Abstract
Gut microbiota is established to be associated with the diversity of gastrointestinal conditions, but information on the variation associated with music and gut microbes is limited. Current study revealed the impacts of music intervention during feeding on the growth performance and gut microbes of mice by using clinical symptoms and 16S rRNA sequencing techniques. The results showed that feeding mice with music had a significant increase in body weight after the 25th day. The Firmicutes and Proteobacteria were the most dominant phylum in the gut microbiota. Also, the relative abundance of the dominant bacteria was variable after musical intervention. In contrast to the control group, a significant decrease in alpha diversity analysis of gut bacterial microorganisms and Metastats analysis showed a significant increase in the relative abundance of 5 genera and one phylum after the music intervention. Moreover, the musical intervention during feeding caused modifications in the gut microbial composition of mice, as evidenced by an increase in the level of Firmicutes and Lactobacillus, while decreases the richness of pathogenic bacteria, e.g. Proteobacteria, Cyanobacteria and Muribaculaceae, etc. In summary, music intervention increased body weight and enhanced the abundance of beneficial bacteria by reducing the prevalence of pathogenic bacteria in gut microbiota of mice.
Similar content being viewed by others
Introduction
Gut microbiota (GM) is a diverse population of microbes that inhabitant in the host digestive system1. Animal gut microbiota form an ecosystem, and the healthy animals GM prevails in a dynamic balance2. Abundant, active, and stable GM promoted human health, and new evidence revealed its an indispensable role in the human body3. The number of microorganisms in the gut is enormous, up to one hundred trillion4. The type, distribution, function and characteristics of the GM vary in different animals and even in different ages of the same individual5. An inevitablelinkage was revealed between centeral nervous system (CNS) and gut microbiota6. The theory of the gut-brain axis, suggests that the CNS-regulated gut microbiota to influences brain activity7. A healthy GM can positively regulate the neuroimmune response to the CNS; conversely, the dysregulation of GM increased the risk of neurodegenerative diseases and exacerbated the disease response to neurological disorders, which in most cases were accompanied by psychiatric, psychological abnormalities and disorders of the GM8,9,10,11.
Music, the sound wave stimulus, affected the physiology and psychology of animals. Previous studies have demonstrated that feeding animals with music can affect their growth performance and animal production12. The rhythm of music encouraged muscle activity and induced vitality in the organism. When the rhythm of music is close to the heart rhythm of the organism, it stimulates the organism to excrete regulating hormones13. A study with music therapy was developed in the United States of America, revealed that it could be applied to unhealthy and injured people to relieve emotions, improve digestion, balance the mind stats, and promotes recovery from disease14. Since China introduced the five-elements music therapy (FEMT) into the medical field in the Pre-Qin Dynasty, the Chinese FEMT had a long history and a complete system15. Scientists observed that FEMT could relieve anxiety symptoms, improve spatial cognition, regulate intestinal microbiota, and assist in drug therapy16. It was hypothesized that the interaction of the bidirectional pathways between the CNS and the gastrointestinal system influenced primarily by the GM, indicating the importance of GM in the treatment of neurological diseases17. Although there are limitations in the mechanisms of music therapy regarding the treatment of disease at present, music therapy was used as a common assistant clinical treatment18. However, there are few reports on the effects of listening music during the feeding process on GM. Therefore, the primary objective of this study was to evaluate the effect of feeding with music on GM and growth performance in mice to provide theoretical support for music therapy.
Results
Clinical symptoms of animal experiments
No animal suffered from depression, illness, or passed away during the entire study. Neither necropsy nor electron microscopic imaging revealed macroscopic and microscopic pathologies. The mice weight addition is shown in Table 1. The weight of the CG group was higher than the MG group (music intervention during feeding, Fig. 1) during the initial 16 days out of the 30-days feeding period, with a significant difference between the two groups on the 4th day (P < 0.05). However, the mice weight between the two groups was overturned after the 19th day, the weight of MG group was higher than that of the CG group between days 19–30 days. Among them, on 25th (38.444 versus 39.600), 28th (39.120 versus 40.238) and 30th (40.120 versus 40.838) day, there was a statistical difference between the groups (P < 0.01; P < 0.05; P < 0.05). Interestingly, during the feeding period we found that the mice following the music intervention were more active than the control mice regarding their emotional condition and activity status.
DNA sequence analyses
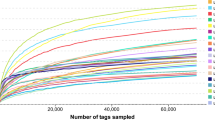
Following microbial composition assignment, a total of 400,333 and 399,471 primitive sequences were found from Jcon and Jm groups that based on 97% nucleotide sequence similarity (Table 2). After removing the unqualified reads, a total of 797,069 reads were available in this study, including Jcon 398,941 and Jm 398,128 reads. The total effective reads used for resulting analysis were 776,089, with an average of 77,609 per sample, ranging from 75,336 to 78,401 reads of all samples. There were 1021 OTUs detected in this test (Fig. 2C), among which Jcon (Fig. 2A) had 926 OTUs (Jcon2 has 318 OTUs; Jcon3 had 458 OTUs; Jcon4 had 685 OTUs; Jcon5 had 380 OTUs and Jcon6 had 412 OTUs) and Jm (Fig. 2B) had 788 OTUs, among them Jm1 had 348 OTUs, Jm2 had 458 OTUs, Jm3 had 432 OTUs, Jm4 had 310 OTUs and Jm5 had 336 OTUs, among which the two groups commonly have 693 OTUs. The Feature curve (Fig. 2D) and Shannon (Fig. 2E) curve tended to be horizontal and stable characteristics, indicating that the sequencing volume and depth justify the analysis requirements. Furthermore, the relative abundance curve was wide and decreased gently, showing significant uniformity and abundance (Fig. 2F).
Operational taxonomic units (OTUs) and sample feasibility analysis. (A) The total number of OTUs in Jcon groups (Jcon2, Jcon3, Jcon4, Jcon5 and Jcon6); (B) the total number of OTUs in Jm groups (Jm1, Jm2, Jm3, Jm4 and Jm5); (C) venn diagrams show specific and common bacteria OTUs in Jcon group and Jm group; (D) rarefaction curves; (E) rank abundance curves; (F) species accumulation curve.
Alteration of gut microbial diversity
We matched the qualified sequences gained by seizure and performed α-diversity analysis to assess the differences in the diversity and abundance of intestinal communities between control and music intervened groups. The results showed that the microbial diversity and abundance were higher in the Jcon group than that of Jm, with a significant chao1 (Fig. 3A) index difference among both groups (P < 0.05). The other three indices (Shannon index, ACE index and Simpson index), didn’t show statistical differences (Fig. 3B–D). Taken together, it could be seen that the feeding process with music decreases the abundance of intestinal microflora. The β diversity analysis was calculated using QIIME software to compare the similarity in the variety of species diversity between samples. The PCoA analysis was based on the unweighted—binary jaccard (Fig. 3E, P > 0.05) and weighted—bray curtis (Fig. 3F, P > 0.05) showed no statistical difference between the two groups. The results showed no difference, indicating a high similarity and low species diversity among both groups.
Microbial composition of the mouse intestines
The relative proportions of dominant samples at the phylum and genus levels were assessed by classifying microbial taxa from different species using QIIME2 software. The results showed that the dominant bacteria at the phylum level in the Jcon were Firmicutes (42.19%), Proteobacteria (19.57%), Cyanobacteria (13.05%) and Bacteroidetes (9.85%), which accounted for 84.66% of the gut microbiota; in Jm, the dominant bacteria at the phylum level were Firmicutes (61.42%), Proteobacteria (16.00%), Cyanobacteria (7.07%) and Bacteroidetes (4.46%), which accounted for 88.94% of the microorganisms (Fig. 4A). The microbial composition (genus level) of both groups showed Lactobacillus as the dominant community. Besides, uncultured_bacterium_f_Enterobacteriaceae (3.97% versus 5.83%) and uncultured_bacterium_f_Muribaculaceae (3.47% versus 1.62%) were further dominant strains in both groups (Fig. 4B). From the above results, it could be concluded that musical intervention during the feeding of mice could alternate the composition of intestinal microorganisms. The heat map indicates the similarities and differences in the gut microbial papulation of multiple samples through the color ramp and degree of closeness (Fig. 4C,D). The results showed that low similarity in microbial abundance between samples of the two groups both at the bacterial phylum level (Fig. 4C) and at genus level (Fig. 4D), but less variation between samples within the groups and high similarity in microbial abundance.
Using Metastats analysis, statistically different classifications of phylum and genus level composition of the two groups of microorganisms were investigated (Fig. 5). The results showed significant differences in relative abundance of Firmicutes (Jcon 0.422 ± 0.056 versus Jm 0.614 ± 0.051, P < 0.05) at the phylum level. Moreover, eleven statistically significant taxa at genus level were found, where Jcon was higher in uncultured_bacterium_o_Microtrichales (P < 0.0001), uncultured_bacterium_f_Micromonosporaceae (P < 0.05), Pseudolabrys (P < 0.05), Methylobacterium (P < 0.05), uncultured_bacterium_f_Muribaculaceae (P < 0.05) and Ruminococcaceae_UCG-005 (P < 0.05) than in the Jm group. Contrary to this, the Jm was higher in uncultured_bacterium_f_Atopobiaceae (P < 0.01), Ileibacterium (P < 0.01), Lachnospiraceae_FCS020_group (P < 0.05), Serratia (P < 0.05) and Dietzia (P < 0.05) than the Jcon group. Considering the limitations of the Metastats analysis for difference in bacterial relative abundance between two groups, the LEfSe analysis (Fig. 6) was performed to search for statistically different biomarkers between Jcon and Jm. The results showed that fifteen statistically different biomarkers were recovered, in addition to the above mentioned significantly different bacteria, that the most dominant groups in Jcon were Verrucomicrobia, Deltaproteobacteria and Acidimicrobiia, while Lleibacterium and Oceanobacillus were significantly expressed in Jm group.
Web-based correlation analysis
In the field of ecology, correlations are often used to construct network models that could be implemented to analyze community data of species (co-occurrence pattern) or to combine multiple data sets for analysis. The taxonomic analysis of all species in this study was shown in supplementary information (Table S1). Top 50 genera were used as base on python to create a web-based correlation, which indicated 78 nodes, 237 edges and 6 communities (Fig. 7). We found that Lactobacillus was the most abundant bacteria among the two groups regarding web-based correlation analysis. Besides, abundance of Lactobacillus in mouse after music intervention (Jm group) was higher than that of control group (47.34% versus 28.03%), while the Phaseolus_acutifolius_tepary_bean richness was lower than control group (6.24% versus 12.36%). The Lactobacillus showed negative correlation with Parabacteroides (0.6364), Faecalibacterium (0.7455), Helicobacter (0.7720), RB41 (0.6485), Blauti (0.8182) and Staphylococcus (0.7538). The Phaseolus_acutifolius_tepary_bean showed positive correlation with Candidatus_Koribacter (0.7173) and Breznakia (0.7195), while it showed negative correlation with Pantoea (0.66060), Enterococcus (0.6809) and Ruminococcaceae_UCG-014 (0.6606).
Discussion
In recent years, increasing attention has been absorbed on environment and animal welfare-associated research19. The enrichment and diversity of environmental factors was an important research parameter to improve animal welfare20. The environmental factors referred to a normal environment in which animals have been provided with the environmental incentives to make beneficial enhancements that allow them to express their behavior and mental activities normally, improving health status and growth performance21,22. Music, as an emotion-reflecting art, could play a positive role in reducing stress in animals23. The hypothalamic–pituitary–adrenal (HPA) axis is an important part of the neuroendocrine system that plays a vital role in regulating the stimulation response of the animals, and its activity could reflect the stress intensity of the animals, in addition, corticosterone (Cort) is a marker of the excitability of the HPA axis24. Previous studies have shown that a mixture of music and inspected sounds significantly reduces the feed-to-meat ratio of piglets and improves their growth performance. However, there are some music (e.g. heavy metal, frequency < 20 Hz, 95–105 dB) that have the opposite effect25,26. This study was performed on El Condor Pasa music, which was natural in style and not overly layered27. In the 30-days feeding experiment (with music intervention), it was found that the body weight of the mice was significantly higher than control group at day 25. Moreover, Gao et al.28 also revealed that the animals consumed more diet and gained significantly more weight after the musical intervention. Some research has found that music can promote immune systems and gut microbial nutrition absorption while also mitigating the negative impacts of noise29. Overall, this study indicated that the music could improve body weight and promote the growth and physical development of mice.
Jejunum is the primary digestive and absorption site of animals, it is essential for animal growth and development30. The nutrients and water in the jejunum pass through the intestinal mucosal epithelium, eventually enter the blood and lymph31. Among them, the intestinal mucosal epithelium is composed of simple columnar epithelial cells, goblet cells and a few endocrine cells32. However, research shows that the absorption of these substances could not be achieved with jejunal mucosal epithelium but the jejunal microbiota33. Music have been found to have a positive effect on the body, inducing a coordinated resonance and promoting the harmony of organ rhythms, along with a series of endocrine transformations34. Furthermore, it influence the coordinated resonance of the digestive system, the secretion of growth hormones, and the growth of beneficial bacteria35. The analysis of DNA sequences of GM showed that the number of OTUs and Reads of music group were lower than those of the control group, indicating that the musical intervention at feeding environment reduced the abundance of microorganisms in the intestinal habitat. Moreover, the mice GM α-diversity and PCoA analyses showed that the music group was modified in comparison to the control group. Interestingly, the whole diversity analysis revealed a high aggregation of intra-group samples in music group and a significant enhancement of intra-group similarity. Sangkyu et al. suggested that a decrease in gut microbial diversity was associated with a decrease in specific bacteria or overgrowth of individual strains36. This showed that musical intervention during feeding can reduce discrete organism microbiota and contribute to the relative stability.
Gut microbiota, the microbial community that resides in the animal intestine, an increasing research trend have been observed in gut microbiota associated fields e.g. microbiology, medicine and genetics, etc37. Analysis of the gut microbiota in the Jcon and Jm groups revealed that Firmicutes, Proteobacteri, Cyanobacteria, Bacteroidetes and Lactobacillus were the most dominant bacteria at the phylum and genus level. Homeostasis of the gut microbiota found to be an essential barrier for resident invasion and colonization by external disease agents, and alterations to the gut microbiota may be linked to a variety of diseases38. Behera et al. indicated that the occurrence of the disease was inextricably linked to the GM39. Apart from gastrointestinal diseases and metabolic diseases, the intestinal microbiota was also associated with a variety of systemic diseases, such as neurological, respiratory, cardiovascular, and oncological diseases40. Moreover, previous studies have shown that FEMT reduced depression and enhance the activity and diversity of the intestinal flora41. We found that musical interventions during feeding, statistically increased the richness of Firmicutes and Lactobacillus, while exponentially reduced the production of Cyanobacteria, Bacteroidetes, Phaseolus_acutifolius_tepary_bean and uncultured_bacterium_f_Muribaculaceae in intestinal microorganisms. Talib et al. revealed that Firmicutes and Lactobacillus were predominant bacteria, indicating a good intestinal health condition42. Furthermore, the Metastats analysis (P < 0.05) with two groups showed significant differences at the genus level with an extra 11 bacterial species, namely: uncultured_bacterium_o_Microtrichales, uncultured_bacterium_f_Micromonosporaceae, Pseudolabrys, Methylobacterium, uncultured_bacterium_f_Muribaculaceae, Ruminococcaceae_UCG-005, uncultured_bacterium_f_Atopobiaceae, Ileibacterium, Lachnospiraceae_FCS020_group, Serratia and Dietzia.
Firmicutes were the most numerous bacterium, most of which are Gram-positive and appear as spherical or rod-shaped, and many members of Firmicutes are beneficial bacteria such as Lactobacillus, Bacillus, Bifidobacterium, Clostridium butyricum, etc43. It has been shown that microorganisms such as Lactobacillus could produce acetate (a short-chain fatty acid), lactate and antibacterial substances that can prevent pathogens from interfering with health44. Moreover, they can help in maintaining the micro-ecological balance of the intestines, prevent and inhibit the occurrence of tumors, enhance animal immunity, promote digestion, synthesize amino acids and vitamins45. Interestingly, it was found that feeding with music could increase the papulation of Lactobacillus. Additionally, it was also found that feeding with in the presence of music caused significant differences in some bacteria among the intestinal microbiota of mice, indicating that such variations may play an important role in the intestinal ecosystem and function. Studies showed that increased gene levels of Lactobacillus in the intestinal microbiota promote growth in mice, trigger the production of interferon and enhance the resistance to animal disease46. Moreover, LEfSe analysis revealed the recovery of 15 statistically different biomarkers among two groups. Specifically, the relative abundance of 5 bacteria significantly increased in the music group, and the relative abundance of 10 bacteria (Beijerinckiaceae, uncultured_bacterium_g_Staphylococcus, Staphylococcaceae, Staphylococcus, Acidimicrobiia, Deinococci, Deinococcus_Thermus, Deltaproteobacteria, Verrucomicrobia, Verrucomicrobiae) significantly increased in the Jcon group.
Materials and methods
Ethical certification
We followed the guidelines of ARRIVE47. The animal experiments were performed in conformity with the Committee on Animal Ethics Code of Operations, Huazhong Agricultural University (license number HZAUMO-2022-0011). All methods were conducted in accordance with the relevant guidelines and regulations.
Animals and sample collection
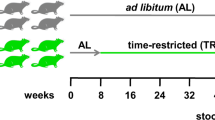
A total of 20 male SPF KM (Kunming) mice were purchased from the Experimental Animal Center of Huazhong Agricultural University (Permits No: SYXK 2020-0084), and all mice weighed 18.0 ± 2.0 g (4 weeks old) and were raised in the Experimental Animal Center (Wuhan, China). During the entire feeding period, sufficient diet (LAD3001G, TROPHIC Animal Feed High-Tech Co. Ltd, China) and water (distilled water) were guaranteed ad libitum for all mice. Moreover, the 12 h of normal light, room temperature of 24.0 ± 2.0 °C, and suitable humidity in the feeding environment was provided. Acclimatization feeding for 2 days in the above-mentioned feeding environment. Subsequently, mice (30 days old) were randomly divided into 4 cages with 5 mice per cage marked as Jcon, Jcont, Jm and Jmt groups. The mice in the Jcon and Jcont groups (CG group) were reared in a normal environment; the Jm and Jmt groups (MG group) were fed in the same environment except for 6 h of music (El Condor Pasa, DanieL alomia roblesas shown in Fig. 1)48 at 10:00 am every day throughout 30 days. Day 1 after the acclimation feeding completed had seen the start musical intervention. Throughout the feeding period, we employ the clinical symptoms to detect the physical signs and survival rate, the mental status was tested with open field test (OFT) method and daily body weights were recorded. After 30 days of feeding, mice were processed with the cervical dislocation method, and the mice were necropsied with reference to the Lorna’s method49. The jejunum and jejunal contents of the mice in the Jcon and Jm groups were taken immediately into sterile 1.0 mL lyophilized tubes and stored at − 80 °C for microbiological studies. To investigate the effect of musical intervention in the mice growth environment on the CNS, the hippocampus and jejunum of the Jcont and Jmt groups were cleaned with sterile saline and then stored frozen at − 80 °C. Fresh tissue samples from both groups (Jcont and Jmt groups), including the hippocampus and jejunum, were immediately fixed in 4% paraformaldehyde at ambient temperature. The fixed tissues were conducted with the assistance of the company (Pinuofei Biological Technology Co, Wuhan). Tissue samples were evaluated under an electron microscope to assess histological changes.
DNA extraction and sequencing
Extraction of microbial genomic DNA from jejunal contents samples were used in the FOREGENE DNA Mini Kit (Chengdu, China) according to the recommended guidelines. The recovered DNA was electrophoresed on a 0.8% agarose gel to confirm completeness and magnitude, and DNA concentration was calculated using a UV–visible spectrophotometer (Alpha-1506, China). Amplification of 16 s rRNA V3/V4 to the conserved region by polymerase chain reaction (PCR) using bacterial general primers 338F (50-ACTCCTACGGGAGGCAGAG-30) and 806R (50-GGACTA CHVGGGTWTCTAAT-30). Amplified PCR products were extracted with QIAquick Gel Extraction Kit (Qiagen, USA) for recovery of target sequences. Based on the preliminary ionization results of electrophoresis, the purified PCR products were fluorescently determined on a Microplate reader (PHERAstar FSX, Germany). Afterward, following the fluorescent quantification and sequencing analysis quality measurements, each sample was blended in the appropriate ratio. Purified PCR was used to produce sequencing libraries using Illumina TruSeq (Illumina, United States) according to the manufacturer's statement. Spliced and screened only single peaks and raw data at concentrations ≥ 2 nM, filtering out polluted data such as chimeric sequences, nucleotide mismatches, and indistinct reads to get exact and reliable sufficient data.
Bioinformatics and data analysis
The Trimmomatic (0.33, https://github.com/usadellab/Trimmomatic) was utilized to filter raw data based on the quality of single nucleotides. Cutadapt (1.9.1, http://cutadapt.readthedocs.org/) was used to identify and remove primer sequences. The USEARCH (10.0.240, http://drive5.com/usearch) was used to construct the PE readings collected in earlier phases, which were then chimera removed using UCHIME (4.1, http://drive5.com/uchime/uchime_download.html). Classify-consensus-blast in QIIME2 (2020.6.0, https://qiime2.org) was a blast-based annotation technique, as the name suggests. It found the annotation with the greatest consensus among N outstanding hits. USEARCH (10.0.240, http://drive5.com/usearch) was used for four main parts that were 97% similar and the conservative OTU filtration cutoff is 0.005%. All sequences were divided into OTUs for various levels of similarity among samples, and each OTU corresponds to one representative sequence. Alpha diversity (Chao1, Ace, Shannon and Simpson) reflected the abundance and diversity among species, and its analysis was performed on QIIME2 (2020.6.0, https://qiime2.org) software. The PCoA analysis was based on the unweighted—binary jaccard and weighted—bray curtis to assume that there exist data that could measure the difference or distance between samples, the approach generated a rectangular coordinate system. Moreover, the T-tests on microbial diversity statistics comparing two groups were performed using Metastats (http://metastats.cbcb.umd.edu/), as the species richness is a continuous variable, meaning it is measured on an interval or ratio scale. The non-parametric factorial Kruskal–Wallis (KW) sum-rank test available for LefSe analysis (Lefse 1.1.1 software, https://github.com/SegataLab/lefse/tree/master/lefse). We used plain package in R language (v3.0.3, https://cran-archive.r-project.org/bin/windows/base/old/3.0.3/), Python (3.8.1, https://www.python.org/downloads/release/python-381/) and GraphPad (6.0, https://www.graphpad.com/dl/96314/10B92408/) for network-wide correlation and statistical analysis. The heat map was produced using the pheatmap (1.0.12) software, of which the link was https://cran.r-project.org/web/packages/. The Spearman (default method) rank correlation analysis was performed based on the quantity of each sample bacterium. The correlation network was built using correlations more than 0.1 and p-values less than 0.05. For statistical analysis SPSS 7.0 software (https://en.freedownloadmanager.org/users-choice/Free_Download_Spss_7.0.html) was used for calculations and P-values were labeled.
Conclusion
Overall, present study described variation of the gut microbiota in mice listening music during feeding. The results showed significant alterations in the gut microbiota after musical intervention, characterized by a decrease in intestinal bacteria diversity and alteration in mice gut microbiota composition. In addition, the papulation of beneficial bacteria increased, while the pathogenic or conditionally pathogenic bacteria were decreased during feeding mice with music. These results contribute in understanding of the relationship between music and the gut microbiota, as well as the essential information that gut microbiota could alter with accordance to different feeding environment. This study also provides a theoretical base for musical feeding to enhance animal welfare through improving the animal growth environment.
Data availability
The datasets supporting the original study inferences are included in the study and as additional files. The original sequence data have been uploaded to the Sequence Read Archive (SRA) (NCBI, USA) with the Accession Number: PRJNA901480.
References
Nagata, N. et al. Human gut microbiota and its metabolites impact immune responses in COVID-19 and its complications. Gastroenterology 164, 272–288 (2023).
Järbrink-Sehgal, E. & Andreasson, A. The gut microbiota and mental health in adults. Curr. Opin. Neurobiol. 62, 102–114 (2020).
Adak, A. & Khan, M. R. An insight into gut microbiota and its functionalities. Cell. Mol. Life Sci. 76, 473–493 (2019).
Chen, X. et al. Pregnancy-induced changes to the gut microbiota drive macrophage pyroptosis and exacerbate septic inflammation. Immunity 56, 336-352.e9 (2023).
Ticinesi, A. et al. Gut microbiota, muscle mass and function in aging: A focus on physical frailty and sarcopenia. Nutrients 11, 1633 (2019).
Kang, J. N. et al. Alterations in gut microbiota are related to metabolite profiles in spinal cord injury. Neural Regen. Res. 18, 1076–1083 (2023).
Kennedy, P. J., Cryan, J. F., Dinan, T. G. & Clarke, G. Kynurenine pathway metabolism and the microbiota-gut-brain axis. Neuropharmacology 112, 399–412 (2017).
Cignarella, F. et al. intermittent fasting confers protection in CNS autoimmunity by altering the gut microbiota. Cell Metab. 27, 1222-1235.e6 (2018).
Chen, H. et al. Gut Microbiota interventions with clostridium butyricum and norfloxacin modulate immune response in experimental autoimmune encephalomyelitis mice. Front. Immunol. 10, 1662 (2019).
Chu, F. et al. Gut microbiota in multiple sclerosis and experimental autoimmune encephalomyelitis: Current applications and future perspectives. Mediat. Inflamm. 2018, 17 (2018).
Kostic, M., Stojanovic, I., Marjanovic, G., Zivkovic, N. & Cvetanovic, A. Deleterious versus protective autoimmunity in multiple sclerosis. Cell. Immunol. 296, 122–132 (2015).
Solanki, M. S., Zafar, M. & Rastogi, R. Music as a therapy: Role in psychiatry. Asian J. Psychiatr. 6, 193–199 (2013).
Raglio, A., Giambelluca, E., Balia, G., Imbriani, C. & Panigazzi, M. Music as support to occupational therapy. G. Ital. Med. Lav. Ergon. 42, 133–136 (2020).
Park, J.-I. et al. Effects of music therapy as an alternative treatment on depression in children and adolescents with ADHD by activating serotonin and improving stress coping ability. BMC Complement. Med. Ther. 23, 73 (2023).
Hao, J., Jiang, K., Wu, M., Yu, J. & Zhang, X. The effects of music therapy on amino acid neurotransmitters: Insights from an animal study. Physiol. Behav. 224, 113024 (2020).
Lin, F., Huang, D., He, N., Gu, Y. & Wu, Y. Effect of music therapy derived from the five elements in Traditional Chinese Medicine on post-stroke depression. J. Tradit. Chinese Med. 37, 675–680 (2017).
Wei, H. et al. Butyrate ameliorates chronic alcoholic central nervous damage by suppressing microglia-mediated neuroinflammation and modulating the microbiome-gut-brain axis. Biomed. Pharmacother. 160, 114308 (2023).
Liao, J. et al. Progressive muscle relaxation combined with Chinese medicine five-element music on depression for cancer patients: A randomized controlled trial. Chin. J. Integr. Med. 24, 343–347 (2018).
da Silva Lourenço, M. I. et al. Behaviour and animal welfare indicators of broiler chickens housed in an enriched environment. PLoS ONE 16, e0256963 (2021).
Mellor, D. J. Positive animal welfare states and encouraging environment-focused and animal-to-animal interactive behaviours. N. Z. Vet. J. 63, 9–16 (2015).
Guyomard, H. et al. Review: Why and how to regulate animal production and consumption: The case of the European Union. Animal 15, 100283 (2021).
Koknaroglu, H. & Akunal, T. Animal welfare: An animal science approach. Meat Sci. 95, 821–827 (2013).
Nardone, V. et al. Music therapy and radiation oncology: State of art and future directions. Complement. Ther. Clin. Pract. 39, 101124 (2020).
Oyola, M. G. & Handa, R. J. Hypothalamic–pituitary–adrenal and hypothalamic–pituitary–gonadal axes: Sex differences in regulation of stress responsivity. Stress 20, 476–494 (2017).
Crone, C. et al. Environmental enrichment for pig welfare during transport. J. Appl. Anim. Welf. Sci. https://doi.org/10.1080/10888705.2021.1983725 (2021).
Lippi, ICd. C. et al. Effects of music therapy on neuroplasticity, welfare, and performance of piglets exposed to music therapy in the intra- and extra-uterine phases. Animals 12, 2211 (2022).
Triadafilopoulos, G. E. Cóndor pasa: Reflections on the condor trial. Gastroenterology 140, 1095–1097 (2011).
Gao, J., Chen, S., Lin, S. & Han, H. Effect of music therapy on pain behaviors in rats with bone cancer pain. J. BUON 21, 466–472 (2016).
Zhang, A. et al. The immune system can hear noise. Front. Immunol. 11, 619189 (2021).
Zhao, J., Liao, D., Yang, J. & Gregersen, H. Biomechanical remodelling of obstructed guinea pig jejunum. J. Biomech. 43, 1322–1329 (2010).
Lema, I., Araújo, J. R., Rolhion, N. & Demignot, S. Jejunum: The understudied meeting place of dietary lipids and the microbiota. Biochimie 178, 124–136 (2020).
Taatjes, D. J. & Roth, J. Glycosylation in intestinal epithelium. Int. Rev. Cytol. 126, 135–193 (1991).
Villmones, H. C. et al. Investigating the human jejunal microbiota. Sci. Rep. 12, 1682 (2022).
Valero-Cantero, I. et al. Complementary music therapy for cancer patients in at-home palliative care and their caregivers: Protocol for a multicentre randomised controlled trial. BMC Palliat. Care 19, 1 (2020).
Nijs, L. & Nicolaou, G. Flourishing in resonance: Joint resilience building through music and motion. Front. Psychol. 12, 666702 (2021).
Kim, S. & Jazwinski, S. M. The gut microbiota and healthy aging: A mini-review. Gerontology 64, 513–520 (2018).
Dinan, T. G. & Cryan, J. F. Brain-gut-microbiota axis and mental health. Psychosom. Med. 79, 920–926 (2017).
Gao, J. et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol. 8, 13 (2018).
Behera, J., Ison, J., Tyagi, S. C. & Tyagi, N. The role of gut microbiota in bone homeostasis. Bone 135, 115317 (2020).
Ma, Q. et al. Research progress in the relationship between type 2 diabetes mellitus and intestinal flora. Biomed. Pharmacother. 117, 109138 (2019).
Zhang, Z., Cai, Z., Yu, Y., Wu, L. & Zhang, Y. Effect of Lixujieyu recipe in combination with five elements music therapy on chronic fatigue syndrome. J. Tradit. Chin. Med. 35, 637–641 (2015).
Talib, N. et al. Isolation and characterization of Lactobacillus spp. From kefir samples in Malaysia. Molecules 24, 2606 (2019).
Koliada, A. et al. Association between body mass index and firmicutes/bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. https://doi.org/10.1186/s12866-017-1027-1 (2017).
So, D. et al. Dietary fiber intervention on gut microbiota composition in healthy adults: A systematic review and meta-analysis. Am. J. Clin. Nutr. 107, 965–983 (2018).
Du, J. et al. Dietary betaine prevents obesity through gut microbiota-drived microRNA-378a family. Gut Microbes 13, 1–19 (2021).
Rahayu, E. S. et al. Effect of probiotic Lactobacillus plantarum Dad-13 powder consumption on the gut microbiota and intestinal health of overweight adults. World J. Gastroenterol. 126, 107–128 (2021).
du Sert, N. P. et al. Reporting animal research: Explanation and elaboration for the arrive guidelines 2.0. PLoS Biol. 18, e3000411 (2020).
Angelucci, F., Ricci, E., Padua, L., Sabino, A. & Tonali, P. A. Music exposure differentially alters the levels of brain-derived neurotrophic factor and nerve growth factor in the mouse hypothalamus. Neurosci. Lett. 429, 152–155 (2007).
Rasmussen, L. & McInnes, E. Necropsy of the Mouse. In A Practical Guide to the Histology of the Mouse: Scudamore 1–24 (Wiley, 2013). https://doi.org/10.1002/9781118789568.ch1.
Funding
The study was supported by Start-up fund of Industry-university-research Project of Ministry of Education in 2022: Research on the Application of Singing Therapy in community Psychological Rehabilitation (220504108271304); 2022 Innovative Training Project of Hubei Province: Study on the effect of singing Therapy on the Mental health of college students under the background of COVID-19 (S202210524047); 2022 University-level Laboratory Research Project of South-Central University for Nationalities: An Empirical Study on College Students’ Music Therapy (SYYJ2022026); 2022 South-Central University for Nationalities Graduate Student Case Database Project (YJS22060); South-central University for Nationalities University-level team: Music therapy Research.
Author information
Authors and Affiliations
Contributions
J.N. conceived and designed the experiments. G.Z., Z.Z., and H.X. contributed sample collection and reagents preparation. B.R. and S.N. revised the manuscript. J.N. wrote the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Niu, J., Xu, H., Zeng, G. et al. Music-based interventions in the feeding environment on the gut microbiota of mice. Sci Rep 13, 6313 (2023). https://doi.org/10.1038/s41598-023-33522-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-33522-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.