Abstract
Gastric cancer (GC) is one of the major causes of cancer deaths with 5-year survival ratio of 20%. RNU12 is one of long noncoding RNAs (lncRNAs) regulating the tumor progression. However, how RNU12 affecting GC is not clear. qRT-PCR was utilized for determining the RNU12 expression in cell lines, 113 cases of paired gastric cancer (GC) and their adjacent normal gastric tissues. The biofunction alterations of RNU12 were assessed by its overexpression or knockdown in GC cells. MTT and cloning assay were assayed for the cell proliferation, the flow cytometry for the detection of cell cycle and the wound healing assay (WHA) and transwell invasion assay (TIA) for examining the migration and invasion of cells. The expressions of a set of genes related proliferation and migration were investigated with the Western Blotting (WB). RNA immunoprecipitation (RIP), biotinylated RNA pull-down and dual luciferase reporter tests were used to detect the interactions of RNU12 with miR-575/BLID. The in vivo proliferation and migration ability of RNU12 infected cells were determined in zebrafish system. This study revealed that RNU12 inhibited proliferation, invasion and metastasis by sponging of miR-575 and regulating the downstream BLID and modulated EMT of GC cells. The RNU12/miR-575/BLID axis is likely to be the prognosis biomarkers and drug targets of GC.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is a common type of cancer worldwide, ranking the third of cancer death following lung cancer and liver cancer1. Due to the early stages of GC are clinically silent, the most patients were clinically diagnosed at the advanced stage. For this reason, its 5-year survival rate (SR) < 20%2. Although statistics show a gradual decrease of GC incidence rates, an unexplained increase in GC incidence of younger individuals from developed regions is emerging3. In addition, risk factors for gastric cancer are diverse4; Although the primary cause is considered to directly related to Helicobacter pylori4,5. Previous findings showed that tobacco use, familial predisposition, alcohol consumption, pernicious anemia, high dietary salt consumption, previous gastric surgery, processed meat consumption, age, and low socio-economic status are all associated with the occurrence of GC6,7,8,9,10. The complexity of the GC causes promotes us to understand the molecular mechanisms behind the GC, which could lead to advancements in diagnosis and treatment.
Long noncoding RNAs (lncRNAs) are usually considered to be a length over 200 nt RNA and can not code protein11. LncRNAs feature abundant quantity, diversity, and action mechanisms related with various biological cancer processes, such as carcinogenesis, apoptosis, differentiation, proliferation, invasion, metastasis, etc.12,13. The lncRNAs exert the function of “sponges” chelating miRNAs from target mRNAs to antagonize the biological function possessed by miRNAs as competitive endogenous RNAs (ceRNAs)14. For example, SNHG8 as a lncRNA presents an abnormal upregulation in GC and sponges to the miR-5125-p for promoting GC cell invasion15. LncRNA ZNF667-AS1 sponges miR-1290 and promotes ABLIM1 expression to suppress nasopharyngeal cancer (NPC)16. Some evidence has indicated that closely associations between expression levels of lncRNAs in GC tissues17. Accumulating evidence has shown that lncRNAs play a critical role in the progress of GC.
RNU12 is in the 22q12-q13 chromosomal region, and associated with many diseases, such as psoriatic arthritis (PSA)18, the MOPD I phenotype19,20, CDAGS Syndrome21 and congenital cerebellar ataxia22. Recent study indicates knockdown of lnc-RNU12 can influence cell cycle by altering the expression of protein-coding genes related to the cell cycle and apoptosis in immune T cells23. However, the relationship between RNU12 and GC progression has been poorly studied. Our previous results have discovered that RNU12 was a decreased expression level in GC, suggesting that RNU12 is an essential regulator in suppressing tumor genesis. Here, we explored the function of RNU12 in controlling the tumor growth and progression and found that RNU12, as a sponge of miR-575, reduced miR-575 expression, regulated BLID expression and reduced GC cell proliferation and metastasis.
Results
Characterization of RNU12 in GC and its association with clinicopathological features in GC patients
The real-time PCR detected the expression level of RNU12 in 113 cases of GC and their adjacent tissues. The RNU12 expression in GC tissues was 0.0008 ± 0.428 (log10RQ, RQ = 2−ΔΔCT) while that of the corresponding adjacent tissue was 1.744 ± 0.458 (P = 0.001, Fig. 1A). Similarly, a real-time PCR assay was performed in 1 normal human gastric mucosa cell line (Ges-1) and 4 GC tumor lineage cells (AGS, MKN45, MKN28 and MGC803). The result (Fig. 1B) showed that the much lower RNU 12 expression in the 4 GC tumor cell lines than that in normal gastric cell line, Ges-1.
RNU12 was down-regulated in GC. (A) Expression of RNU12 in 113 paired GC and adjacent tissues; The data is analyzed to conform to a normal distribution and presented as the mean ± SD value (N = 113), y-axis units (log10RQ, RQ = 2−ΔΔCt). (B) Expression of RNU12 in GES-1 and 4 human GC cell lines (AGS, MKN45, MKN28 and MGC803). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
The clinical data demonstrated a negative relationship between RNU12 expression and clinic pathologic feature-lymph node metastasis in GC patients (Table 1, P < 0.05).
Overexpression of RNU12 inhibits GC proliferation, migration, and invasiveness
To determine the way if RNU12 overexpression affected GC behaviors, AGS and MGC803 cell lines were infected with HBLV-NC or HBLV- RNU12-OE lentivirus, respectively. RT-qPCR analysis illustrated the obvious increase in RNU12 expression in RNU12-OE relative to NC in AGS or MGC803 cell lines (Fig. 2A). MTT assay (Fig. 2B,C) and colony assay (Fig. 2D,E) demonstrated the overexpression of RNU12 reduced the cell proliferation and colony formation. In addition, S phase of cell cycle was remarkably decreased in the RNU12 overexpressed AGS or MGC803 cells as compared to control AGS/NC or MGC803/NC cells (Fig. 2F,G). The results of WHA and TIA showed that upregulation of RNU12 weakened the capacities of wound healing (Fig. 2L–O) and the transwell invasiveness (Fig. 2P,Q). Furthermore, the results of RT-qPCR and WB indicated that there were downregulations of development-related genes (CCND1, PCNA), EMT-associated genes (N-cadherin, Vimentin) and anti-apoptotic BCL-2, while E-cadherin was upregulated in AGS/RNU12-OE or MGC803/RNU12-OE cells (Fig. 2H–K,R,S); These findings showed that RNU12 might inhibit the GC cell aggressiveness in vitro (the original blots in Fig. 2I,K,R,S are shown in Supplementary file 1).
RNU12 overexpression inhibited GC cells in terms of the proliferation, migration, and invasion. (A) Real-time PCR on RNU 12 expression in AGS and MGC 803 cells; (B,C) The impact of RNU 12 overexpression on the cell vitality under the measurement of MTT assay; (D,E)The impact of RNU 12 overexpression on cell colony formation; (F,G) The impact of RNU 12 overexpression on cell cycle distribution under FACS; (L–O) The impact of RNU 12 overexpression on cell migration under WHA; (P,Q) The impact of RNU 12 overexpression on cell invasiveness under transwell invasive assay (TIA); (H–K) qRT-PCR and WB detected the expressions regarding proliferation-related-genes of Bcl-2, CCND1, and PCNA; (R,S) The expression exhibited by invasion/metastasis-related-genes, E-cadherin, VIMENTIN and N-cadherin according to WB. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
RNU12 silencing facilitates GC proliferation, migration, and invasiveness
For evaluating whether silencing of RNU12 could enhance GC malignant behaviors, AGS and MGC803 were transfected with HBLV-NC or HBLV-RNU12-SH lentivirus, respectively. Based on RT-qPCR analysis, the above treatments could remarkably decrease the RNU12 expression in RNU12-SH compared with NC in AGS or MGC803 cell lines (Fig. 3A). RNU12 silencing significantly promotes AGS or MGC803 in terms of the proliferation and the colony formation (Fig. 3B–E). In addition, relative to AGS/NC or MGC803/NC, an obvious increase was observed in the number of cells arrested at the S phase of AGS/RNU12-SH or MGC803/RNU12-SH as measured by a Flow Cytometer (Thermo Fisher Scientific, USA), (Fig. 3F,G). Furthermore, downregulation of RNU12 enhanced the wound healing migratory capacity and the transwell invasiveness (Fig. 3L–Q). The results of RT-qPCR and WB showed the upregulation of proliferation-related genes (CCND1, PCNA), EMT-associated genes (N-cadherin,Vimentin) and anti-apoptotic BCL-2, while downregulation of E-cadherin in AGS/RNU12-SH cells (Fig. 3H–K,R,S). The above findings indicated that the ability of RNU12 silencing could enhance the proliferation, migration, and invasiveness of GC cells in vitro (the original blots in Fig. 3I,K,R,S are shown in Supplementary file 2).
RNU12 silencing enhanced the proliferation, migration, and invasiveness of AGS and MGC 803 cells. (A) Real-time PCR on RNU12 expression in the two types of cells; (B,C) The impact of RNU12 silencing on various AGS and MGC 803 cells under the measurement of MTT assay; (D,E) The impact of RNU12 silencing on cell colony formation; (F,G) The impact of RNU12 knockdown on the cell cycle distribution under FACS; (L–O) The impact of RNU12 silencing on cell migration under WHA; (P,Q) The impact of RNU12 silencing on cell invasiveness under TIA; (H–K) qRT-PCR and WB detected the expression exhibited by BCL-2, CCND1, and PCNA; (R,S) WB confirmed the expression regarding EMT-associated genes, E-cadherin, VIMENTIN and N-cadherin. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
RNU12 sponges hsa-miR-575 and regulates BLID
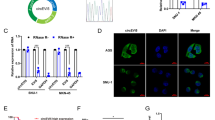
The crosstalk between Marianas and RNU12 in GC cells was detected, as lncRNAs mainly played a role as miRNA spongers24,25. We used the lncRNASNP2 database to conclude that hsa-miR-575 is a potent target of RNU12. Using the TargetScan and miRDB databases, we further found that BLID was also a potent target specific to hsa-miR-575. Predicted binding sites regarding hsa-miR-575 and wt regions of RNU12 and BLID were presented in DNA sequences (Fig. 4A). RIP assay revealed the co-enrichment of RNU12 and hsa-miR-575 in a manner depending on Ago2 (Fig. 4B). RT-qPCR analysis revealed the ability of RNU12 overexpression or silencing to remarkably lower or improve hsa-miR-575 expression in AGS (Fig. 4C,D), respectively. In addition, according to results of RT-qPCR and WB, the expressions of BLID compared to hsa-miR-575 were positively correlated with that of RNU12 (Fig. 4C–E). Parallelly, hsa-miR-575 overexpression lowered the RNU12 and BLID expressions. Consistently, hsa-miR-575 knockdown contributed to an increase in RNU12 and BLID expressions (Fig. 4F–H). The dual-luciferase reporter assay was used to analyze whether RNU12 and BLID were functional targets specific to hsa-miR-575. The results showed that the luciferase activity presented an obvious decline in that hsa-miR-575 mimics were transfected into cultured RNU12-Wt or BLID-Wt-co-transfected HEK293T and AGS cells. However, the same cell receiving the transfection of RNU12-Mut or BLID-Mut vector had no effect (Fig. 4I–L). The integral results suggested that RNU12 and BLID’s 3′ UTR possessed hsa-miR-575-binding sites. Hsa-miR-575 acted as the RNU12 inhibitor and the BLID blocker in the GC progression. (The original blots in Fig. 4E are shown in Supplementary file 3).
RNU12 sponged hsa-miR-575 and regulated BLID. (A) Sequence alignment of hsa-miR-575 which had putative binding sites in RNU12 and BLID wt and mt regions; (B) After detecting ago2 or IgG RIP assay, qRT-PCR assisted in determining the RNU12 and miR-575 levels; (C,D) MiR-575 expression presented a decrease and increase in AGS /RNU 12-OE cells and AGS /RNU 12-SH cells under qRT-PCR, respectively; (C-E) AGS /RNU 12-OE and AGS /RNU12-SH cells presented increased and decreased BLID expression under qRT-PCR and WB, respectively; (F) qRT-PCR analyzed the miR-575 expression in AGS cells receiving the transfection of miR-575 mimics or inhibitors; (G,H) qRT-PCR analyzed RNU12 and BLID expression in AGS cells receiving the transfection of miR-575 mimics or inhibitors. (I–L) Based on Double Luciferase Report analysis, hsa-miR-575 mimics weakened the fluorescence intensity exhibited by 293 T or AGS cells under the transfection of RNU12-Wt or BLID-Wt rather than RNU12-Mut or BLID-Mut vector. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
BLID enhancement critically affects malignant behaviors mediated by RNU12
For investigating whether BLID attenuated malignant behaviors mediated by RNU12, BLID was silenced in AGS /RNU12-OE in a rescue assay. The qRT-PCR and WB showed that AGS/RNU12-OE/BLID-SH presented obviously lower BLID compared to AGS/RNU12-OE (Fig. 5A,B). The silencing of BLID could enhance the proliferation and the colony formation in AGS/RNU12-OE cells determined by MTT assay (Fig. 5C) and colony assay (Fig. 5D). By silencing BLID, the cell cycle arrest was remarkably enhanced at the S phase of AGS compared to AGS/RNU12-OE (Fig. 5E). In addition, as revealed by qRT-PCR and WB assay, silence of BLID in AGS/RNU12-OE cells elevated the expression of genes associated with proliferation or apoptosis, (BCL-2, CCND1, and PCNA) (Fig. 5F,G). The silence of BLID also assisted in remarkably increasing the migratory and invasive abilities exhibited by AGS/RNU12-OE cell lines, based on the WHT (Fig. 5H,I) and TIA (Fig. 5J). The results of qRT-PCR and WB showed an upregulation of downstream molecules (N-cadherin, and Vimentin) related to metastasis, and the downregulation of E-cadherin (Fig. 5K). Taken together, silencing BLID critically affects the malignant behaviors mediated by RNU12 in GC cells in vitro (The original blots in Fig. 5B,K are shown in Supplementary file 4).
BLID enhancement critically inhibited malignant behaviors mediated by RNU12. (A,B) BLID expression in AGS cells in qRT-PCR and WB rescue assay. (C,D) MTT assay and colony formation regarding AGS cells. (E) AGS cell cycle distribution by FACS; (H,I) WHA of AGS cells; (J) TIA of AGS cells; (F,G) The expression of genes related to proliferation: BCL-2, CCND1, and PCNA under the measurement of qRT-PCR and WB; (K) The expressions of genes related to EMT: E-cadherin, N-cadherin, VIMENTIN based on WB. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
RNU12 promotes growth of GC tumor in zebrafish
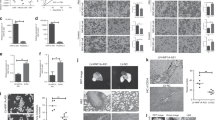
The GC tumor xenograft in zebrafish model was used for exploring the association of the RNU12 biological function in vivo. The experiment (Fig. 6A,B) on the zebrafish that received AGS cell injection at various time points traced the fluorescence distribution exhibited by GC cells in vivo with GFP (Green fluorescent protein). The results revealed the obviously decreased GFP-AGS/RNU12-OE cells (P < 0.001) relative to the AGS/NC in zebrafish at 72 h, indicating that the overexpression of RNU12 attenuated GC cells’ proliferation in vivo as that in vitro. In addition, AGS/RNU12-OE possessed lower tail GFP area relative to AGS/NC at 72 h, indicating that the overexpression of RNU12 inhibited migration of GC cell (P < 0.001, Fig. 6C,D). All above results demonstrated the inhibitory effects of RNU12 on GC proliferation, migration, and invasiveness in vivo.
RNU12 inhibited GC tumor growth in zebrafish. (A) Observation of fluorescence distribution exhibited by GC cells with GFP in whole body by stereomicroscope and confocal microscope at various time points; (B) The fluorescence area of whole body of GC cells at various time points; (C) The GC cell tail fluorescence area at various time points; (D) Observation of the fluorescence distribution exhibited by GC with GFP in tail by stereomicroscope together with confocal microscope at various time points; (E) Schematic diagram of the mechanism: RNU12 down-regulation in gastric cancer cell modulates EMT and acts as a miR-575 sponge to promote its function and down-regulates BLID, forming RNU12/miR-575/BLID axis to promote progression and migration of gastric cancer. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
This study revealed the gastric cancer-associated lncRNA RNU12 is a tumor-suppressor lncRNA that inhibits cancer progression via miR-575/BLID axis, which plays crucial role in gastric cancer progression and suggest that RNU12 is potential GC marker and target for GC therapy.
Discussion
LncRNAs affect cancer progression26,27,28,29, However, the effects of RNU12 on GC are unreadable21,22. In this study, we revealed for the first time the function of RNU12 and its related molecules in GC: (1) RNU12 was downregulated in GC with negative relation to lymph node metastasis; (2) RNU12 could sponge miR-575 and upregulate BLID, then BLID functioned as a tumor suppressor; (3) RNU12 could control proliferation, colony formation, migratory, invasion through RNU12/miR-575/BLID axis; (4) RNU12 exerted the tumor-suppressing effect via regulating the expression regarding CCND 1, PCNA, N-cadherin, E-cadherin, Vimentin and anti-apoptosis BCL-2. Our results aid to the comprehensive understanding of the RNU12 functionality (Fig. 6E).
LncRNAs are a species of ncRNA, with the length usually over 200 nt and having tissue-specific expression as exonic, intronic, overlapping, and intergenic30,31. Basic and clinical researchers have focused on comprehensively investigating the adoption of lncRNAs as new biomarkers and proper candidates for different cancer types32. Evidences indicated that lncRNAs could regulate the expression of gene at various genomic, transcriptomic and post transcriptomic levels, and could serve as the putative biomarkers and the therapeutic targets for GC31,33. LINC00589 promoted hnRNPA1 protein ubiquitination and proteasomal degradation for suppressing tumor progression, meanwhile restricting the peritoneal metastasis in GC, hence achieving the suppression of tumors34. LncRNA ZFAS1 modulated LIN28 and CAPRIN1 expression for enhancing the invasion and proliferation of GC cells, which suggested the possibility of ZFAS1 being a marker for GC diagnosis and prognosis35. Our present data supported the function of RNU12 in inhabiting GC tumor progression through suppressing the cell growth, migration, invasion, as well as the proliferative ability expression with CCND1, PCNA, N-cadherin, E-cadherin, Vimentin and BCL-2.
Clearly, lncRNA competitively bound to proper miRNA to sponge miRNA, and prevented miRNA targeting mRNA for weakening the gene suppression mediated by miRNA36,37,38, regulating cellular proliferation, migration, and angiogenesis in cancer38,39. Our results confirmed that RNU12 sponged miR-575p with evidences: (i) endogenous RNU12 enriched in the AGO2-FLAG IP fraction and could possibly incorporate into RISC complexes (silencing complex induced by RNA); (ii) RNA pull-down assays revealed the enrichment of RNU12 in the miR-575 biotin IP fraction; (iii) luciferase reporter assays demonstrated the binding of RNU12 with miR-575; (iv) miR-575 knockdown elevated BLID expression; and (v) miR-575 over-expression inhibited RNU12 and BLID expression. Our data demonstrated that RNU12 could sponge miR-575 and weaken miRNA mediated gene BLID function.
MicroRNAs (miRNAs) refer to a series of non-coding RNAs and can remarkably help to regulate protein expressions40. The previous reports have confirmed the miRNA profiles can reflect differentiation state of the tumor and regulate mRNA interpretation41. MicroRNA-575 was first reported in 200942. MiR-575 is oncogene in many tumors. And miR-575 targets PTEN for regulating the proliferation and apoptosis of cancer cells, thereby critically affecting GC43. Furthermore, the regulatory relationship between miR-575 and BLID has been reported in NSCLC44, which is consistent with our results defined in GC for first time.
BLID is a BH3-like motif that contains apoptotic member belonging to the Bcl-2 family. According to previous studies, BLID could suppress tumor and was associated with tumor prognosis45,46. BLID downregulated AKT pathway for inhibiting the cell growth and metastasis of breast cancer47. BLID could also assist in independently predicting the prognosis of breast cancer in terms of the overall survival and the distant metastasis-free survival48,49. In our study, we indicated BLID suppressed cell proliferation and invasion of GC. We first confirmed that miR-575 could regulate the expression of BLID in GC.
Methods
Tissue specimen collection
113 pairs of GC and their adjacent normal gastric tissues came from tumor biobank of Fujian Cancer Hospital. From June 2014 to June 2019. The patients undergone surgical treatment prior to any chemotherapy or radiotherapy in Fujian Cancer Hospital (Fuzhou, China). All samples were collected and analyzed after we obtained patients’ written informed consents. The Ethics Board of the Fujian Cancer Hospital (Fuzhou, China) approved all experiments. These fresh tissues were immediately submerged into liquid nitrogen and frozen at − 80 °C. The experiments were conducted following the Helsinki declaration.
Cell lines and cell culture
Cell lines, including human GC cell lines: AGS, MGC803, MKN45, MKN28 and human normal gastric epithelial cell line GES-1, came from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences, confirmed by short tandem repeat DNA finger printing (STR) and free of mycoplasma contamination. The cell culture was performed in RPMI-1640 (GIBCO, Grand Island, NY, USA) with 100 IU/ml penicillin, 100 µg/ml streptomycin, 10% FBS (GIBCO) and with 5% CO2 at 37 °C.
Construction of overexpression or knockdown RNU12 cell lines
To perform experiments on RNU12 overexpression or knockdown, the cells were infected with the virus vectors (Hanheng Biotechnology CoLtd). To establish the stable cell lines with overexpressed RNU12, HBLV-RNU12-OE (pHBLV-CMV-mcs-3flag-EF1-ZsGReen-T2A-PURO inserted with RNU12 gene) was used and HBLV-NC as the vector alone control. Similarly, the virus vectors inserted with shRNA were used to establish stable RNU12 knockdown cell lines, HBLV-RNU12-shrna1, HBLV-RNU12-shrna2, HBLV-RNU12-shrna3 and their parental vector pHBLV-U6-MCS-CMV-Zs/mcherry as the vector alone control. 10 MOI of viral vectors were used for infecting AGS or MGC 803 cells then seeded to six-well plates (5 × 105/well) with 6 μg/ml of polyamine. 48 h later, the medium was added with puromycin (2 μg/ml). The cells were maintained in the above medium for 2 weeks for obtaining steady infected cells. The stable cell lines were named as AGS/NC, AGS/RNU12-OE, AGS/RNU12-SH, MGC803/NC, MGC803/RNU12-OE, MGC803/RNU12-SH. The symbol -OE represents overexpression and –SH represents knockdown. (The target-specific primer sets and primary antibodies are described in Supplementary Table S1).
Overexpression BLID AGS cell construction
AGS cells in 5 × 105/well plates underwent infection of 10 MOI of HBLV-BLID-Null-Zs Green-PURO virus vectors (Hanheng Biotechnology Co Ltd). 48 h of transfection later, we named AGS cells with steady BLID overexpression AGS/BLID-OE and those possessing empty vector as AGS-NC.
RT-qPCR
The total RNAs were extracted with RNeasy Mini Kit. The Revert Aid First Strand cDNA Synthesis Kit together with miScript Reverse Transcription Kit were used for the reverse transcription of total RNAs into the complementary DNA (cDNA) of RNU12, hsa-miR-575, PCNA, BCL-2, CCND1, E-cadherin, N-cadherin and Vimentin in AGS cells. The miScript SYBR Green PCR Kit and LightCycler 480 SYBR Green I Master were used to assess the gene expressions using the resulted cDNAs as templates. The housekeeping gene β-actin was the internal control for PCNA, BCL-2, CCND1, E-cadherin, N-cadherin and Vimentin, and U6 small nuclear RNA was the internal reference for hsa-miR-575 expression. The 2−ΔΔCt method served for calculating the relative gene expression16,50. The comparative 2−ΔΔCt method helped to quantify the relative mRNA expression regarding RNU12 in all GC samples. β-actin assisted in the normalization of RNU12 expression50,51.
MTT assay and cell cycle analysis
Cells were cultured overnight after transfection, then the collected cells were resuspended with the medium, and added into 96-well plates (5000 cells in 100 μl/well). The cell proliferation at 24 h, 48 h, 72 h, and 96 h later was measured by adding 20 μl/well of MTT solution (Promega, Madison, WI, USA) at each time point. After that, cells underwent four hours of incubation at 37 °C. then adding 150 μl of dimethyl sulfoxide into each well and shaking 10 min for completely dissolving the MTT. Model 680 reader was used for determining the absorbance exhibited by each well. The Flow Cytometer (Thermo Fisher Scientific, USA) assisted in analyzing the cell cycle, following the instructions of the manufacturer52.
Transwell invasion assay (TIA)
The transwell chambers was used for assessing the cell migration. The cells were seeded in serum-free medium of the Matrigel-precoated upper-side of chambers (BD Biosciences, USA). The bottom chambers were added with the serum-free medium containing DMEM/F-12 without FBS as a chemoattractant. After 24 h, a cotton swab was employed for removing the residual non-invasive cells in the upper side of chambers. The cells moved to the low side of chambers were fixed with 100% methanol, then stained with 0.5% crystal violet. The transwelled cells were pictured under the inverted microscope.
Colony formation assay
For colony-formation assay, 200 cells/well were seeded into six-well plates and cultured for 2 weeks, then the colonies were fixed in methanol, stained with 0.2% crystal violet and colonies with over 50 cells were photographed with Image Scanner (GE, USA) and analyzed by ImageJ (NIH, MD, USA).
Wound healing assay (WHA)
The transfected cells were detached with Trypsin, suspended in DMEM/F12-10% FBS and seeded in 6 well plate in triplicates. 24 h later, a wound was inflicted in the midline of each well by a 10 μl sterile pipette tip. PBS was used for carefully washing the removed cells twice. 24 h later, the cells migrated in the midline were photographed under microscope and analyzed with Image J.
Prediction of target genes of RNU12 and hsa-miR-575
LncRNASNP2 database assisted in predicting the binding site for hsa-miR-575 in RNU12. The miRDB (http://mirdb.org/) and Target Scan (http://www.targetscan.org/vert_72/) were used for the analysis.
Dual-luciferase reporter
113 pairs of GC and their adjacent normal gastric tissues came from tumor biobank. The wild-type (wt) RNU12 and mutant (mut) RNU12 were compared to predict hsa-miR-575-binding site which then chemically synthesized (Hanheng Biotechnology Co. Ltd) and inserted into pSI-check2 luciferase reporter plasmids for generating the pMIR-RNU12-wt (RNU12-wt) and pMIR- RNU12-mut (RNU12-mut) reporter plasmids. Similarly, BLID-wt and BLID-mut were also generated.
For reporter assay, cells in the 24-well plates cultured to 70% confluence, the reporter plasmids and hsa-miR-575 mimics or miR-NC were co-transfected with Lipofectamine 2000 reagent. 48 h later, Dual-Luciferase Reporter Assay System was used to detect the luciferase activity exhibited by the transfected cells. Normalization between relative luciferase activity and Renilla luciferase activity was achieved.
RNA immunoprecipitation (RIP)
The RNA immunoprecipitation (RIP) was performed with EZ-Magna RIP Kit. Briefly, 1 × 107AGS cells were lysed using RIP lysis buffer, mixed with magnetic beads coated with Anti-IgG or Anti-Ago2. After 6 h, these beads enriched RNAs of RNU12 and miR-575 complex which were extracted and quantified with RT-qPCR.
Western blot analysis
Cells harvested and lysed with RIPA buffer that contained 0.1% SDS and protease inhibitor cocktail. BCA protein assay was used for measuring the protein concentration of supernatants. 25 μg of proteins were run on SDS–PAGE (sodium dodecyl sulfate and 10–12% polyacrylamide gels) and transferred 1.5 h onto a nitrocellulose membrane (Millipore A) at 100 V. After blocking with 3% BSA in TBST (TBS-1% Tween 20) at room temperature, and the membranes incubated overnight at 4 °C with 1:1,000 primary antibodies, BLID(Abcam, Cambridge, MA), PCNA (Cell Signaling Technology,CA, USA), BCL-2 (Cell Signaling Technology,CA, USA), CCND1 (Cell Signaling Technology,CA, USA), E-cadherin (Cell Signaling Technology,CA, USA), N-cadherin (Cell Signaling Technology,CA, USA) and Vimentin (Cell Signaling Technology,CA, USA), respectively. After washed three times using TBS-N, the membranes were incubated with secondary horseradish peroxidase conjugated anti-rabbit IgG (1:2000; Cell Signaling Technology, CA, USA) for one hour at room temperature and stained with Immobilon ECL Ultra Western HRP Substrate. The Image Station 4000MM Pro imaging platform was used to reveal the bands. Protein levels relative to the load control β-actin were quantified by Image J.
Transplantation of zebrafish with RNU12 knockdown GC Cell AGS
Transgenic zebrafish TG (apo14-GFP) came from the Institute of Hydrobiology (Chinese Academy of Sciences). Adult fish were raised at 26.5 °C, with the light/dark ratio of 12 h:12 h. The IVF Development Zone of apo14-EGFP zebrafish yolk sac was injected with AGS/RNU12-SH cells and AGS/NC cells on the 2nd day following fertilization. Following the injection, embryos received 24 h of culturing at 33 °C, followed by being transferred to an incubator at 35 °C. A laser scanning confocal microscope (LSM 710) assisted in observing in detail how red fluorescence was distributed and expressed in zebra’s abdominal cavity. ImageJ 1.48v was used for analyzing red fluorescence expression in the abdominal cavity. An inverted SP5 STED confocal microscope was used for imaging and quantifying the results. Each group had no less than 40 zebrafishes, with three representative images being utilized. All the tests were parallelly carried out three times.
Statistical analysis
SPSS 19.0 software assisted in data analysis. Pearson analysis was used for correlation. The two tailed t test served for examining group difference. Paired or nonparametric Kruskal–Wallis test served for evaluating how RNU12 level was related to other characteristics. The Kaplan–Meier method calculated the survival curve. P value less than 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001) denoted statistical significance.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Xia, C. et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 135, 584–590. https://doi.org/10.1097/cm9.0000000000002108 (2022).
Correa, P. Gastric cancer: Overview. Gastroenterol. Clin. North Am. 42, 211–217. https://doi.org/10.1016/j.gtc.2013.01.002 (2013).
Wong, M. C. S. et al. Global incidence and mortality of gastric cancer, 1980–2018. JAMA Netw. Open 4, e2118457. https://doi.org/10.1001/jamanetworkopen.2021.18457 (2021).
Smyth, E. C., Nilsson, M., Grabsch, H. I., van Grieken, N. C. & Lordick, F. Gastric cancer. Lancet 396, 635–648. https://doi.org/10.1016/s0140-6736(20)31288-5 (2020).
Amieva, M. & Peek, R. M. Jr. Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology 150, 64–78. https://doi.org/10.1053/j.gastro.2015.09.004 (2016).
Praud, D. et al. Cigarette smoking and gastric cancer in the Stomach Cancer Pooling (StoP) Project. Eur. J. Cancer Prev. 27, 124–133. https://doi.org/10.1097/cej.0000000000000290 (2018).
Gaddy, J. A. et al. High dietary salt intake exacerbates Helicobacter pylori-induced gastric carcinogenesis. Infect. Immun. 81, 2258–2267. https://doi.org/10.1128/iai.01271-12 (2013).
Raei, N., Behrouz, B., Zahri, S. & Latifi-Navid, S. Helicobacter pylori infection and dietary factors act synergistically to promote gastric cancer. Asian Pac. J. Cancer Prev. 17, 917–921. https://doi.org/10.7314/apjcp.2016.17.3.917 (2016).
Hu, J., La Vecchia, C., Morrison, H., Negri, E. & Mery, L. Salt, processed meat and the risk of cancer. Eur. J. Cancer Prev. 20, 132–139. https://doi.org/10.1097/CEJ.0b013e3283429e32 (2011).
Lippi, G., Mattiuzzi, C. & Cervellin, G. Meat consumption and cancer risk: A critical review of published meta-analyses. Crit. Rev. Oncol. Hematol. 97, 1–14. https://doi.org/10.1016/j.critrevonc.2015.11.008 (2016).
Ma, L., Bajic, V. B. & Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 10, 925–933. https://doi.org/10.4161/rna.24604 (2013).
Rudnicka, K., Backert, S. & Chmiela, M. Genetic polymorphisms in inflammatory and other regulators in gastric cancer: Risks and clinical consequences. Curr. Top. Microbiol. Immunol. 421, 53–76. https://doi.org/10.1007/978-3-030-15138-6_3 (2019).
Li, X., Wu, Z., Fu, X. & Han, W. lncRNAs: Insights into their function and mechanics in underlying disorders. Mutat. Res. Rev. Mutat. Res. 762, 1–21. https://doi.org/10.1016/j.mrrev.2014.04.002 (2014).
Paraskevopoulou, M. D. & Hatzigeorgiou, A. G. Analyzing MiRNA-LncRNA interactions. Methods Mol. Biol. 1402, 271–286. https://doi.org/10.1007/978-1-4939-3378-5_21 (2016).
Zou, C. et al. SNHG8 promotes the progression of Epstein–Barr virus-associated gastric cancer via sponging miR-512–5p and targeting TRIM28. Front. Oncol. 11, 734694. https://doi.org/10.3389/fonc.2021.734694 (2021).
Chen, X. et al. LncRNA ZNF667-AS1 promotes ABLIM1 expression by adsorbing micro RNA-1290 to suppress nasopharyngeal carcinoma cell progression. Onco Targets Ther. 13, 4397–4409. https://doi.org/10.2147/ott.S245554 (2020).
Esfandi, F. et al. Brain-derived neurotrophic factor downregulation in gastric cancer. J. Cell Biochem. 120, 17831–17837. https://doi.org/10.1002/jcb.29050 (2019).
Yue, T. et al. Comprehensive analyses of long non-coding RNA expression profiles by RNA sequencing and exploration of their potency as biomarkers in psoriatic arthritis patients. BMC Immunol. 20, 28. https://doi.org/10.1186/s12865-019-0297-9 (2019).
He, H. et al. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science 332, 238–240. https://doi.org/10.1126/science.1200587 (2011).
Edery, P. et al. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science 332, 240–243. https://doi.org/10.1126/science.1202205 (2011).
Xing, C. et al. Biallelic variants in RNU12 cause CDAGS syndrome. Hum. Mutat. 42, 1042–1052. https://doi.org/10.1002/humu.24239 (2021).
Elsaid, M. F. et al. Mutation in noncoding RNA RNU12 causes early onset cerebellar ataxia. Ann. Neurol. 81, 68–78. https://doi.org/10.1002/ana.24826 (2017).
Mo, X. B. et al. A novel long noncoding RNA, lnc-RNU12, influences the T-cell cycle via c-JUN and CCNL2 in rheumatoid arthritis. Rheumatology https://doi.org/10.1093/rheumatology/keac553 (2022).
Fan, C. N., Ma, L. & Liu, N. Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer. J. Transl. Med. 16, 264. https://doi.org/10.1186/s12967-018-1640-2 (2018).
Huang, Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J. Cell Mol. Med. 22, 5768–5775. https://doi.org/10.1111/jcmm.13866 (2018).
Ferrè, F., Colantoni, A. & Helmer-Citterich, M. Revealing protein-lncRNA interaction. Brief. Bioinform. 17, 106–116. https://doi.org/10.1093/bib/bbv031 (2016).
Charles Richard, J. L. & Eichhorn, P. J. A. Platforms for investigating LncRNA functions. SLAS Technol. 23, 493–506. https://doi.org/10.1177/2472630318780639 (2018).
Ma, Y., Zhang, J., Wen, L. & Lin, A. Membrane-lipid associated lncRNA: A new regulator in cancer signaling. Cancer Lett. 419, 27–29. https://doi.org/10.1016/j.canlet.2018.01.008 (2018).
Schmitz, S. U., Grote, P. & Herrmann, B. G. Mechanisms of long noncoding RNA function in development and disease. Cell Mol. Life Sci. 73, 2491–2509. https://doi.org/10.1007/s00018-016-2174-5 (2016).
Bhan, A., Soleimani, M. & Mandal, S. S. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 77, 3965–3981. https://doi.org/10.1158/0008-5472.Can-16-2634 (2017).
Fattahi, S. et al. LncRNAs as potential diagnostic and prognostic biomarkers in gastric cancer: A novel approach to personalized medicine. J. Cell Physiol. 235, 3189–3206. https://doi.org/10.1002/jcp.29260 (2020).
Peng, W. X., Koirala, P. & Mo, Y. Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 36, 5661–5667. https://doi.org/10.1038/onc.2017.184 (2017).
Ghafouri-Fard, S. & Taheri, M. Long non-coding RNA signature in gastric cancer. Exp. Mol. Pathol. 113, 104365. https://doi.org/10.1016/j.yexmp.2019.104365 (2020).
Wang, S. et al. Delivery of LINC00589 via mesoporous silica nanoparticles inhibits peritoneal metastasis in gastric cancer. Cancer Lett. 549, 215916. https://doi.org/10.1016/j.canlet.2022.215916 (2022).
Zhuo, Z. L. et al. Long noncoding RNA ZNFX1-AS1 promotes the invasion and proliferation of gastric cancer cells by regulating LIN28 and CAPR1N1. World J. 28, 4973–4992. https://doi.org/10.3748/wjg.v28.i34.4973 (2022).
Militello, G. et al. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief. Bioinform. 18, 780–788. https://doi.org/10.1093/bib/bbw053 (2017).
Jalali, S., Bhartiya, D., Lalwani, M. K., Sivasubbu, S. & Scaria, V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS ONE 8, e53823. https://doi.org/10.1371/journal.pone.0053823 (2013).
Salmena, L., Poliseno, L., Tay, Y., Kats, L. & Pandolfi, P. P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language?. Cell 146, 353–358. https://doi.org/10.1016/j.cell.2011.07.014 (2011).
Du, Z. et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat. Commun. 7, 10982. https://doi.org/10.1038/ncomms10982 (2016).
Bartel, D. P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297. https://doi.org/10.1016/s0092-8674(04)00045-5 (2004).
Doench, J. G. & Sharp, P. A. Specificity of microRNA target selection in translational repression. Genes Dev. 18, 504–511. https://doi.org/10.1101/gad.1184404 (2004).
Yao, Y. et al. MicroRNA profiling of human gastric cancer. Mol. Med. Rep. 2, 963–970. https://doi.org/10.3892/mmr_00000199 (2009).
Wang, Y. N. et al. MicroRNA-575 regulates development of gastric cancer by targeting PTEN. Biomed. Pharmacother. 113, 108716. https://doi.org/10.1016/j.biopha.2019.108716 (2019).
Wang, H. et al. MicroRNA-575 targets BLID to promote growth and invasion of non-small cell lung cancer cells. FEBS Lett. 589, 805–811. https://doi.org/10.1016/j.febslet.2015.02.013 (2015).
Yu, X. & Li, Z. B. L. I. D. A novel tumor-suppressor gene. Oncol. Res. 22, 333–338. https://doi.org/10.3727/096504015x14410238486568 (2014).
Xu, Y. C. et al. A novel mechanism of doxorubicin resistance and tumorigenesis mediated by microRNA-501-5p-suppressed BLID. Mol. Ther. Nucl. Acids 12, 578–590. https://doi.org/10.1016/j.omtn.2018.06.011 (2018).
Li, X., Kong, X., Wang, Y. & Yang, Q. BRCC2 inhibits breast cancer cell growth and metastasis in vitro and in vivo via downregulating AKT pathway. Cell Death Dis. 4, e757. https://doi.org/10.1038/cddis.2013.290 (2013).
Zhao, F. et al. Evaluation of BLID and LOC399959 as candidate genes for high myopia in the Chinese Han population. Mol. Vis. 16, 1920–1927 (2010).
Cavalli, L. R. et al. Frequent loss of the BLID gene in early-onset breast cancer. Cytogenet. Genome Res. 135, 19–24. https://doi.org/10.1159/000330265 (2011).
Ribet, M. et al. Congenital cystic adenomatoid malformation of the lung. Eur. J. Cardiothorac. Surg. 4, 403–405. https://doi.org/10.1016/1010-7940(90)90067-a (1990) (discussion 406).
Yuan, J. S., Reed, A., Chen, F. & Stewart, C. N. Jr. Statistical analysis of real-time PCR data. BMC Bioinform. 7, 85. https://doi.org/10.1186/1471-2105-7-85 (2006).
Bavelloni, A. et al. Cell cycle arrest and apoptosis induced by kinamycin F in human osteosarcoma cells. Anticancer Res. 37, 4103–4109. https://doi.org/10.21873/anticanres.11797 (2017).
Funding
This study was supported by Natural Science Foundation of Fujian Province (No. 2019J01196, 2020J011109), Medical Innovation Program of Fujian Province (No. 2019-CX-5), the National Clinical. Fujian Provincial Clinical Research Center for Cancer Radiotherapy and Immunotherapy (No. 2020Y2012), Fujian Clinical Research Center for Radiation and Therapy of Digestive, Respiratory and Genitourinary Malignancies (2021Y2014), Joint Funds for the innovation of science and Technology, Fujian province (2021Y9191, 2021Y9195).
Author information
Authors and Affiliations
Contributions
X.L. (Xiandong Lin), T.H. (Tao Huang) and J.L. designed the research. S.W., C.Z., X.L., and D.H. performed experiments. T.H., X.Z., C.Z., X.L. (Xinyi Lin) and H.H. analyzed data. D.H., X.Z., X.L. (Xinyi Lin) and L.Z. conducted the histological/pathological analysis. X.L. (Xiandong Lin), S.W. and Y.S. wrote the paper. L.Z., J.L. and X.L. (Xinyi Lin) edited the paper. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, S., Zou, C., Lin, X. et al. RNU12 inhibits gastric cancer progression via sponging miR-575 and targeting BLID. Sci Rep 13, 7523 (2023). https://doi.org/10.1038/s41598-023-34539-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-34539-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.