Fig. 13A). Findings exhibited that METTL5 expression was negatively correlated with a large number of immunoinhibitors of BTLA, CD96, CD274, CSF1R, HAVCR2, KDR, PDCD1LG2, TGFBR1, and VTCN1 (Fig. 13B). The aforementioned results demonstrated the function of METTL5 as an immunoregulatory factor for HCC.
The expression of METTL5 is associated with immunomodulators in hepatocellular carcinoma. (A) Correlation between METTL5 expression and immunostimulators in hepatocellular carcinoma at TISIDB database. (B) Correlation between METTL5 expression and immunoinhibitors in hepatocellular carcinoma at TISIDB database.
Correlation between METTL5 expression and chemokines in HCC
METTL5 expression presented a strongly positive association with several chemokines of CCL15, CCL16, CCL20, CXCL17, and XCL1 (Fig. 14A). Moreover, METTL5 expression was positively linked with the chemokine receptors of CCR4 and CX3CR1 (Fig. 14B). These findings indicated that METTL5 played a critical function in the immune interaction in HCC.
Correlation between the expression of METTL5 and chemokines in hepatocellular carcinoma. (A) Correlation between METTL5 expression and chemokines in hepatocellular carcinoma at TISIDB database. (B) Correlation between METTL5 expression and chemokine receptors in hepatocellular carcinoma at TISIDB database.
Discussion
HCC is a widely recognized prevalent malignant tumor with highly aggressive and fatal biological behavior across the world10. An increasing body of evidence revealed that exposure to chronic hepatitis virus infection11, persistent alcohol addiction11, and host tumor immune microenvironment12 were principally responsible for the initiation and development of HCC. Surgical resection is a widely accepted optimal therapy for HCC patients at the earliest stages; however, even after the surgery of a patient, HCC is still accompanied by a high recurrence rate13. Although massive advancement was achieved in the aspect of therapeutic strategies in recent years, the clinical survival of HCC patients was far from satisfactory. Thus, promising diagnostic and prognostic biomarkers in HCC are needed to enhance the potential of precision treatments.
Cancer cells are typically characterized by the malignant biological behavior of unrestrained cellular growth14. Recent studies reported that cancer cells exhibit the molecular characteristic of misregulated ribosome biogenesis15. m6A modification of rRNAs at the sixth adenine site at position 1832 of 18S rRNA (m6A1832) is emerging as a crucial oncogenic signal that facilitates carcinogenesis and rapid progression16. METTL5 was widely recognized as the methyltransferase that took charge of specifically catalyzing 18S rRNA m6A1832 modification17. A growing body of evidence displayed that the aberrant expression of METTL5 existed in a wide variety of human malignant tumors. A previous study reported that METTL5 is crucial for breast cancer cell growth7. Previous researchers clearly proposed that METTL5 expression considerably upregulated in HCC cells, targeted the knockdown of METTL5 suppressed proliferation and invasion abilities of HCC cells, and further induced cancer cell apoptosis in vitro tests18. Recent studies emphasized that METTL5 stimulated the proliferation and invasion activity of tumor cells in pancreatic cancer19. The study was conducted to evaluate the influence of METTL5 on the tumorigenesis and survival of HCC.
The correlation between METTL5 gene and HCC expressions was investigated using various databases. Our analysis exhibited that METTL5 gene expression was markably elevated in HCC. Furthermore, the METTL5 gene expression gradually increased with the development of the severity of liver cancer. Recent studies revealed that METTL5 functioned as an oncogene that produced a marked effect on the hyperactivation of tumor cell proliferation, migration, and invasion19. HCC patients had the worst survival in the presence of enhanced METTL5. Our study revealed that intensified mRNA expression of METTL5 were observed in HCC specimens in comparison with adjacent healthy tissues. METTL5 mRNA in HCC specimens was up-regulated in N1, stage 4, and grade 4 diseases. Our study reported that the overexpression of METTL5 mRNA had a close association with poor OS, PFS, and RFS among HCC patients. Increased METTL5 mRNA expression was strongly linked with unfavorable OS and PFS in stage 3, stage 3 + 4, and AJCC T3 diseases and male HCC patients. METTL5 protein expression was substantially elevated in liver cancer when compared with paired healthy tissues. The overexpression of the METTL5 protein had adverse effects on the prognosis of HCC. The trend of METTL5 expression in the aspects of transcript and protein was substantially in line with that of the gene, that is, METTL5 gene, transcript, and protein have the same influence on the prognosis of HCC. These observations strongly supported that METTL5 might be taken as a potential diagnostic and prognostic indicator for HCC patients. The final results exhibited that the hypermethylation of METTL5 indeed enhanced the expression of METTL5 in HCC samples.
Related functional networks of METTL5 gene are significantly implicated in ribosome signaling, oxidative phosphorylation, spliceosome, proteasome, RNA transport, and mismatch repair, which were consistent with the previous findings that METTL5 alterations engaged in post-transcriptional regulation and ribosome biogenesis20. Existing literature reports indicated that METTL5 played an essential role in the promotion of translation initiation7, and previous studies revealed that METTL5 participated in the modulation of mismatch repair21.
Immune infiltration played a pivotal role in the carcinogenesis and rapid progression of HCC22. The infiltration of immune cells was an essential component of the immune microenvironment and contributed to the mediation of tumor proliferation and metastasis23. TIMER tools were further adopted to investigate the underlying correlation of immune infiltration and HCC. METTL5 expression showed positive correlation with the infiltration degrees of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophil, and dendritic cells in HCC. Accumulative evidence suggested that the interplay between neutrophils and circulating tumor cells could facilitate distant metastasis of tumor cells24, and macrophages were proven to enhance tumor cell migration and invasion by promoting the EMT process25. Our data revealed that METTL5 expression had an intimate link with the majority of the examined marker genes of B cells, CD8+ T cells, M1 macrophage, neutrophil, dendritic cell, Th1 cells, Th2 cells, and other known immune cells. Moreover, increased METTL5 expression showed a significant correlation with immunostimulators, immunoinhibitors, chemokines, and chemokine receptors. The aforementioned results proved the potential immune function of METTL5 in HCC, implying that its overexpression resulted in HCC by mediating the immune contexture.
Numerous studies identified PD-1 as a principally expressed in CD4+ T cells, and other types of immunocyte and the high abundance of PD-1 contributed to the modulation of immune evasion26,27. The interaction of PD-1 and PD-L1 was regarded as a crucial mechanism for evading anti-tumor immunity28. CTLA-4 served as the immunoinhibitory molecule universally produced by highly activated Treg cells29. PD1/PDL1 and CTLA-4 were indispensable components of immune checkpoint inhibitors; PD1/PDL1 or CTLA-4 checkpoint blockade therapy exhibited potential therapeutic effects on HCC patients30. Our results suggested that METTL5 expression had a positive link with PD1 (PDCD1) and CTLA4 in HCC specimens. The aforementioned results reflected that METTL5 might be used as a promising target in antitumor immunotherapy.
Our study suggested that METTL5 expression in HCC specimens had a close link with kinase networks, such as myosin light chain kinase (MLCK), aurora kinase B (AURKB), ribosomal protein S6 kinase A4 (RPS6KA4) functioned on modulating mitosis, cell cycle checkpoint, DNA damage response, cell growth, and cell proliferation31,32. AURKB produced marked effects on tumorigenesis and genomic instability31. MLCK participates in the regulation of cellular processes for cell adhesion and migration33. MLCK was identified as a critical modulator of mitotic division34, and the decline in MLCK expression or MLCK activity inhibition highly attenuated the proliferation capacity of different kinds of cancerous cells35,36. RPS6KA4 was considered to play the cancer-promoting effects of HCC32.
MiRNAs refer to short noncoding RNAs (20–24 nucleotides) that serve as key molecular components of post-transcriptional regulation in controlling gene expressions to facilitate human oncogenic transformation37. Our results demonstrated several miRNAs with positive link with METTL5, and accumulative evidence supported that MIR-127 and MIR-423 could act as potential diagnostic and prognostic biomarkers of HCC, respectively38,39. The hyperactivation of MIR-127 regulates NF-κB signaling to inhibit HCC cell proliferation40. miR-423 was discovered to contribute to HCC progression by modulating the BP of cell growth and cell cycle41. MIR-510 also participated in tumor development42. The aforementioned results implied that the dysregulation of these miRNAs led to the initiation and progression of HCC.
Several limitations existed in our study. Firstly, most of our data were principally extracted on the basis of online platform databases. Secondly, there were few data from public databases, which were utilized to analyze the relationship between chronic liver diseases and METTL5 expression, so we couldn’t further investigated whether METTL5 expression contributed to HCC occurrence and progression by affecting the process of chronic liver diseases. Hence, further experimental validation must be conducted in subsequent studies. Finally, the assessment of “low” and “high” METTL5 expression was not elaborated in these online public databases, a more precise analysis should be conducted if a detailed and accurate cut-off level of METTL5 expression is set.
Conclusions
METTL5 expressions were markedly upregulated among HCC patients. Increased METTL5 expression had detrimental effects on the survival of HCC patients. METTL5 overexpression affects the carcinogenic effect by taking part in the regulation of tumor immunity. Thus, targeting METTL5 might shed new light on the improvement of immunotherapy effectiveness.
Materials and methods
GEPIA analysis
GEPIA (http://gepia.cancer-pku.cn/) obtained the gene expression and clinical data of malignancy samples from the TCGA databases43. We used GEPIA to examine the METTL5 gene expression among multiple types of malignancies. We also assessed the effects of METTL5 expression on the clinical outcomes of liver HCC patients through the “Survival” module. Statistical significance was represented by a cutoff value of p < 0.05.
UALCAN
UALCAN (http://ualcan.path.uab.edu/) provided the extensive expression information of mRNA, promoter methylation, and protein and survival parameters of multiple malignancy type samples from the TCGA database44. The original expression data of the METTL5 gene, transcript, protein, and methylation in cancer were compared with healthy samples, and the prognostic effect of METTL5 was evaluated through the UALCAN database. Statistical significance was set as a p-value < 0.05.
Kaplan–Meier (KM) Plotter
The KM plotter (http://kmplot.com/analysis/)45 was applied to estimate the effects of METTL5 genes on the survival outcomes among liver cancer patients. Statistical significance was set as a p-value < 0.05.
HPA
HPA databases46 were used to illustrate the effect of METTL5 protein on the survival outcomes of liver cancer patients. A p-value < 0.05 was considered statistically significant.
cBioPortal
cBioPortal (http://www.cbioportal.org/)47 was utilized to visualize the frequency of the genetic alteration of METTL5 in the HCC samples via OncoPrint module.
LinkedOmics
Various analyses of functional enrichment, kinase-target enrichment, and miRNA-target enrichment were conducted by analyzing cancer-associated datasets through the LinkedOmics websites (http://linkedomics.org/admin.php)48. The meta p-value was analyzed by utilizing “RankCriteria”, the “Minimum Number of Genes Size” was set as 3 and the “Simulations” was set as 500.
Metascape
Metascape (http://metascape.org/gp/index.html) was comprehensively used to generate interactive METTL5 genes49.
GeneMANIA
Protein–protein interaction (PPI) networks were constructed through the publicly available platform of GeneMANIA (http://genemania.org/)50. Interactive METTL5 genes derived from the Metascape dababase were inputted into the GeneMANIA for the assessment of gene functions.
TIMER
TIMER (https://cistrome.shinyapps.io/timer/) was applied to systematically assess the correlation of METTL5 with immune cell infiltration in various malignancies and evaluate the impact of immune infiltration on survival outcomes51. The relationship between METTL5 expressions and the levels of immune cell infiltration in LIHC were assessed by utilizing the “Gene” module. The correlation between the copy number alterations of METTL5 and the levels of immune cell infiltration through the “SCNA” module.
TISIDB
TISIDB (http://cis.hku.hk/TISIDB/index.php)52 was applied to explore the interaction between human tumors and immunoregulatory factor for cancer immunology research.
Ethics approval and consent to participate
Ethics approval from the Institutional Review Board and written informed consent from eligible patients were not required because the data are publicly accessible and patient’s private information was deleted.
Data availability
Publicly available datasets were analyzed during the present study. This data can be found here: http://gepia.cancer-pku.cn/.
References
Villanueva, A. Hepatocellular carcinoma. N. Engl. J. Med. 380, 1450–1462 (2019).
Ferlay, J. et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359-386 (2015).
Aldrighetti, L. et al. Liver resection with portal vein thrombectomy for hepatocellular carcinoma with vascular invasion. Ann. Surg. Oncol. 16, 1254 (2009).
Allemani, C. et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): Analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 391, 1023–1075 (2018).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Natchiar, S. K., Myasnikov, A. G., Kratzat, H., Hazemann, I. & Klaholz, B. P. Visualization of chemical modifications in the human 80S ribosome structure. Nature 551, 472–477 (2017).
Rong, B. et al. Ribosome 18S m(6)A methyltransferase METTL5 promotes translation initiation and breast cancer cell growth. Cell Rep. 33, 108544 (2020).
Sun, S. et al. Construction and comprehensive analyses of a METTL5-associated prognostic signature with immune implication in lung adenocarcinomas. Front. Genet. 11, 617174 (2020).
Yan, X., Zhao, X., Yan, Q., Wang, Y. & Zhang, C. Analysis of the role of METTL5 as a hub gene in lung adenocarcinoma based on a weighted gene co-expression network. Math. Biosci. Eng. 18, 6608–6619 (2021).
Viveiros, P., Riaz, A., Lewandowski, R. J. & Mahalingam, D. Current state of liver-directed therapies and combinatory approaches with systemic therapy in hepatocellular carcinoma (HCC). Cancers (Basel) 11, 258 (2019).
Ding, B. et al. In silico analysis excavates potential biomarkers by constructing miRNA-mRNA networks between non-cirrhotic HCC and cirrhotic HCC. Cancer Cell Int. 19, 186 (2019).
Chen, Y. & Tian, Z. HBV-induced immune imbalance in the development of HCC. Front. Immunol. 10, 2048 (2019).
Famularo, S. et al. Recurrence patterns after anatomic or parenchyma-sparing liver resection for hepatocarcinoma in a western population of cirrhotic patients. Ann. Surg. Oncol. 25, 3974–3981 (2018).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011).
Pelletier, J., Thomas, G. & Volarević, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 18, 51–63 (2018).
Maden, B. E. Identification of the locations of the methyl groups in 18 S ribosomal RNA from Xenopus laevis and man. J. Mol. Biol. 189, 681–699 (1986).
van Tran, N. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47, 7719–7733 (2019).
Xu, W., Liu, S., Zhang, G., Liu, J. & Cao, G. Knockdown of METTL5 inhibits the Myc pathway to downregulate PD-L1 expression and inhibits immune escape of hepatocellular carcinoma cells. J. Chemother. 2022, 1–10 (2022).
Huang, H. et al. Ribosome 18S m(6)A methyltransferase METTL5 promotes pancreatic cancer progression by modulating c-Myc translation. Int. J. Oncol. 60, 1 (2022).
Peng, H. et al. N(6)-methyladenosine (m(6)A) in 18S rRNA promotes fatty acid metabolism and oncogenic transformation. Nat. Metab. 4, 1041–1054 (2022).
Liu, X., Ma, H., Ma, L., Li, K. & Kang, Y. The potential role of methyltransferase-like 5 in deficient mismatch repair of uterine corpus endometrial carcinoma. Bioengineered 13, 5525–5536 (2022).
Rohr-Udilova, N. et al. Deviations of the immune cell landscape between healthy liver and hepatocellular carcinoma. Sci. Rep. 8, 6220 (2018).
Ding, W. et al. Prognostic value of tumor-infiltrating lymphocytes in hepatocellular carcinoma: A meta-analysis. Med. (Baltim.) 97, e13301 (2018).
Mizuno, R. et al. The role of tumor-associated neutrophils in colorectal cancer. Int. J. Mol. Sci. 20, 529 (2019).
Wei, C. et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol. Cancer 18, 64 (2019).
Agata, Y. et al. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int. Immunol. 8, 765–772 (1996).
Keir, M. E., Butte, M. J., Freeman, G. J. & Sharpe, A. H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 26, 677–704 (2008).
Yamamoto, R. et al. PD-1-PD-1 ligand interaction contributes to immunosuppressive microenvironment of Hodgkin lymphoma. Blood 111, 3220–3224 (2008).
Peggs, K. S., Quezada, S. A., Chambers, C. A., Korman, A. J. & Allison, J. P. Blockade of CTLA-4 on both effector and regulatory T cell compartments contributes to the antitumor activity of anti-CTLA-4 antibodies. J. Exp. Med. 206, 1717–1725 (2009).
Ouyang, T., Kan, X. & Zheng, C. Immune checkpoint inhibitors for advanced hepatocellular carcinoma: Monotherapies and combined therapies. Front. Oncol. 12, 898964 (2022).
Marima, R., Hull, R., Penny, C. & Dlamini, Z. Mitotic syndicates Aurora Kinase B (AURKB) and mitotic arrest deficient 2 like 2 (MAD2L2) in cohorts of DNA damage response (DDR) and tumorigenesis. Mutat. Res. Rev. Mutat. Res. 787, 108376 (2021).
Lu, Y. et al. Overexpression of ribosomal protein S6 kinase A4 (RPS6KA4) predicts a poor prognosis in hepatocellular carcinoma patients: A study based on TCGA samples. Comb. Chem. High Throughput Screen 25, 2165–2179 (2022).
Webb, D. J. et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6, 154–161 (2004).
Dulyaninova, N. G., Patskovsky, Y. V. & Bresnick, A. R. The N-terminus of the long MLCK induces a disruption in normal spindle morphology and metaphase arrest. J. Cell Sci. 117, 1481–1493 (2004).
Zhou, X. et al. Myosin light-chain kinase contributes to the proliferation and migration of breast cancer cells through cross-talk with activated ERK1/2. Cancer Lett. 270, 312–327 (2008).
Zou, D. B. et al. Melatonin inhibits the migration of colon cancer RKO cells by down-regulating myosin light chain kinase expression through cross-talk with p38 MAPK. Asian Pac. J. Cancer Prev. 16, 5835–5842 (2015).
Hayes, J., Peruzzi, P. P. & Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 20, 460–469 (2014).
Qin, L. et al. Integrated analysis of clinical significance and functional involvement of microRNAs in hepatocellular carcinoma. J. Cell Physiol. 234, 23581–23595 (2019).
An, Y. et al. Novel serum microRNAs panel on the diagnostic and prognostic implications of hepatocellular carcinoma. World J. Gastroenterol. 24, 2596–2604 (2018).
Huan, L. et al. MicroRNA-127-5p targets the biliverdin reductase B/nuclear factor-κB pathway to suppress cell growth in hepatocellular carcinoma cells. Cancer Sci. 107, 258–266 (2016).
Lin, J. et al. MicroRNA-423 promotes cell growth and regulates G(1)/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis 32, 1641–1647 (2011).
Chen, D. et al. Downregulated microRNA-510-5p acts as a tumor suppressor in renal cell carcinoma. Mol. Med. Rep. 12, 3061–3066 (2015).
Tang, Z. et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 (2017).
Chandrashekar, D. S. et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19, 649–658 (2017).
Lánczky, A. & Győrffy, B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 23, e27633 (2021).
Asplund, A., Edqvist, P. H., Schwenk, J. M. & Pontén, F. Antibodies for profiling the human proteome—The Human Protein Atlas as a resource for cancer research. Proteomics 12, 2067–2077 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, pl1 (2013).
Vasaikar, S. V., Straub, P., Wang, J. & Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 46, D956–D963 (2018).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Warde-Farley, D. et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214-220 (2010).
Li, T. et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77, e108–e110 (2017).
Ru, B. et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 35, 4200–4202 (2019).
Author information
Authors and Affiliations
Contributions
L.W. and J.L.P. conceived and designed the study, J.L.P. and L.W. collected the data. J.L.P. and L.W. performed the statistical analysis. J.L.P. and L.W. was responsible for drawing the figures. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, L., Peng, Jl. METTL5 serves as a diagnostic and prognostic biomarker in hepatocellular carcinoma by influencing the immune microenvironment. Sci Rep 13, 10755 (2023). https://doi.org/10.1038/s41598-023-37807-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-37807-5
This article is cited by
-
Recent advances of m6A methylation in skeletal system disease
Journal of Translational Medicine (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.