Abstract
Intracellular protein–protein interactions in aberrant signaling pathways have emerged as a prime target in several diseases, particularly cancer. Since many protein–protein interactions are mediated by rather flat surfaces, they can typically not be interrupted by small molecules as they require cavities for binding. Therefore, protein drugs might be developed to compete with undesired interactions. However, proteins in general are not able to translocate from the extracellular side to the cytosolic target site by themselves, and thus an efficient protein translocation system, ideally combining efficient translocation with receptor specificity, is in high demand. Anthrax toxin, the tripartite holotoxin of Bacillus anthracis, is one of the best studied bacterial protein toxins and has proven to be a suitable candidate for cell-specific translocation of cargoes in vitro and in vivo. Our group recently developed a retargeted protective antigen (PA) variant fused to different Designed Ankyrin Repeat Proteins (DARPins) to achieve receptor specificity, and we incorporated a receptor domain to stabilize the prepore and prevent cell lysis. This strategy had been shown to deliver high amounts of cargo DARPins fused behind the N-terminal 254 amino acids of Lethal Factor (LFN). Here, we established a cytosolic binding assay, demonstrating the ability of DARPins to refold in the cytosol and bind their target after been translocated by PA.
Similar content being viewed by others
Introduction
Intracellular protein–protein interactions in aberrant signaling pathways have emerged as a prime target in several diseases, particularly cancer1,2,3. Direct cell-specific delivery of highly specific inhibitory molecules would provide an efficient and selective way of targeting only aberrant pathways within a cell in a desired tissue. Most therapies today rely on the inhibitory function of cell-permeable small molecules4,5. Those, however, cannot be made cell-specific and many protein–protein interaction surfaces are large, rather flat and hydrophobic, lacking a binding pocket for small molecules, and thus leaving many protein–protein interaction surfaces undruggable6,7.
Advances in the generation of binding molecules based on alternative binding scaffolds, such as Designed Ankyrin Repeat Proteins (DARPins), have allowed the efficient generation of small binding proteins against virtually any target8. DARPins, unlike antibodies, do not require disulfides for stability, and have been demonstrated to fold well when expressed in the cytoplasm of many cells8,9,10. Since proteins, however, are not able to translocate from the extracellular side to the cytosolic target site by themselves, an efficient protein translocation system, ideally combining efficient translocation with receptor specificity, is in high demand. Bacterial protein toxins have naturally evolved to translocate their toxic cargo protein to the cytosol in a cell-specific manner6,11. Our group and others have adapted such toxins to deliver non-native cargoes to cells expressing various receptors11,12,13,14,15,16,17.
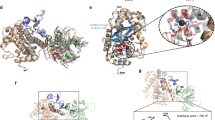
Anthrax toxin, the tripartite holotoxin of Bacillus anthracis, is one of the best studied bacterial protein toxins and has proven to be a suitable candidate for cell-specific translocation of cargoes alternative to its own toxic component in vitro and in vivo12,18,19,20. Our group recently developed a retargeted protective antigen (PA) variant which is fused to a retargeting DARPin binding to the receptor of choice, here EpCAM bound by the DARPin Ac2, to achieve receptor specificity, and on incorporating a receptor domain to stabilize the prepore and prevent cell lysis. Using the translocation domain of anthrax toxin, this system was shown to be able to deliver high amounts of cargoes19,21. Since DARPins can be easily selected to bind to virtually any target and since they have been shown to be effective within the cytosol, they have been used by us and others as alternative cargo molecules by fusing the cargo DARPin behind the N-terminal 254 amino acids of LF (LFN). Since LFN and DARPin are flexibly linked, it is reasonable to assume that the DARPin retains its binding characteristics in this context8,19,22. The two components of the transport system are depicted in Fig. 1.
(a) Schematic diagram and structural model of pore component, consisting of protective antigen (red), a receptor domain to stabilize it and prevent premature activation (green) and the retargeting DARPin (blue), providing cell specificity. (b) Schematic diagram and structural model of translocated payload, consisting of N-terminal 254 residues of lethal factor (green), which interacts with protective antigen, and fused DARPin (blue), which binds to a cytosolic target.
Cargo molecules, translocated via anthrax toxin, have to unfold to penetrate through the narrow PA-channel and once in the cytosol, they must refold to bind to their cytosolic target23. Previously, DARPin translocation has been demonstrated via the BirA assay, a western blot-based assay that shows biotinylation of the cargo DARPin on an avi tag by BirA, a biotin ligase from E. coli, which has been stably expressed in the cytosol of the mammalian cells under study24. As an alternative readout, the toxicity by co-delivering diphtheria toxin (DTA) was reported22. Both methods conclusively show the presence of the cargo but not its function, and a direct readout for cytosolic refolding and activity of the DARPin has been missing19,24.
Since DARPins must be in the folded state to bind their target8, we established here an ELISA-based cytosolic binding assay and thereby prove the ability of DARPins to refold in the cytosol and bind their cytosolic target after having been translocated through the PA channel.
Results
Previously, we showed that the delivery of various DARPin cargoes to the cytosol of Flp-In 293-EpCAM-BirA cells is dependent on thermodynamic stability, probably due to the required unfolding step when traversing the pore. We carried out these experiments using an assay employing cells stably overexpressing the epithelial cell adhesion molecule (EpCAM) as the receptor to be targeted, and E. coli biotin ligase (BirA), stably expressed in the cytosol, to biotinylate the avi tag attached to the cargo once it has reached the cytosol25. We thus confirmed the presence of cytosolic DARPin cargoes, and thereby the successful translocation, by utilizing the BirA assay previously developed24. However, the BirA assay cannot detect whether the translocated DARPin has refolded and is able to bind to its cytosolic target.
Here, we set out to perform a pulldown assay detected by ELISA to confirm cytosolic binding of the translocated DARPins to their cytosolic targets. Since DARPins need to be in the folded state for binding8, we can use this assay to deduce their correct folding. We therefore established a digitonin-based extraction protocol of the cytosolic fraction of targeted cells with a subsequent pulldown of the translocated LFN-DARPin cargo22. The cargo additionally carries both an HA-tag and an avi-tag fused to its C-terminus (Fig. 2). Therefore, cells were incubated with respective delivery components (protective antigen targeting EpCAM and cargo DARPin fused to LFN and the detection tags). Cells are then harvested, and the cytosolic fraction extracted with digitonin extraction buffer (Fig. 2a). To pull down the translocated LFN-DARPin cargo, biotinylated via the resident BirA, streptavidin-carrying magnetic beads were incubated with the cytosolic fraction. After removal of the unbound fraction, the pulldown fraction was used for further analysis (Fig. 2b).
Scheme of anthrax toxin-based delivery with subsequent pulldown to test successful refolding. (a) LFN-DARPin-HA-tag-avi-tag gets delivered to the cytosol and biotinylated by cytosolically present BirA. Protein stuck in the endosome does not get biotinylated. Cells are harvested, extracted with digitonin, and the cytosolic fraction is separated from the residual pellet by centrifugation. (b) The cytosolic fraction is added to streptavidin magnetic beads and the unbound fraction is removed. The pulldown fraction is then analyzed further via western blotting and ELISA.
First, we needed to determine a suitable digitonin concentration for cytosolic extraction, which is efficient but leaves endosomal compartments fully intact, as their leakage would distort the determination of location and thus the binding assay. The western blot (Fig. 3a) and the quantification of it (Fig. 3b) show cytosolic fractions of Flp-In 293-EpCAM-BirA cells incubated with increasing concentrations of digitonin. We found that a concentration of 50 µg/mL digitonin or higher is needed for efficient cytosolic extraction. Furthermore, we tested the digitonin incubation time with 50 µg/mL and found no differences between 10–60 min of incubation, enabling us to perform the assay with 10 min incubation on ice (Fig. 3c). Since higher concentrations than 50 µg/mL of digitonin did not tremendously increase the cytosolic extraction, we stained a western blot of the cytosolic fraction extracted with 50 µg/mL digitonin for Rab5A, an established endosomal marker26, and confirmed that 50 µg/mL of digitonin did not permeabilize the endosomal membrane (Fig. 3d).
Western blot analysis of Flp-In 293-EpCAM-BirA for cytosolic extraction with digitonin. (a) Western blot of GAPDH from Flp-In 293-EpCAM-BirA cells, incubated with 10–100 µg/mL digitonin for 10 min, 4 °C. Cytosolic fractions (lane 1–5) were separated from other cellular compartments (lane 7–11) by centrifugation. Cells labelled “total cell lysis” (lane 12) were incubated directly in Laemmli sample buffer. All samples were analyzed via western blot, stained with anti-GAPDH. (b) Quantification of western blot bands from (a). (c) Western blot of GAPDH from Flp-In 293-EpCAM-BirA cells incubated with 50 µg/mL for 10–60 min on ice without shaking (lane 3–5, 10–12) or with shaking at 4 °C (lane 6–8, 13–15). Cytosolic extracts (lane 3–8) and other cellular compartments (lane 10–15) were analyzed and stained with anti-GAPDH. Cells labelled “total cell lysis” were directly incubated in 1 × Laemmli sample buffer. Lanes 2 and 9 were loaded with protein MW marker. (d) Western blot of GAPDH and Rab5A from Flp-In 293-EpCAM-BirA cells incubated in 50 µg/mL digitonin extraction buffer for 10 min and analyzed via western blot, staining for GAPDH (cytosolic fraction) and anti-Rab5A (endosomal fraction). (e) Western blot analysis of HA-tagged DARPins in Flp-In 293-EpCAM-BirA incubated with 50 nm PAwt-sANTXR-Ac2 and 500 nM LFN-NI1C (lanes 2, 6, 11) or LFN-J1/2_2_25 (lanes 3, 7, 12). DARPin cargoes were pulled down from the cytosolic fraction via streptavidin magnetic beads (lanes 5–7) and the beads were treated with Laemmli buffer. The remaining cytosolic fraction after pulldown (lanes 10–12), as well as the residual cell pellet of the digitonin extraction (lanes 1–3) are shown in addition. Lanes 4, 8–9 and 13 were loaded with protein ladder. The western blot was stained with anti-HA-tag for LFN-DARPin detection. High concentrations of digitonin in samples 10–12 lead to a different running behavior of analyzed samples, resulting in protein bands appearing at lower molecular weight.
We then performed a delivery assay, based on the previously established protocol of the BirA assay24; however, instead of generating whole cell lysates, we extracted the cytosolic fraction with the digitonin extraction buffer as described above and separated all other cellular compartments, including endosomes, by centrifugation. The cytosolic fraction was then incubated with streptavidin magnetic beads for 2 h for LFN-DARPin cargo pulldown (Fig. 2). To confirm the presence of successfully delivered LFN-DARPin cargoes in the pulled-down fraction constituting the digitonin extract of the cytosol, we tested two DARPins, NI1C and J1/2_2_25, which previously showed highly efficient cytosolic delivery25. DARPin NI1C is a consensus designed DARPin with a single internal repeat without target binding27, J1/2_2_25 is a target selected NI2C DARPin binding to JNK19.
The pulldown of the biotinylated DARPins by streptavidin magnetic beads from the cytosol of targeted cells confirmed the presence of both DARPins in western blots by detecting their HA tag (Fig. 3e), with the DARPin NI1C being delivered in much greater quantities than J1/2_2_25. This higher delivery of NI1C, found here with the digitonin-based solubilization of the cytosolic fraction, is consistent with the previously found results based on total lysis and detection via avi-tag biotinylation (BirA assay)25. It is also consistent with an ELISA-based detection of the HA-tag of pulled down LFN-DARPins (Supplementary Fig. S1). Therefore, all three quantification methods confirmed the highly efficient delivery of LFN-NI1C, being 2.8–4.5 × more efficiently delivered to the cytosol than LFN-J1/2_2_25 (Supplementary Fig. S1c). Conversely, this agreement shows that all assays are sufficiently reliable for quantification of translocation.
In order to detect interaction of DARPin J1/2_2_25, we first stained the western blot in Fig. 3 for its target JNK1, but we could not detect JNK1 pulled down with J1/2_2_25 on this blot (Supplementary Fig. S2). We propose that the signal for JNK1 is below the detection limit on a western blot, taking the weak signal of the western blot band of the LFN-J1/2_2_25 construct itself into account and the respective quantification of LFN-DARPin cargo translocated to the cytosol (Supplementary Fig. S3). We therefore set out to analyze the pulled down fraction via a more sensitive ELISA-based assay, similar to a recently published delivery assay28. Following the pulldown, we incubated the beads, containing the biotinylated DARPin bound to streptavidin, with primary antibody against the DARPin’s target and detected this antibody with an HRP-labelled secondary antibody (Fig. 4a). We measured the absorbance at 450 nm of the LFN-JNK1 binding construct (LFN-J1/2_2_25) and the non-binding LFN-NI1C as a control. A cells-only control was included to determine the background absorbance for normalization, while the LFN fused to the binding DARPin, i.e. J1/2_2_25 was set as the maximum absorbance.
Pulldown ELISA of LFN-J1/2_2_25 and LFN-008_C6. (a) Assay scheme of pulldown ELISA. LFN-DARPin is pulled down with streptavidin magnetic beads via the avi-tag biotinylated by cytoplasmic BirA. An anti-JNK1 (b) or anti-BCL2 (c) primary antibody and an HRP-labelled secondary antibody are used for quantification of pulled down target. (b, c) Delivered LFN-J1/2_2_25 (b) and LFN-008_C6 (c) show a higher absorbance compared to a non-binding DARPin control when stained with anti-JNK1 (b) or anti-BCL2 (c) antibody. Values were normalized between 0 (cells only) and 1 (binding DARPin). Statistical analysis: two-tailed unpaired t test; *p < 0.05; **p < 0.01. Error bars reflect SEM (n = 3).
The JNK1-targeted DARPin showed increased absorbance in ELISA, compared to the non-binding control, confirming the pulldown of the target, JNK1, and thus it can be reasonably concluded that the DARPin successfully refolded and thus binds its target in the cytosol (Fig. 4b). We then tested a second DARPin-target combination to verify these results. We chose the BCL2 binder 008_C6, which has previously shown cytosolic activity and efficient cytosolic translocation25,29. Similar to the LFN-J1/2_2_25, the LFN-008_C6 construct showed an increase in absorbance compared to the non-binding control (Fig. 4c), consistent with its successful cytosolic refolding and binding to its cognate target BCL2.
Discussion
In this study, we show for the first time the successful DARPin cargo refolding after translocation to the cytosol with PA via a cytosolic binding assay that is dependent on the folding of the DARPins. We delivered two different target-selected DARPins to the cytosol and tested their capability of binding their cytosolic target in an indirect pulldown assay, which requires the interaction of the folded DARPin and the folded target, as demonstrated from the crystal structures of the complexes30,31. With this assay, we indirectly confirmed successful refolding based on the reasonable assumption that the pulldown only occurs for cytosolically active DARPins, as only folded DARPins bind their targets, and DARPins fold cooperatively8.
We first developed a cytosolic extraction protocol based on digitonin, which leaves the endosomal membranes intact and does not solubilize endosomal markers, and found similar extraction conditions as previously published in other delivery assays22. We then proved in an ELISA-based assay that DARPins are capable of binding their target in the cytosol after being translocated by anthrax toxin, confirming the concept of DARPin-based cytosolic targeting. The assay developed here is very sensitive, but the particular format used requires the use of BirA-expressing cells. While the current format of the assay does not allow for a direct measurement of the binding-active fraction in the cytosol, we assume, due to the beneficial biophysical characteristics of the DARPins8, that the level of active DARPins in the cytosol is high.
Previously, our group had shown that DARPins are active in cells in which they have been cytosolically expressed, can bind and inhibit their targets9,10. However, DARPin activity after cytosolic translocation via anthrax toxin, comprising unfolding, traversing a narrow pore and refolding and binding to their targets, had not yet been directly shown.
Several other LFN-cargoes have been successfully tested for their cytosolic activity. These cargoes were mostly based on other naturally occurring bacterial protein toxins, e.g., Pseudomonas exotoxin A or diphtheria toxin A chain and they were tested for their cellular toxicity13,32,33. However, the toxic function is a limitation for a generic assay and toxicity might also arise from other sources. Other alternative protein cargoes also showed cytosolic activity after translocation by anthrax toxin13,34,35,36,37,38. Nonetheless, direct interaction is the most generically useful assay for a wide variety of targets. In line with these results, it has been shown here that DARPins belong to the proteins that can be delivered to the cytosol via anthrax toxin and regain their cytosolic activity after translocation and refolding the cytosol.
Conclusion
Here, we confirmed the cytosolic activity of two DARPins after cytosolic translocation with anthrax toxin. With the highly beneficial biochemical characteristics of DARPins we now generated an anthrax toxin-based targeting platform comprising a retargeted PA and LFN-cargo fusions, allowing us to target any cytosolic protein with two layers of specificity, one for the cell surface receptor, the other for the cytosolic target.
Methods
Cell lines
Flp-In 293 cells, stably overexpressing EpCAM and BirA (Flp-In 293-EpCAM-BirA)19 were cultured using DMEM. The medium was supplemented with 10% fetal calf serum and 100 IU/mL penicillin and 100 µg/mL streptomycin.
Protein expression and purification
The production of His6-MBP-PAwt-sANTXR-Ac2 and His6-MBP-LFN-DARPin-HA-tag-avi-tag cargo constructs has been described before19,21. Briefly, protective antigen and lethal factor fusion proteins were expressed in soluble form in the cytoplasm of E. coli BL21. Purification was achieved via immobilized metal ion affinity chromatography (IMAC) for all constructs. Fusions between MBP and LF-DARPin constructs were cleaved with TEV protease and further purified via reverse IMAC and size-exclusion chromatography. Fusion proteins containing protective antigen were purified directly via size-exclusion chromatography after IMAC19,21.
Delivery of LFN-DARPins
For cytosolic delivery of LFN-DARPin constructs, the first steps of the BirA assay, previously developed in our lab, were performed up to the cell harvest after delivery24. Briefly, 3 × 105 Flp-In 293-EpCAM-BirA cells were seeded in 24-well plates 24 h prior to a 4 h incubation of cells with delivery components. Delivery components, PAwt-sANTXR-Ac2 (50 nM) and LFN-DARPin (500 nM), were premixed in DMEM medium containing 10% fetal calf serum, 100 IU/mL penicillin and 100 µg/mL streptomycin, 100 µM biotin, and 50 µM MG132. The medium in which cells were seeded in was replaced by the medium containing the delivery components.
Cytosolic extraction by digitonin
After 4 h incubation of cells with PAwt-sANTXR-Ac2 and LFN-DARPin, cells were washed with PBS and detached with trypsin–EDTA. For cytosolic extraction, cell pellets were washed with PBS and suspended in 50 µL digitonin extraction buffer (PBS, supplemented with 1% BSA, 50 µg/mL digitonin, 20 µM avi-tag peptide, 0.1 mg/mL DNase, 0.4 mM 4-(2-aminoethyl)benzolsulfonyl fluoride (AEBSF), 10 mM Leupeptin, 1 mM Pepstatin-A), incubated for 10 min on ice, and then centrifuged for 5 min at 16,000g at 4 °C. Supernatants were collected for further processing, i.e. western blotting, streptavidin pulldown and target detection ELISA.
LFN-DARPin pulldown via streptavidin magnetic beads
Streptavidin MagBeads (GenScript) were washed 3 × with PBS containing 1% BSA (PBS-B). 10 µL of beads was added to each cytosolic fraction and rotated for 2 h at 4 °C. Then, beads were washed 3 × with PBS-B and either prepared for western blotting or further used for pulldown ELISA.
Western blot
Cytosolic fractions of the digitonin extraction were analyzed by adding 4 × Laemmli sample buffer (Bio-Rad). Pelleted fractions of digitonin extraction were resuspended in 20 µL 1 × Laemmli sample buffer. Streptavidin MagBeads were resuspended after pulldown in 20 µL of 1 × Laemmli sample buffer, the supernatant of the bead incubation was diluted with 4 × Laemmli sample buffer. Translocated LFN-DARPin amounts were quantified via the BirA assay, by titrating known concentrations of fully biotinylated avi-tagged MBP (cytosolic uptake) or LFN-J1/2_25-HA-tag-avi-tag (total cellular uptake) on the same blot.
All samples were heated for 10 min at 96 °C. Samples were separated by SDS-PAGE (4–20% Mini-PROTEAN TGX Stain Free Gels, Bio-Rad) and further transferred to PVDF-FL membranes. Membranes were blocked (4 °C, ON) with 1 × Casein Blocking Buffer (Sigma), incubated for 1 h at RT in PBS containing 0.01% Tween 20 (PBS-T) with primary antibodies, rabbit anti-HA (1:1000; Sigma), mouse anti-JNK1 (F-3) (1:1000; SantaCruz), mouse anti-GAPDH (1:1000; SantaCruz) or rabbit anti-Rab5 (1:1000, Cell Signaling). Antibody staining was followed by 3 × 5 min washing with PBS-T and secondary antibody staining with goat anti-rabbit IgG AF680 (1:5000; Invitrogen) or sheep anti-mouse IgG DyLight 800 (1:5000; Rockland) with a final 3 × 5 min washing step before imaging. Western blots were imaged with a LI-COR Odyssey CLx instrument and band intensities were quantified using the Image Studio Lite (LI-COR). Uncropped blots are shown in Supplementary Fig. S4 and summarized in Supplementary Table ST1.
Pulldown ELISA
After pulldown, beads were incubated with rabbit anti-HA (Sigma), mouse anti-BCL2 (Cell Signaling) or mouse anti-JNK1 (F3) (SantaCruz) antibodies 1:200 overnight at 4 °C while rotating. Then, beads were washed 3 × with PBS-B and incubated with horseradish peroxidase coupled goat anti-mouse antibodies (Pierce) or goat anti-rabbit antibodies (Cell Signaling Technology) 1:5000 for 2 h at 4 °C while rotating. Beads were then washed 3 × with PBS-B and incubated with 75 µL 1-Step Ultra TMB-ELISA Substrate Solution (Thermo Scientific) for 15–30 min at RT while shaking. 75 µL of 2 M sulfuric acid was added and the absorbance at 450 nm was measured on a Tecan plate reader.
Data availability
All data is included in this manuscript and its supplementary files.
Abbreviations
- PA:
-
Protective antigen
- DARPin(s):
-
Designed ankyrin repeat protein(s)
- LFN :
-
Lethal factor 1–254
- EpCAM:
-
Epithelial cell adhesion molecule
- BirA:
-
Biotin ligase derived from E. coli
- AEBSF:
-
4-(2-Aminoethyl) benzenesulfonyl fluoride
References
Hopkins, A. L. & Groom, C. R. The druggable genome. Nat. Rev. Drug Discov. 1, 727–730 (2002).
Lage, K. Protein–protein interactions and genetic diseases: The interactome. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 1842, 1971–1980 (2014).
Ivanov, A. A., Khuri, F. R. & Fu, H. Targeting protein–protein interactions as an anticancer strategy. Trends Pharmacol. Sci. 34, 393–400 (2013).
Mosquera, J., García, I. & Liz-Marzán, L. M. Cellular uptake of nanoparticles versus small molecules: A matter of size. Acc. Chem. Res. 51, 2305–2313 (2018).
Huck, B. R., Kötzner, L. & Urbahns, K. Small molecules drive big improvements in immuno-oncology therapies. Angew. Chem. Int. Ed. 57, 4412–4428 (2018).
Deprey, K., Becker, L., Kritzer, J. & Plückthun, A. Trapped! A critical evaluation of methods for measuring total cellular uptake versus cytosolic localization. Bioconjug. Chem. 30, 1006–1027 (2019).
Scott, D. E., Bayly, A. R., Abell, C. & Skidmore, J. Small molecules, big targets: Drug discovery faces the protein–protein interaction challenge. Nat. Rev. Drug Discov. 15, 533–550 (2016).
Plückthun, A. Designed ankyrin repeat proteins (DARPins): Binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 55, 489–511 (2015).
Parizek, P. et al. Designed ankyrin repeat proteins (DARPins) as novel isoform-specific intracellular inhibitors of c-Jun N-terminal kinases. ACS Chem. Biol. 7, 1356–1366 (2012).
Kummer, L. et al. Structural and functional analysis of phosphorylation-specific binders of the kinase ERK from designed ankyrin repeat protein libraries. Proc. Natl. Acad. Sci. 109, E2248–E2257 (2012).
Beilhartz, G. L., Sugiman-Marangos, S. N. & Melnyk, R. A. Repurposing bacterial toxins for intracellular delivery of therapeutic proteins. Biochem. Pharmacol. 142, 13–20 (2017).
Bachran, C. & Leppla, S. Tumor targeting and drug delivery by anthrax toxin. Toxins 8, 197 (2016).
Rabideau, A. E. & Pentelute, B. L. Delivery of non-native cargo into mammalian cells using anthrax lethal toxin. ACS Chem. Biol. 11, 1490–1501 (2016).
Beer, L.-A. et al. The binary toxin CDT of Clostridium difficile as a tool for intracellular delivery of bacterial glucosyltransferase domains. Toxins 10, 225 (2018).
Mechaly, A., McCluskey, A. J. & Collier, R. J. Changing the receptor specificity of anthrax toxin. MBio 3, e00088-12 (2012).
McCluskey, A. J., Olive, A. J., Starnbach, M. N. & Collier, R. J. Targeting HER2-positive cancer cells with receptor-redirected anthrax protective antigen. Mol. Oncol. 7, 440–451 (2013).
Roderer, D., Schubert, E., Sitsel, O. & Raunser, S. Towards the application of Tc toxins as a universal protein translocation system. Nat. Commun. 10, 5263 (2019).
Lu, Z. et al. IgG-engineered protective antigen for cytosolic delivery of proteins into cancer cells. ACS Cent. Sci. 7, 365–378 (2021).
Verdurmen, W. P. R., Luginbühl, M., Honegger, A. & Plückthun, A. Efficient cell-specific uptake of binding proteins into the cytoplasm through engineered modular transport systems. J. Control. Release 200, 13–22 (2015).
Friebe, S., van der Goot, F. & Bürgi, J. The ins and outs of anthrax toxin. Toxins 8, 69 (2016).
Becker, L., Verdurmen, W. P. R. & Plückthun, A. Reengineering anthrax toxin protective antigen for improved receptor-specific protein delivery. BMC Biol. 18, 100 (2020).
Liao, X., Rabideau, A. E. & Pentelute, B. L. Delivery of antibody mimics into mammalian cells via anthrax toxin protective antigen. ChemBioChem 15, 2458–2466 (2014).
Machen, A. J., Fisher, M. T. & Freudenthal, B. D. Anthrax toxin translocation complex reveals insight into the lethal factor unfolding and refolding mechanism. Sci. Rep. 11, 13038 (2021).
Verdurmen, W. P. R., Mazlami, M. & Plückthun, A. A biotin ligase-based assay for the quantification of the cytosolic delivery of therapeutic proteins. Meth. Mol. Biol. 1575, 223–236 (2017).
Becker, L., Singh Badwal, J., Brandl, F., Verdurmen, W. P. R. & Plückthun, A. Thermodynamic stability is a strong predictor for the delivery of DARPins to the cytosol via anthrax toxin. Pharmaceutics 13, 1285 (2021).
Zerial, M. & McBride, H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2, 107–117 (2001).
Devi, V. S. et al. Folding of a designed simple ankyrin repeat protein. Protein Sci. 13, 2864–2870 (2009).
Lucchino, M. et al. Absolute quantification of drug vector delivery to the cytosol. Angew. Chem. Int. Ed. 60, 14824–14830 (2021).
Schilling, J., Schöppe, J. & Plückthun, A. From DARPins to LoopDARPins: Novel LoopDARPin design allows the selection of low picomolar binders in a single round of ribosome display. J. Mol. Biol. 426, 691–721 (2014).
Wu, Y., Honegger, A., Batyuk, A., Mittl, P. R. E. & Plückthun, A. Structural basis for the selective inhibition of c-Jun N-terminal kinase 1 determined by rigid DARPin–DARPin fusions. J. Mol. Biol. 430, 2128–2138 (2018).
Schilling, J., Schöppe, J., Sauer, E. & Plückthun, A. Co-crystallization with conformation-specific designed ankyrin repeat proteins explains the conformational flexibility of BCL-W. J. Mol. Biol. 426, 2346–2362 (2014).
Arora, N., Klimpel, K. R., Singh, Y. & Leppla, S. H. Fusions of anthrax toxin lethal factor to the ADP-ribosylation domain of Pseudomonas exotoxin A are potent cytotoxins which are translocated to the cytosol of mammalian cells. J. Biol. Chem. 267, 15542–15548 (1992).
Arora, N. & Leppla, S. H. Fusions of anthrax toxin lethal factor with shiga toxin and diphtheria toxin enzymatic domains are toxic to mammalian cells. Infect. Immun. 62, 4955–4961 (1994).
Wesche, J., Elliott, J. L., Falnes, P. Ø., Olsnes, S. & Collier, R. J. Characterization of membrane translocation by anthrax protective antigen. Biochemistry 37, 15737–15746 (1998).
Sheahan, K.-L. & Satchell, K. J. F. Inactivation of small Rho GTPases by the multifunctional RTX toxin from Vibrio cholerae. Cell. Microbiol. 9, 1324–1335 (2007).
Antic, I., Biancucci, M. & Satchell, K. J. F. Cytotoxicity of the Vibrio vulnificus MARTX toxin effector DUF5 is linked to the C2A subdomain. Proteins 82, 2643–2656 (2014).
Antic, I., Biancucci, M., Zhu, Y., Gius, D. R. & Satchell, K. J. F. Site-specific processing of Ras and Rap1 Switch I by a MARTX toxin effector domain. Nat. Commun. 6, 7396 (2015).
Cordero, C. L., Kudryashov, D. S., Reisler, E. & Satchell, K. J. F. The actin cross-linking domain of the Vibrio cholerae RTX toxin directly catalyzes the covalent cross-linking of actin. J. Biol. Chem. 281, 32366–32374 (2006).
Funding
This research was funded by the Schweizerische Nationalfonds, grant 310030_192689.
Author information
Authors and Affiliations
Contributions
L.B.: Conceptualization, methodology, formal analysis, investigation, writing—original draft preparation, writing—review and editing, visualization, project administration; A.P.: writing—review and editing, supervision, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Becker, L., Plückthun, A. DARPins bind their cytosolic targets after having been translocated through the protective antigen pore of anthrax toxin. Sci Rep 13, 8048 (2023). https://doi.org/10.1038/s41598-023-34647-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-34647-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.