Abstract
With the widespread use of Integrase strand transfer inhibitors (INSTIs), surveillance of HIV-1 pretreatment drug resistance is critical in optimizing antiretroviral treatment efficacy. However, despite the introduction of these drugs, data concerning their resistance mutations (RMs) is still limited in Ethiopia. Thus, this study aimed to assess INSTI RMs and polymorphisms at the gene locus coding for Integrase (IN) among viral isolates from ART-naive HIV-1 infected Ethiopian population. This was a cross-sectional study involving isolation of HIV-1 from plasma of 49 newly diagnosed drug-naive HIV-1 infected individuals in Addis-Ababa during the period between June to December 2018. The IN region covering the first 263 codons of blood samples was amplified and sequenced using an in-house assay. INSTIs RMs were examined using calibrated population resistance tool version 8.0 from Stanford HIV drug resistance database while both REGA version 3 online HIV-1 subtyping tool and the jumping profile Hidden Markov Model from GOBICS were used to examine HIV-1 genetic diversity. Among the 49 study participants, 1 (1/49; 2%) harbored a major INSTIs RM (R263K). In addition, blood specimens from 14 (14/49; 28.5%) patients had accessory mutations. Among these, the M50I accessory mutation was observed in a highest frequency (13/49; 28.3%) followed by L74I (1/49; 2%), S119R (1/49; 2%), and S230N (1/49; 2%). Concerning HIV-1 subtype distribution, all the entire study subjects were detected to harbor HIV-1C strain as per the IN gene analysis. This study showed that the level of primary HIV-1 drug resistance to INSTIs is still low in Ethiopia reflecting the cumulative natural occurrence of these mutations in the absence of selective drug pressure and supports the use of INSTIs in the country. However, continues monitoring of drug resistance should be enhanced since the virus potentially develop resistance to this drug classes as time goes by.
Similar content being viewed by others
Introduction
Globally antiretroviral therapy (ART) has been scaled up with an estimated 26 million people accessing at the end of 2020 that led to the dramatic decrease of mortality and morbidity due to Human Immunodeficiency virus-1 (HIV-1) associated diseases1,2. Despite this, however, the emergence of HIV-1 pre-treatment drug resistance (PDR) is posing a threat to the success of ART3. PDR reduces the treatment options available to newly HIV-1 infected patients, and it is associated with an increased risk of virological failure2. Therefore, surveillance of drug resistance mutations (RMs) among ART-naive patients is critical in optimizing ART efficacy4.
Due to an increasing trend of HIV drug resistance (HIVDR) to non-nucleoside reverse-transcriptase inhibitor (NNRTI), nucleoside reverse transcriptase inhibitor (NRTI), and protease inhibitors (PI), integrase strand transfer inhibitor (INSTI)-based regimens are now recommended as first-line combination ART in several updated treatment guidelines4. Hence, they have become an essential component of ART in use in many countries including Ethiopia. These latest antiretroviral (ARV) drug class works via the inhibition of DNA strand transfer and are clinically effective against HIV-1 strains that had previously exhibited resistance-associated mutations against other ARV drugs5. To date, five INSTIs are approved for clinical use by the US food and drug administration, namely, raltegravir (RAL), elvitegravir (EVG), bictegravir (BIC), dolutegravir (DTG), and cabotegravir (CAB)4,5,6,7. Among these INSTI drugs, DTG, which is a second generation drug, is preferable due to some notable advantages including having a limited or no-cross resistance to early generation INSTI drugs, and being responsible for the higher inhibition potency8. However, drug RMs like Q148H/R and G140S in combination with mutations L74I/M, E92Q, T97A, E138A/K, G140A, or N155H are associated with 5-fold to 20-fold reduction in DTG effectiveness9. In addition, the R263K, which is the most commonly DTG-selected mutation, is associated with a 2-fold susceptibility reduction when it occurs alone10,11.
DTG was introduced as a preferred first-line drug combination in Ethiopia in 201812. However, data concerning INSTI drug RMs is still limited13 in the country. The aim of this study was thus to generate updated information regarding the prevalence of INSTI associated drug RMs among ART-naive Ethiopian populations before the introduction of DTG.
Materials and methods
Study population
This is a cross-sectional health facility-based study involving drug-naive HIV-1 infected study participants. Study participants who were asymptomatic at the time of recruitment, above the age of 18, and willing to take part in the study were sequentially enrolled. This research did not include pregnant women, those with known chronic conditions, or anyone who had ever used ART. For detailed patients screening and enrollment, please refer to our previous publication14. Accordingly, forty-nine volunteered adult (≥ 18 years old) study participants from voluntary VCT centers in Addis Ababa were consecutively enrolled in this study from June to December 2018. Blood samples (10 ml) were collected at the time of HIV diagnosis before initiation of ART for genotypic assay of the IN gene region from these participants using an EDTA tube. Plasma was then separated by centrifugation for 10 min at 3000 rpm and stored in deep freeze (− 80 °C) till required for laboratory investigation.
Nucleic acid extraction, PCR amplification, and sequencing
Viral RNA extraction and viral load were performed consecutively from 200 μL of thawed participants' plasma sample input using the Abbott Real-time HIV-1 M2000 system (Abbott Laboratories, Abbott Park, USA). For sequencing purpose, cDNA was synthesized from the extracted RNA in a 20 microliter (µL) reaction mixture using Superscript IV Reverse Transcriptase enzyme and HIVpcrRev1 primer (Table 1). The mixture constituted; 1 µL of HIVpcrRev1 outer primer, 1 µL of dNTPs, 1 µL of DDT, 1 µL of RiboLock, 4 µL of 5X superscript IV buffer, 1 µL of superscript IV (Invitrogen Carlsbad, CA, USA), 5 µL of RNA, and 6 µL of molecular grade water. The thermal cycling for this cDNA synthesis was; 50 °C for 1 h13.
This was followed by a first-round PCR, which was performed in a 50 µL reaction mix composed of 1 µL of each outer primer (HIVpcr For1 and HIVpcr Rev1; Table 1), 27.8 µL of nuclease‐free water, 10 µL of 5X GoTaq buffer, 4 µL of 25 mM MgCl2, 1 µL of 10 mM dNTPs, 0.2 µL of GoTaq polymerase (Promega, USA), and 5 µL of cDNA. The PCR cycling conditions were 98 °C for 2 min followed by 39 cycles of 10 s at 98 °C, 25 s at 64 °C, 40 s at 72 °C, and a final extension of 72 °C for 5 min13.
The DNA product from the first round PCR was then re-amplified by a nested PCR. This nested PCR was carried out in another 50 µL reaction mix utilizing 1 µL of the first-round PCR product, 1 µL of each inner primer (HIVpcr For2 and HIVpcr Rev2; Table 1), 31.8 µL of nuclease‐free water, 10 µL of 5X GoTaq buffer, 4 µL of 25 mM MgCl2, 1 µL of 10 mM dNTPs, and 0.2 µL of GoTaq polymerase (Promega, USA). This final re-amplified PCR product (793 bp) was verified by agarose gel electrophoresis (using 1.5% agarose gel) and was then purified consecutively using the GenepHlow™ Gel/PCR Kit (Geneaid biotech ltd., Taiwan), following the manufacturer’s instruction. Purified amplicons were then sequenced with Sanger DNA sequencing using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), and run on the ABI PRISM® 3500 xL automated Genetic Analyzer. At each step of the laboratory procedures, both positive and negative controls were incorporated in order to keep the procedures quality assurance.
Primary INSTI resistance mutation analysis and HIV-1 subtype determination
Sequences were first collected from the ABI PRISM® 3500 xL automated Genetic Analyzer (Applied Biosystems) and transferred to separate personal computer. Sequence edition and alignment were then made using Geneious prime® software v.2020.2.1 (https://www.geneious.com/academic/). The quality of the sequence data was checked using an online sequence quality control tool found in the Los Alamos HIV sequence database (http://hiv-web.lanl.gov). Calibrated Population Resistance (CPR) tool version 8.0, which is available at Stanford University HIV Drug Resistance Database (HIVdb) (http://cpr.standford.edu/cpr/index.html) was used to assess the drug RMs. The susceptibility of HIV to ARV drugs was determined using the HIVdb program (http://hivdb.stanford.edu).
All sequences were thoroughly examined for the presence of primary mutations, nonpolymorphic [NoP] and polymorphic mutations associated with resistance to INSTI. Each aminoacid prevalence at each IN position was calculated and compared to the HIV-1 subtype B reference sequence (GenBank accession number: K03455). For this study, NoP was defined as substitutions within the HIV-1 IN that occurred in ≥ 1% of the sequences. Positions with ≥ 20% substitutions were considered as highly polymorphic, while those with ≤ 0.5% substitution were considered highly conserved.
HIV-1 subtype determination was performed using two online HIV-1 subtyping tools; the REGA version 3 online HIV-1 subtyping tool from the Stanford HIVdb (http://hivdb.stanford.edu) and the jumping profile Hidden Markov Model (jpHHM) at GOBICS (http://jphmm.gobics.de/). The subtypes were also further confirmed by phylogenetic analysis using reference sequences from Los Alamos National Laboratory HIV Sequence Database (http://hiv-web.lanl.gov). The analysis was made using Molecular Evolutionary Genetics Analysis (MEGA) v.10.0.5 software15.
Statistical analyses
Epi data v3.1 software was used for data entry while SPSS version 25.0 (SPSS Inc. the United States) was used for analysis. A logistic regression model was used to determine the presence of any associations between drug RMs and sociodemographic variables. Thus, variables having a p value < 0.05 were considered to have a statistically significant association. All isolates are deposited in GenBank with accession number MW560010 to MW560058.
Ethics approval and consent to participate
This study was approved by the institutional ethical review committee of the Armauer Hansen Research Institute (Protocol Number: PO16/18). All participants provided their written informed consent to participate in this study and that this study was conducted following the declaration of Helsinki.
Results
Characteristics of the study population
Near full-length HIV-1 IN region, (263 codons) of the 49 study participants were successfully sequenced and analyzed. Of these, 59.2% (n = 29) were females (Table 2). The mean ± SD age of the participants was 32 ± 1.2 years ranging from 20 to 57 years old. The median (IQR) viral load was 69,625 copies/mL (32,219–208,299). The detailed demographic and clinical information of the study participants is summarized in Table 2.
The level of primary INSTI resistance mutation and HIV-1 Subtype distribution
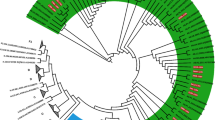
As per the INSTI RMs analysis, one (n = 1/49; 2%) participant harbored one major INSTIs RM R263K (Fig. 2.). Concerning the HIV-1 subtype distribution, both the REGA version 3 online HIV-1 subtyping tool and the jumping profile Hidden Markov Model from GOBICS indicated that the entire 49 subjects were harboring HIV-1C virus as per the IN region analysis. This was further confirmed by a maximum likelihood phylogenetic analysis along with the reference sequences (A–K) and CRFs from the Los Alamos National Library database (www.lanl.gov) (Fig. 1.). As indicated in the phylogenetic tree, sequences from this study with GenBank accession number (MW560010 to MW560058) were clustered with HIV-1C with a bootstrap value of 93% (Fig. 1).
Maximum-likelihood phylogenetic tree of the HIV-1 integrase sequences from the 49 drug-naive HIV-infected individuals. The ML tree was constructed using MEGA version 10.0.5 with the Kimura 2 parameter. Bootstrapping was performed with 1,000 replicates and only those that have a bootstrap value > 70% are shown on the tree. HIV-1 reference sequences (A—K and CRFs) that were retrieved from the HIV-1 LANL HIV Sequence Database (green color) are indicated by subtype followed by the corresponding country name, year, and accession number. All of the sequences from this study that are indicated in blue color (MW560010–MW560058) were clustered with HIV-1 subtype C strain with a bootstrap value of 93%. The one sequence (MW560010) that harbored a major INSTI RM R263K is shown with a red rectangular node. INSTI Integrase strand transfer inhibitor, LANL Los alamos national library, MEGA Molecular evolutionary genetics analysis, ML Maximum likelihood.
Accessory INSTI RMs and/or polymorphisms
Besides the major INSTI RM observed, 14 (14/49; 28.5%) patients in the current study harbored at least one accessory RMs. Among these, almost 93% of the accessory mutations observed were M50I (13/49; 28.3%) followed by L74I (1/49; 2%), S119R (1/49; 2%), and S230N (1/49; 2%) (Fig. 2.). Two patients had two simultaneous accessory RMs (M50I and S119R; M50I and S230N). Furthermore, other naturally occurring polymorphisms that indicate variability between HIV-1 subtypes were also observed.
Discussions
INSTIs have rapidly become an important class in the arsenal of ARV drugs due to their higher genetic barrier6,16. The world health organization (WHO) recommended one of the INSTI drugs, DTG, as a preferred first-line ART in 201717. This drug classes is now becoming the best choice in many countries due to an increase in resistance rates against NNRTIs, NRTIs as well as PIs in the past years18.
In Ethiopia, where no routine genotyping testing nor routine drug resistance surveillance is present, this drug was introduced in 2018 following the aforementioned WHO recommendation12. However, studies on primary HIV-1 drug resistance related to these drugs are limited in the country. So far, only a single study that indicated a lack of a major INSTI RM was reported13. Therefore, the current study was intended to provide an update on INSTI RM prevalence among untreated HIV-1 infected people in Ethiopia after 10 years of a previous study13. Accordingly, this study found out that the prevalence of baseline major INSTI RM among untreated patients is 2%. This observation of low level resistance is in line with the previous studies from Ethiopia13, Brazil19, South Africa18,20, India21, Cameroon22, Europe23, and Taiwan4.
The only major INSTI RM detected in the current study is a non-polymorphic mutation R263K that can be selected in vivo during a treatment with DTG and RAL, where it is capable of causing a 2 to 5-fold reduction in susceptibility to DTG, EVG, and RAL6. In addition, R263K has been observed to be selected in vitro under pressure with BIC and the INSTI investigational drug CAB6.
Compared to high-income settings, where it is primarily associated with subtype B-infected individuals treated with ABC/3TC/DTG, this R263K mutation, along with other DTG RMs, is reported to be more common in low-middle-income settings, where patients with treatment failure use an alternative NRTI drug class, leading to the accumulation of multi-NRTI resistance24.
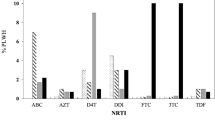
In this study, we have also further detected accessory RMs that have little effect unless they present with other major mutations. The overall magnitude of these polymorphic accessory RMs was 28.5%, which is higher than detection rate in studies from Cameroon 8.1–10%22,25, India 10.1%26, and sub-Saharan Africa 8.7%27. Among the minor RMs detected in the current study, M50I was the most common accessory RMs (13 of the 14 (93%) minor mutations detected from 13/49 (28.3%) participants, which is in line with the magnitude of its global distribution28 (Fig. 3). Moreover, this detection rate is higher relative to the report from previous study in Ethiopia13, indicating the increases this variant's circulation in the country. M50I polymorphic mutation is selected in vitro by DTG and BIC in combination with R263K, a combination that appears to reduce DTG susceptibility6.
The other minor RMs detected were L74I (1/49; 2%), S119R (1/49; 2%), and S230N (1/49; 2%). L74M alone has minimal, if any, effect on INSTI susceptibility. However, they contribute to reduced susceptibility to each of the INSTIs when present in combination with major mutations6. S119R is a polymorphic mutation that is weakly selected by INSTIs usually in combination with several major INSTI-associated drug RMs. Alone, it has little, if any effect, on INSTI susceptibility28. While all these three accessory RMs were previously reported from other countries like South Africa18 and India28 in line with the current study, only the former (L74I) was detected previously in Ethiopia13. The overall comparative summary of these accessory mutations detected in the current study with HIV-1 isolates from untreated patients from South Africa18, India28, Ethiopia13, and worldwide28 is presented in Fig. 3.
Regarding the HIV-1 integrase diversity observed, this study found out that all 49 (100%) study subjects harbored HIV-1C subtype, which is in line with previous reports from Ethiopia. This confirms that the HIV-1 strain circulation in Ethiopia is still dominated by HIV-1C despite the early introduction of the clade29.
Conclusions and recommendations
In conclusion, this study confirmed homogeneity in the circulating HIV-1 clade C and indicated that the level of primary HIV-1 resistance to INSTIs among treatment naive population is still low in Ethiopia, which supports the use of INSTIs in the country. However, the observation from this study does not support the need for performing INSTI resistance testing at baseline, as the prevalence is low. Nevertheless, further larger studies are necessary to assess the impact of accessory INSTI RMs on INSTI-based ART regimens.
Data availability
All nucleotide sequences from this study have been deposited in the Genbank (ncbi) repository with accession numbers from MW560010 to MW560058.
Abbreviations
- AHRI:
-
Armauer hansen research institute
- AIDS:
-
Acquired immuno deficiency syndrome
- ART:
-
Antiretroviral treatment
- ARV:
-
Antiretroviral
- HIV:
-
Human immunodeficiency virus
- HIVDR:
-
HIV drug resistance
- IN:
-
Integrase
- INSTI:
-
Integrase strand transfer inhibitor
- NRTI:
-
Nucleoside reverse-transcriptase inhibitor
- NNRTI:
-
Non-nucleoside reverse transcriptase inhibitor
- PCR:
-
Polymerase chain reaction
- PDR:
-
Primary drug resistance
- PI:
-
Protease inhibitor
- PR:
-
Protease
- RM:
-
Resistance mutation
- RT:
-
Reverse transcriptase
- NoP:
-
Nonpolymorphic
- WHO:
-
World health organization
References
World Health Organization. HIV drug resistance strategy. (2021).
Tsai, H. C. et al. HIV-1 integrase strand-transfer inhibitor resistance in southern Taiwan. Oncotarget 9(38), 24927–24935 (2018).
Unaids, Global HIV & AIDS Statistics—2018 Fact Sheet, 2019. http:// www.unaids.org/en/resources/fact‐sheet. Accessed Sep (2019).
Chang, S. Y. et al. Prevalence of Integrase strand transfer inhibitors (INSTI) resistance mutations in Taiwan. Sci. Rep. 6, 35779 (2016).
Alaoui, N. et al. Prevalence of resistance to integrase strand-transfer inhibitors (INSTIs) among untreated HIV-1 infected patients in Morocco. BMC. Res. Note 11(1), 369 (2018).
Wensing, A. M. et al. 2019 update of the drug resistance mutations in HIV-1. Top. Antivir. Med. 27(3), 111 (2019).
Markham, A. Cabotegravir plus rilpivirine: First approval. Drugs 80, 915–922 (2020).
Richetta, C., Tu, N. Q. & Delelis, O. Different pathways conferring integrase strand-transfer inhibitors resistance. Viruses 14(12), 2591 (2022).
Wensing, A. et al. 2022 update of the drug resistance mutations in HIV-1. Top. Antivir. Med. 30(4), 559–574 (2023).
Smith, S. J. et al. Integrase strand transfer inhibitors are effective anti-HIV drugs. Viruses 13(2), 205 (2021).
Rhee, S. Y. et al. A systematic review of the genetic mechanisms of dolutegravir resistance. J. Antimicrob. Chemother. 74(11), 3135–3149 (2019).
Federal Ministry of Health (FMOH). National guidelines for comprehensive HIV prevention, care and treatment. (2018).
Mulu, A., Maier, M. & Liebert, U. G. Lack of integrase inhibitors associated resistance mutations among HIV-1C isolates. J. Transl. Med. 13, 377 (2015).
Kiros, M. et al. Increased HIV-1 pretreatment drug resistance with consistent clade homogeneity among ART-naive HIV-1 infected individuals in Ethiopia. Retrovirology 17(1), 1–10 (2020).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Bradley-Stewart, A., Urcia, C., MacLean, A., Aitken, C. & Gunson, R. HIV-1 integrase inhibitor resistance among treatment naïve patients in the West of Scotland. J. Clin. Virol. Off. Pub. Pan Am. Soc. Clin. Virol. 92, 7–10 (2017).
Mikasi, S. G. et al. HIV-1 drug resistance mutation analyses of cameroon-derived integrase sequences. AIDS Res. Hum. Retrovir. 37(1), 54–56 (2021).
Brado, D. et al. Analyses of HIV-1 integrase sequences prior to South African national HIV-treatment program and available of integrase inhibitors in Cape Town, South Africa. Sci Rep. 8(1), 4709 (2018).
Torres, M. K. et al. Low prevalence of HIV-1 integrase resistance among antiretroviral-naive patients newly diagnosed with HIV-1 from Belém, Pará, Amazon region of Brazil. AIDS Res. Hum. Retrovir. 36(2), 97–98 (2020).
Onoriode Digban, T., Chucks Iweriebor, B., Chikwelu Obi, L., Nwodo, U. & Ifeanyi, O. A. Analyses of HIV-1 integrase gene sequences among treatment-naive patients in the Eastern Cape South Africa. J. Med. Virol. 92(8), 1165–1172 (2020).
Karade, S., Sen, S. & Sashindran, V. K. Absence of integrase strand transfer inhibitor associated resistance in antiretroviral therapy naïve and experienced individuals from Western India. AIDS Res. Hum. Retrovir. 35(6), 567–571 (2019).
Mikasi, S. G. et al. HIV-1 drug resistance mutation analyses of Cameroon derived Integrase sequences. AIDS Res. Hum. Retrovir. 37(1), 54–56 (2020).
Casadellà, M. et al. Primary resistance to integrase strand-transfer inhibitors in Europe. J. Antimicrob. Chemother. 70(10), 2885–2888 (2015).
Ahmed N., Flavell S., Ferns B., Frampton D., Edwards S., Miller R., et al., editors. Development of the R263K mutation to dolutegravir in an HIV-1 subtype D virus harboring 3 class-drug resistance. Open forum infectious diseases; 2019: Oxford University Press US.
Mikasi, S. G. et al. HIV-1 integrase diversity and resistance-associated mutations and polymorphisms among integrase strand transfer inhibitor-naive HIV-1 patients from cameroon. AIDS Res. Hum. Retrovir. 36(5), 450–455 (2020).
Wenk, B. M. et al. Prevalence of integrase strand transfer inhibitor resistance mutations in antiretroviral-naive HIV-1-infected individuals in Cameroon. J. Antimicrob. Chemother. 76(1), 124–129 (2021).
Inzaule, S. C. et al. Primary resistance to integrase strand transfer inhibitors in patients infected with diverse HIV-1 subtypes in sub-Saharan Africa. J. Antimicrob. Chemother. 73(5), 1167–72 (2018).
Liu T.F., Shafer R.W. (2006). Web resources for HIV type 1 genotypic-resistance test interpretation. Clin. Infect. Dis. 42(11):1608–18. Epub 2006 Apr 28. [Accessed Feb 08, 2021].
Abebe, A. et al. Timing of the introduction into Ethiopia of subcluster C′ of HIV type 1 subtype C. AIDS Res. Hum. Retrovir. 17(7), 657–61 (2001).
Acknowledgements
We would like to thank the Armauer Hansen Research Institute for the provision of financial support to this study. We thank Ethiopian Public Health Institute (EPHI), for availing the ABI PRISM® 3500 xL automated Genetic Analyzer for use in this study. We would like also extend our acknowledgment to all subjects who participated in this study and all staffs of the Armauer Hansen Research Institute for their unreserved help.
Funding
This research project was funded by the Armauer Hansen Research Institute (AHRI).
Author information
Authors and Affiliations
Contributions
Conceptualization: M.K, A. M.; Laboratory investigation and data acquisition: M.K, D.A.T, A.T, T.S.W, E.K., and D.H.A; Data analysis: M.K, H.A, A.G, A. M., and A. M.; Writing—original draft preparation: M.K; Writing—review and editing: M.K, D.A.T, H.A, A.G, A.T, T.S.W, E.K., A. M., M.M, D.H.A, W.E.A, and A. M.; Supervision: A. M., M.M, D.H.A, W.E.A, and A. M., Project administration: A. M. and A. M. All authors have read and approved the final version of the manuscript. All authors have approved the manuscript for publication.
Corresponding author
Ethics declarations
Competing of interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kiros, M., Tefera, D.A., Andualem, H. et al. Low level of HIV-1C integrase strand transfer inhibitor resistance mutations among recently diagnosed ART-naive Ethiopians. Sci Rep 13, 6546 (2023). https://doi.org/10.1038/s41598-023-33850-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-33850-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.