Abstract
A growing of evidence has showed that patients with osteoarthritis (OA) had a higher coronavirus 2019 (COVID-19) infection rate and a poorer prognosis after infected it. Additionally, scientists have also discovered that COVID-19 infection might cause pathological changes in the musculoskeletal system. However, its mechanism is still not fully elucidated. This study aims to further explore the sharing pathogenesis of patients with both OA and COVID-19 infection and find candidate drugs. Gene expression profiles of OA (GSE51588) and COVID-19 (GSE147507) were obtained from the Gene Expression Omnibus (GEO) database. The common differentially expressed genes (DEGs) for both OA and COVID-19 were identified and several hub genes were extracted from them. Then gene and pathway enrichment analysis of the DEGs were performed; protein–protein interaction (PPI) network, transcription factor (TF)-gene regulatory network, TF-miRNA regulatory network and gene-disease association network were constructed based on the DEGs and hub genes. Finally, we predicted several candidate molecular drugs related to hub genes using DSigDB database. The receiver operating characteristic curve (ROC) was applied to evaluate the accuracy of hub genes in the diagnosis of both OA and COVID-19. In total, 83 overlapping DEGs were identified and selected for subsequent analyses. CXCR4, EGR2, ENO1, FASN, GATA6, HIST1H3H, HIST1H4H, HIST1H4I, HIST1H4K, MTHFD2, PDK1, TUBA4A, TUBB1 and TUBB3 were screened out as hub genes, and some showed preferable values as diagnostic markers for both OA and COVID-19. Several candidate molecular drugs, which are related to the hug genes, were identified. These sharing pathways and hub genes may provide new ideas for further mechanistic studies and guide more individual-based effective treatments for OA patients with COVID-19 infection.
Similar content being viewed by others
Introduction
Osteoarthritis (OA) is a degenerative joint disease, exerting a substantial health burden on individuals, society and governments1,2. With women and old people disproportionately affected, OA affects more than 500 million people worldwide1,3. The etiology of OA is complicated and is currently believed to be related to a combination of factors such as biomechanical processes, trauma, chronic inflammation, and immune response, etc., which still needs to be explored1,4,5,6,7.
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged at the end of 2019 and has been ravaging the world from then on8,9. There have been 760,360,956 confirmed cases and 6,873,477 deaths of COVID-19 globally up to 16 March 2023 (https://covid19.who.int/).
COVID-19 has placed a severe health burden on individuals, especially those with underlying medical conditions, such as OA9,10. According to a cross-sectional study in Barcelona, OA patients had a higher percentage of COVID-19 infection and a poorer prognosis if infecting COVID-19 in the long term comparing to the general health population11. Furthermore, Fredi et al. reported that seniority and comorbidities were associated with poorer prognosis of COVID-19 infection in patients with rheumatic and musculoskeletal diseases12. In another prospective cohort study, COVID-19 outcomes were worse in inflammatory arthritis patients treated with glucocorticoids13. Moreover, it has been reported that COVID-19 patients would develop some symptoms such as joint pain, muscle discomfort and bone demineralization which resembled clinical manifestations of early OA14.
Some possible mechanism might account for the epidemiological phenomenon. As is known to all, OA often coexists with other chronic diseases such as obesity, cardiovascular disease and diabetes which are all risk factors for COVID-19 infection. For example, in OA individuals with obesity, the pre-existing inflammatory state could accelerate or potentiate SARS-CoV-2 infection via the NLRP3 inflammasome activation and the release of pro-inflammatory cytokines15. Additionally, patients suffering from COVID-19 often presented with hypocalcemia, vitamin D deficiency, and immobility due to the diseases all contributing to bone demineralization which is the typical symptoms of early OA14,16. Scientists also found that the systematic inflammation, the overstimulation of the immune response contributing to endothelial and adipose tissue dysfunction, and neuronal sensitization could be the trigger for OA-like changes in COVID-19 patients14. Additionally, a common neutrophil activation characteristics, renin-angiotensin system (RAS) perturbation and the onset of “cytokine storm” shared by severe COVID-19 and other acute inflammatory states might also be the underlined mechanism which need to be clarified in the future17,18.
Investigating the common gene profiles of both OA and COVID-19 patients may shed light on the underlying common pathogenesis of these two diseases. Our study was the first research aiming to identify key genes associated with the pathogenesis of OA complicated with COVID-19. Integrated bioinformatics and system biology analysis were used to determine common differentially expressed genes (DEGs) and identify hub genes of both COVID-19 and OA. Function annotation, PPI network, transcription factor (TF)-gene regulatory network, microRNA (miRNA)-gene regulatory network and gene-disease association network were constructed based on the DEGs and hub genes. These hub genes may provide new ideas for further biological mechanistic studies and help discover novel therapeutic targets for OA patients with COVID-19 infection.
Materials and methods
Datasets preparation
GSE147507 and GSE51588 datasets were downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database. GSE147507 dataset includes 23 COVID-19 lung biopsy samples and 55 non-COVID-19 controls applying high throughput sequencing Illumina NextSeq 500 platform for extracting RNA sequence19. GSE51588 dataset contains 40 OA subchondral bone samples and 10 healthy controls which used Agilent microarray platform20.
Identification of DEGs and shared DEGs between COVID-19 and OA
We used “limma” package of R software (version 4.1.1) to select DEGs between COVID-19 and non-COVID-19, and between OA and normal21. The criteria for screening out DEGs were set as P < 0.05 and |log fold change (FC)|> 121,22. “Pheatmap”, “EnhancedVolcano” and “ggplot2” packages of R software (version 4.1.1) were applied to generate heatmaps and volcano plots23,24,25. Then the common DEGs of GSE147507 and GSE51588 were acquired using jvenn, an online VENN analysis tool (http://jvenn.toulouse.inra.fr/app/example.html)26.
Gene ontology (GO) and pathway enrichment analysis
GO (biological processes, cellular component, and molecular functions) and pathway enrichment analysis (WikiPathways, Reactome, BioCarta, and Kyoto Encyclopedia of Genes and Genomes (KEGG)) were conducted using EnrichR online tool (https://maayanlab.cloud/Enrichr/) to specify the shared function and pathways between COVID-19 and OA27. The P value < 0.05 was considered significantly enriched.
PPI network analysis
STRING (http://string-db.org) (version 11.5) is a database for the study of protein–protein association networks with increased information coverage on more than 14,000 species, 67 million proteins and 20 billion interactions, supporting functional discovery in genome-wide experimental datasets28. We constructed the PPI network of proteins derived from shared DEGs utilizing the STRING repository with an interaction score > 0.15.
Identification and analysis of hub genes
In the PPI network which consists of nodes, edges and their connections, nodes with the most entanglement are considered as hub genes. Cytohubba (http://apps.cytoscape.org/apps/cytohubba) is a novel plugin of Cytoscape for extracting central elements of a biological network29. The hub genes were selected by applying seven algorithms (Closeness, MCC, Degree, MNC, Radiality, Stress and EPC) and intersecting them.
GeneMANIA (https://genemania.org)30, a flexible user-friendly web site for analyzing gene function, was utilized to construct a co-expression network of identified hub genes.
Construction of TF-gene and miRNA-gene regulatory network
TFs govern gene transcription information and miRNAs post-transcriptionally control gene expression; hence, their activity is essential for gaining molecular insights31,32. NetworkAnalyst is a broad online platform for statistics, visualization, and meta-analysis of web-based gene expression data33. JASPAR (http://jaspar.genereg.net) is a public resource, provides across six classification group of TF combining spectrum of multiple species34. MirTarbase is a tool which helps researchers filter top miRNAs and detect biological functions and features that facilitate the development of biological hypotheses35. We topologically located credible TFs which tend to bind to our hub genes from the JASPAR database on the networkAnalyst platform. Then we extracted miRNAs that interacted with our hub genes from mirTarbase via networkAnalyst.
Gene-disease association analysis
DisGeNET is a comprehensive platform that integrates information on genes and variants associated with human diseases and can be used to investigate the molecular basis of specific human diseases and their comorbidities36. We also examined the gene-disease relationship using DisGeNET database via NetworkAnalyst to disclose associated diseases and their complications with hub genes.
Evaluation of applicant drugs
Drug Signatures database (DSigDB) containing 22,527 gene sets was used to generate the small molecules which could downregulate the expression of hub genes37. The access to the DSigDB database is acquired through Enrichr (https://amp.pharm.mssm.edu/Enrichr/) platform38. Drug molecules were identified using the DSigDB via Enrichr based on the selected hub genes.
ROC curves of hub genes
The receiver operating characteristic (ROC) curve was drawn and the area under the curve (AUC) of the ROC curve was calculated using the pROC package in R in order to detect the diagnostic ability of all candidate hub genes, respectively39.
Results
Identification of DEGs of COVID-19 and OA
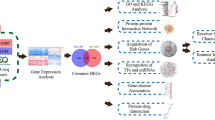
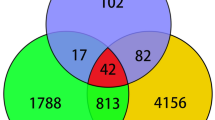
The overall flow chart of this study was shown in Fig. 1. 3569 DEGs were identified in the GSE147507 dataset, and 861 DEGs were figured out in the GSE51588 dataset. Figures 2 and 3 displayed the distribution of DEGs between COVID-19 and non-COVID-19 patients, and between OA patients and normal controls through a heatmap and a volcano plot, respectively. The intersection of DEGs of GSE147507 and GSE51588 datasets was visualized by Venn diagrams, and there were 83 shared DEGs selected (Fig. 4).
Expression characteristics of DEGs in COVID-19 patient. (A) Heat map and (B) Volcano plot present the identified DEGs between COVID-19 patients and normal controls (|logFC|> 1 and P-value < 0.05 were defined as screening standard to obtain DEGs for COVID-19). Blue represents low expression values, and red represents high expression values. DEGs differentially expressed genes, COVID-19 Coronavirus 2019.
Expression characteristics of DEGs in OA. (A) Heat map and (B) Volcano plot present the identified DEGs between OA patients and normal controls (|logFC|> 1 and P-value < 0.05 were defined as screening standard to obtain DEGs for OA). Blue represents low expression values, and red represents high expression values. DEGs differentially expressed genes, OA osteoarthritis.
Gene ontology and pathway enrichment analysis
The most significant terms in the biological process, molecular functions, and cellular components category of GO were positive regulation of muscle contraction, tertiary granule lumen and phosphate iron binding, respectively (Fig. 5). The enriched pathways of the common DEGs between COVID-19 and OA were gathered from four global databases, including KEGG, WikiPathways, Reactome, and BioCarta, and visualized in Fig. 6.
The bar graphs of pathway enrichment analysis of shared DEGs between COVID-19, and OA: (A) wikipathway, (B) biocarta pathway, (C) reactome pathway, and (D) KEGG 2021 human pathway65. DEGs differentially expressed genes, COVID-19 Coronavirus 2019, OA osteoarthritis.
PPI network analysis
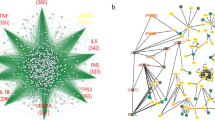
We carefully studied and visualized the PPI network in STRING to predict the interaction and adhesion pathways of common DEGs. The PPI network of common DEGs consisted of 81 nodes and 273 edges, where the PPI enrichment P value was lower than 1.75E−08, as shown in Fig. 7.
Classification and function analysis of hub genes
Seven algorithms of plugin cytoHubba in Cytoscape were applied to screen out the top 30 hub genes. 14 common hub genes were finally identified through the intersection of Venn diagrams, including CXCR4, EGR2, ENO1, FASN, GATA6, HIST1H3H, HIST1H4H, HIST1H4I, HIST1H4K, MTHFD2, PDK1, TUBA4A, TUBB1 and TUBB3 (Fig. 8).
Based on GeneMANIA database, we constructed a complex gene interaction network to decipher the biological functions of these hub genes (Fig. 9), with the co-expression of 63.34%, shared protein domains (17.75%), physical interactions of 17.58%, pathway of 0.75% and co-localization of 0.57%. The results also demonstrated that they were mainly related to the nucleoside binding, purine nucleoside binding, regulation of vacuole organization, purine ribonucleoside binding, guanyl nucleotide binding. These hub genes can be potential biomarkers, which may also lead to new therapeutic strategies for investigated diseases.
Determination of regulatory signatures
TFs and miRNAs interacting with both the DEGs and 14 hub genes were predicted and visualized in Networkanalyst, respectively (Figs. 10, 11). From TFs–genes and miRNA–gene interaction network analysis, it has been ascertained that 71 TFs and 145 miRNAs regulatory signatures regulate with more than one common DEGs. The TF–gene interaction network of 14 hub genes included 53 nodes, 99 edges and 10 genes, while the miRNA–gene interaction network contained 21 nodes, 29 edges and 9 genes.
Identification of disease association
Treatment design strategies for disease open the door to reveal the relationship between genes and diseases. Through the analysis of the gene-disease association by Networkanalyst, we found that arthritis, mammary neoplasms, liver cirrhosis, anemia, autistic disorder, schizophrenia, autosomal recessive predisposition, mental depression, hypertensive disease, bipolar disorder, constipation and cardiac arrhythmia are most related to our hub genes. The association between gene-disease is displayed in Fig. 12.
Identification of candidate drugs
Small molecule drugs regulating the expression of hub genes were collected from the DSigDB database on Enrichr platform. The results from the potential small molecules were generated based on their P values to represent the closeness between the small molecules and genes. Table 1 and Fig. 13 pointed out the top 10 potential small molecule drugs for hub genes.
ROC curves of hub genes
The ROC curve was plotted to evaluate the diagnostic efficacy of 14 key genes (Fig. 14). In the COVID-19 dataset, CXCR4 (AUC:0.733), ENO1 (AUC:0.809), FASN (AUC:0.830), HIST1H4I (AUC:0.755), HIST1H4K (AUC:0.743), TUBA4A (AUC:0.811) and TUBB3 (AUC:0.814) exhibited relatively good diagnostic efficiency for distinguishing the patients with COVID-19 from healthy controls. In the OA dataset, CXCR4 (AUC: 0.998); EGR2 (AUC:0.830), ENO1 (AUC:0.930), FASN (AUC:0.790), GATA6 (AUC:0.902), HIST1H3H (AUC:0.772), HIST1H4H (AUC:0.815), HIST1H4I (AUC:0.820), HIST1H4K (AUC:0.830), MTHFD2 (AUC:0.855), PDK1 (AUC:0.945), TUBA4A (AUC:0.955), TUBB1 (AUC:0.823) and TUBB3 (AUC:0.700) displayed preferable diagnostic performance for differentiating OA from healthy individuals. Specifically, in the COVID-19 dataset, FASN showed the best diagnostic efficiency for differentiating, while CXCR4 showed the best differentiating capability in the OA dataset.
Discussion
There is evidence that patients with osteoarthritis (OA) have a higher prevalence and worse prognosis after COVID-19 infection11,40. COVID-19 infection can cause pathological changes in multiple organs, including the musculoskeletal system18,41. Some relevant researches suggested that some inflammation and immune response might involve in it14. Therefore, we attempted to explore the shared function and pathways between COVID-19 and OA, and to determine the interrelationship between COVID-19 and OA.
In this study, 83 shared DEGs and 14 hub genes (CXCR4, EGR2, ENO1, FASN, GATA6, HIST1H3H, HIST1H4H, HIST1H4I, HIST1H4K, MTHFD2, PDK1, TUBA4A, TUBB1 and TUBB3) have been identified.
Among them, six genes (CXCR4, ENO1, FASN, GATA6, PDK1 and TUBB3) have been reported to be associated with the pathological mechanism of COVID-19 and OA. CXCR4 and PDK1 involved in the pathogenesis of both COVID-19 and OA. CXCR4, shorten for CXC chemokine receptor 4, is a G-protein-coupled receptors (GPCR) which can activate a variety of downstream signaling pathways42. Scientists has found that CXCR4 was highly expressed in COVID-19 patients43. Reasons are probably that CXCR4-positive pre-neutrophils (preNeu) may be released prematurely from bone marrow into blood and infiltrate lung tissue in severe patients44. While in OA tissue, SDF-1/CXCR4 axis has been reported to coordinate the communication between subchondral bone and articular cartilage, which is considered to be a central feature of OA occurrence and development45. PDK1, Phosphoinositide dependent protein kinase-1, has been found to be associated with apoptosis46. In OA, researches demonstrated that PDK1 could promote apoptosis of chondrocytes via modulating MAPK pathway, and has been identified as hub genes of hypermethylation low-expression genes46,47. Another investigation revealed that SARS-CoV-2-encoded nucleocapsid protein N could specifically enhance the M-induced apoptosis via interacting with both M and PDK1, thereby enhancing M-mediated attenuation of PDK1-PKB/Akt interactions48. ENO1 and TUBB3 were found to be probably related to the progression of OA, while FASN and GATA6 may participate in COVID-19. α-Enolase (ENO1) is an enolase isoform widely expressed in almost all mammal cells, characterized as a key glycolytic enzyme and an RNA-binding protein49. In inflammatory arthritis, enhanced surface expression of ENO1 in patient-derived peripheral blood mononuclear cells promotes inflammatory response50. It was also found that ENO1 could promote osteoarthritis progression through interacting with Circular RNA circNFKB1 and sustaining NF-κB signaling49. TUBB3 has been confirmed to be the neuron markers induced by the differentiation process in a model which mimicked a pro-nociceptive microenvironment, which could be further investigated in the field of pain in OA. FASN, Fatty acid synthase, is a key cellular enzyme in palmitate synthesis51. SARS-CoV-2 was expected to increase production of lipid anabolic enzymes including FASN by increasing PI3K/AKT/mTOR/S6K signaling activity52. Scientists also discovered that inhibition of FASN and restoration of lipid catabolism could impede replication of coronaviruses closely related to SARS-CoV-251. GATA6 is a member of a small family of zinc-finger DNA-binding transcription factors, and plays an important role in the regulation of cellular differentiation53. Of note, GATA6 is involved in immune and inflammatory responses by promoting the transcription of SFTPA gene. Scientists discovered that both GATA6 and SFTPA genes were upregulated in SARS-CoV-2-infected lungs54. Genome-wide CRISPR screens also identified GATA6 as a pro-viral host factor for SARS-CoV-2 via modulation of ACE253. The remaining 8 key genes (EGR2, HIST1H3H, HIST1H4H, HIST1H4I, HIST1H4K, MTHFD2, TUBA4A, and TUBB1) were less studied in the roles of COVID-19 and OA, emphasizing their importance in future research.
Enrichment analysis in our study indicated that hypoxia-inducible factor (HIF)-1 signaling pathway is a common pathogenesis of COVID-19 and OA. Since the discovery of HIF-1, numerous studies on the hypoxia signaling pathway have been performed55. The role of HIF stabilization during hypoxia has expanded from the induction of a single gene, erythropoietin, to the upregulation of a couple of hundred downstream targets demonstrating the complexity and importance of HIF signaling pathway55. Several studies have showed that HIF-1 signaling pathway could be a potential target for therapeutic interventions of COVID-1919,52,56. In an integrated proteo-transcriptomics analysis, HIF-1 signaling pathway was markedly modulated during the course of the SARS-CoV-2 infection in vitro52. Codo et al. discovered that elevated glucose levels favored SARS-CoV-2 infection through a HIF-1α/glycolysis-dependent axis56. Moreover, inflammatory and reparative activities of lung macrophages are regulated by β-catenin-HIF-1α signaling, with implications for the treatment of severe respiratory diseases including COVID-1919. HIF-1 signaling pathway is essential in the homeostasis of multiple tissues in OA as well57. For example, Chen et al. found that HIF-1-VEGF-Notch mediated angiogenesis in temporomandibular joint osteoarthritis58. Another in vitro experiment showed that HIF‐1α facilitated osteocyte‐mediated osteoclastogenesis by activating JAK2/STAT3 pathway59. Scientists found that HIF-1α maintained mouse articular cartilage stabilization through suppression of NF-κB signaling60, and the interaction of HIF1α and β-catenin inhibited matrix metalloproteinase 13 expression so that preventing cartilage damage61.
Several chemical agents and drugs have been utilized as potential therapeutic targets against COVID-19 or OA. However, up to now, no drugs were identified to treat individuals with both COVID-19 and OA. In our study, we explored several drugs which could be used as possible targets. Dimethyloxalylglycine is an inhibitor of HIF prolylhydroxylase, which stabilizes and accumulates HIF-1α protein in the nucleus and is an agonist of HIF-1α. Chondrocytes treated with dimethyloxalylglycine could stabilize HIF-1α alleviating osteoarthritis62. Cephaeline acted by repressing HIF-1α ± and disturbing intracellular redox homeostasis. Cephaeline were discovered to inhibit SARS-CoV-2 with EC50 values of low nanomolar levels potently63. Another extracted drug was Cycloheximide, which is an inhibitor of eukaryotic protein synthesis. Cycloheximide inhibits ferroptosis and inhibits autophagy. In clinical trials, cycloheximide showed potent activity against human coronaviruses64.
Although previous studies have respectively explored the pivotal genes associated with each COVID-19 or OA, studies exploring the common molecular mechanisms between the two through bioinformatic approaches are still in vacancy. In this study, for the first time, we explored and identified common DEGs and hub genes of both COVID-19 and OA which may help to further elucidate the pathogenesis of both. However, there are also some limitations in our study. Firstly, the data were downloaded from a public database and input errors could not be assessed. Second, in this study we have used two datasets—one is RNA-seq for COVID-19 and the other one is microarray data for OA. However, microarray data has some limitations over RNA-seq data. Furthermore, the sample sizes, age and sex of COVID-19 and OA are not entirely balanced. Last, this study requires external experimental validation to verify our findings and the function of hub genes needs to be further validated in an in vitro model.
Overall, we identified common DEGs and hub genes for both COVID-19 and OA, and performed multiple bioinformatics analysis based on them. COVID-19 and OA were found to share some common pathogenic mechanism that may be mediated by specific pivotal genes. This study provides a potential horizon for further investigation of the molecular mechanisms, finding novel drugs, developing individual-based therapy for patients with both COVID-19 and OA, and paving the road towards the treatment of long-term COVID-19.
Data availability
The datasets presented in this study can be found in GEO (https://www.ncbi.nlm.nih.gov/geo/), an online repository. GSE147507 and GSE51588 datasets were downloaded from the GEO database.
Abbreviations
- OA:
-
Osteoarthritis
- COVID-19:
-
Coronavirus disease 2019
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- RAS:
-
Renin-angiotensin system
- GEO:
-
Gene Expression Omnibus
- DEGs:
-
Differentially expressed genes
- TFs:
-
Transcription factors
- miRNAs:
-
MicroRNAs
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- DSigDB:
-
Drug Signatures database
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the curve
- CXCR4:
-
CXC chemokine receptor 4
- ENO1:
-
α-Enolase
- FASN:
-
Fatty acid synthase
- AMPK:
-
AMP-activated protein kinase
- PDK1:
-
Phosphoinositide dependent protein kinase-1
- ECM-GC:
-
Glycated extracellular matrix
- DRG:
-
Dorsal root ganglion
- HIF:
-
Hypoxia-inducible factor
References
Hunter, D. J. & Bierma-Zeinstra, S. Osteoarthritis. Lancet 393, 1745–1759 (2019).
Puig-Junoy, J. & Ruiz Zamora, A. Socio-economic costs of osteoarthritis: A systematic review of cost-of-illness studies. Semin. Arthritis Rheum. 44, 531–541 (2015).
Hunter, D. J., March, L. & Chew, M. Osteoarthritis in 2020 and beyond: A Lancet Commission. Lancet 396, 1711–1712 (2020).
Sanchez-Lopez, E., Coras, R., Torres, A., Lane, N. E. & Guma, M. Synovial inflammation in osteoarthritis progression. Nat. Rev. Rheumatol. 18, 258–275 (2022).
Wang, W., Chen, Z. & Hua, Y. Bioinformatics prediction and experimental validation identify a novel cuproptosis-related gene signature in human synovial inflammation during osteoarthritis progression. Biomolecules 13, 127 (2023).
Chen, Z., Wang, W. & Hua, Y. Identification and validation of BCL6 and VEGFA as biomarkers and ageing patterns correlating with immune infiltrates in OA progression. Sci. Rep. https://doi.org/10.1038/s41598-023-28000-9 (2023).
Chen, Z., Wang, W. & Hua, Y. Expression patterns of eight RNA-modified regulators correlating with immune in filtrates during the progression of osteoarthritis. Front. Immunol. 14, 1–12. https://doi.org/10.3389/fimmu.2023.1019445 (2023).
Luo, H. & Zhou, X. Bioinformatics analysis of potential common pathogenic mechanisms for COVID-19 infection and primary Sjogren’s syndrome. Front. Immunol. 13, 1–12. https://doi.org/10.3389/fimmu.2022.938837 (2022).
Alhassan, E., Siaton, B. C. & Hochberg, M. C. Did COVID-19 impact osteoarthritis—Clinical perspective? Curr. Opin. Rheumatol. 34, 68–72 (2022).
Quicke, J. G., Conaghan, P. G., Corp, N. & Peat, G. Osteoarthritis year in review 2021: Epidemiology & therapy. Osteoarthritis Cartil. 30, 196–206 (2022).
Blanch-Rubio, J. et al. Influence of anti-osteoporosis treatments on the incidence of COVID-19 in patients with non-inflammatory rheumatic conditions. Aging (Albany) 12, 19923–19937 (2020).
Ramanathan, K. et al. Since January 2020 Elsevier has Created a COVID-19 Resource Centre with Free Information in English and Mandarin on the Novel Coronavirus COVID 19–21 (Elsevier, 2020).
Haberman, R. H. et al. COVID-19 in patients with inflammatory arthritis: A prospective study on the effects of comorbidities and dmards on clinical outcomes. Arthritis Rheumatol. 72, 1981–1989 (2021).
Lauwers, M., Au, M., Yuan, S. & Wen, C. COVID-19 in joint ageing and osteoarthritis: Current status and perspectives. Int. J. Mol. Sci. 23, 720 (2022).
López-Reyes, A. et al. NLRP3 inflammasome: The stormy link between obesity and COVID-19. Front. Immunol. 11, 1–9 (2020).
Islam, M. B., Nanda, U., Nashiry, A. & Ali, M. Since January 2020 Elsevier has Created a COVID-19 Resource Centre with Free Information in English and Mandarin on the Novel Coronavirus COVID-19 (Elsevier, 2020).
Schimke, L. F. et al. Severe COVID-19 shares a common neutrophil activation signature with other acute inflammatory states. Cells 11, 847 (2022).
Veronesi, F., Contartese, D., Martini, L., Visani, A. & Fini, M. Speculation on the pathophysiology of musculoskeletal injury with COVID-19 infection. Front. Med. 9, 1–7. https://doi.org/10.3389/fmed.2022.930789 (2022).
Brogden, A. K. & Guthmiller, M. J. Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID. Ann. Oncol. 10, 2–5 (2020).
Chou, C. H. et al. Genome-wide expression profiles of subchondral bone in osteoarthritis. Arthritis Res. Ther. 15, 190 (2013).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Sean, D. & Meltzer, P. S. GEOquery: A bridge between the gene expression omnibus (GEO) and BioConductor. Bioinformatics 23, 1846–1847 (2007).
Kolde, R. pheatmap: Pretty Heatmaps. R Packag. version 1.0.8 (2015).
Blighe, K., Rana, S. & Lewis, M. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labelin. R-Package (2019).
Gómez-Rubio, V. ggplot2—Elegant Graphics for Data Analysis 2nd edn, Vol. 77 (Springer, 2017).
Bardou, P., Mariette, J., Escudié, F., Djemiel, C. & Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform. 15, 1–7 (2014).
Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Szklarczyk, D. et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Chin, C. H. et al. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8, S11 (2014).
Franz, M. et al. GeneMANIA update 2018. Nucleic Acids Res. 46, W60–W64 (2018).
Cai, Y., Yu, X., Hu, S. & Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 7, 147–154 (2009).
Lambert, S. A. et al. The human transcription factors. Cell 172, 650–665 (2018).
Xia, J., Gill, E. E. & Hancock, R. E. W. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 10, 823–844 (2015).
Khan, A. et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266 (2018).
Huang, H. Y. et al. MiRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 48, D148–D154 (2020).
Piñero, J. et al. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45, D833–D839 (2017).
Yoo, M. et al. DSigDB: Drug signatures database for gene set analysis. Bioinformatics 31, 3069–3071 (2015).
Kuleshov, M. V. et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, 90 (2016).
Robin, X. et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 12, 77 (2011).
Oh, T. K., Song, I. A., Lee, J., Eom, W. & Jeon, Y. T. Musculoskeletal disorders, pain medication and in-hospital mortality among patients with covid-19 in South Korea: A population-based cohort study. Int. J. Environ. Res. Public Health 18, 6804 (2021).
Wiersinga, W. J., Rhodes, A., Cheng, A. C., Peacock, S. J. & Prescott, H. C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): A review. J. Am. Med. Assoc. 324, 782–793 (2020).
Song, D. et al. PTP1B inhibitors protect against acute lung injury and regulate CXCR4 signaling in neutrophils PTP1B inhibitors protect against acute lung injury and regulate CXCR4 signaling in neutrophils. JCI Insight 7, 158199 (2022).
Daoud, S. & Taha, M. Ligand-based modeling of CXC chemokine receptor 4 and identification of inhibitors of novel chemotypes as potential leads towards new anti-COVID-19 treatments. Med. Chem. 18, 871–883 (2022).
Silvin, A. et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell 182, 1401–1418 (2020).
Qin, H. J. et al. SDF-1/CXCR4 axis coordinates crosstalk between subchondral bone and articular cartilage in osteoarthritis pathogenesis. Bone 125, 140–150 (2019).
Ge, Q. et al. PDK1 promotes apoptosis of chondrocytes via modulating MAPK pathway in osteoarthritis. Tissue Cell 49, 719–725 (2017).
Zheng, L. et al. Identification of abnormally methylated–differentially expressed genes and pathways in osteoarthritis: A comprehensive bioinformatic study. Clin. Rheumatol. 40, 3247–3256 (2021).
Ren, Y. et al. SARS-CoV-2 membrane glycoprotein M triggers apoptosis with the assistance of nucleocapsid protein N in cells. Front. Cell. Infect. Microbiol. 11, 1–12 (2021).
Tang, S. et al. Circular RNA circNFKB1 promotes osteoarthritis progression through interacting with ENO1 and sustaining NF-κB signaling. Cell Death Dis. 13, 1–12. https://doi.org/10.1038/s41419-022-05148-2 (2022).
Bae, S. et al. α-Enolase expressed on the surfaces of monocytes and macrophages induces robust synovial inflammation in rheumatoid arthritis. J. Immunol. 189, 365–372 (2012).
Tanner, J. E. & Alfieri, C. The fatty acid lipid metabolism nexus in COVID-19. Viruses 13, 1–18 (2021).
Gupta, S., Svensson, S., Ambikan, A. T. & Mikaelo, F. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg. Microbes Infect. 9, 1748 (2020).
Israeli, M. et al. Genome-wide CRISPR screens identify GATA6 as a proviral host factor for SARS-CoV-2 via modulation of ACE2. Nat. Commun. 13, 1 (2022).
Islam, A. B. M. M. K. & Khan, M. A. A. K. Lung transcriptome of a COVID-19 patient and systems biology predictions suggest impaired surfactant production which may be druggable by surfactant therapy. Sci. Rep. 10, 1–16 (2020).
Lee, J. W., Ko, J., Ju, C. & Eltzschig, H. K. Hypoxia signaling in human diseases and therapeutic targets. Exp. Mol. Med. 51, 1–13 (2019).
Codo, A. C. et al. Elevated Glucose Levels Favor SARS-CoV-2 (2020).
Fernández-Torres, J., Martínez-Nava, G. A., Gutiérrez-Ruíz, M. C., Gómez-Quiroz, L. E. & Gutiérrez, M. Role of HIF-1α signaling pathway in osteoarthritis: A systematic review. Rev. Bras. Reumatol. 57, 162–173 (2017).
Chen, Y., Zhao, B., Zhu, Y., Zhao, H. Q. & Ma, C. HIF-1-VEGF-notch mediates angiogenesis in temporomandibular joint osteoarthritis. Am. J. Transl. Res. 11, 2969–2982 (2019).
Zhu, J. et al. HIF-1α facilitates osteocyte-mediated osteoclastogenesis by activating JAK2/STAT3 pathway in vitro. J. Cell. Physiol. 234, 21182–21192 (2019).
Okada, K. et al. Hypoxia-inducible factor-1 alpha maintains mouse articular cartilage through suppression of NF-κB signaling. Sci. Rep. 10, 1–11 (2020).
Bouaziz, W. et al. Interaction of HIF1α and β-catenin inhibits matrix metalloproteinase 13 expression and prevents cartilage damage in mice. Proc. Natl. Acad. Sci. U.S.A. 113, 5453–5458 (2016).
Hu, S. et al. Stabilization of HIF-1α alleviates osteoarthritis via enhancing mitophagy. Cell Death Dis. 11, 1 (2020).
Ren, P. et al. A multi-targeting drug design strategy for identifying potent anti-SARS-CoV-2 inhibitors. Acta Pharmacol. Sin. 43, 483–493 (2022).
Khalifa, S. A. M., Yosri, N., El-mallah, M. F. & Ghonaim, R. Since January 2020 Elsevier has Created a COVID-19 Resource Centre with Free Information in English and Mandarin on the Novel Coronavirus COVID-19 (Elsevier, 2020).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Funding
Our research was funded by the China National Key R&D Program (Reference: 2020YFA0803800) and National Natural Science Foundation of China (82072510).
Author information
Authors and Affiliations
Contributions
Z.C. proposed the concept of this research. The first draft of the paper was written by Z.C. All authors contributed equally to the development of the review protocol, data analysis, refinement of the manuscript and approval of the final manuscript. Z.C. reviewed and revised the manuscript before submission for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, Z., Wang, W., Jue, H. et al. Bioinformatics and system biology approach to identify potential common pathogenesis for COVID-19 infection and osteoarthritis. Sci Rep 13, 9330 (2023). https://doi.org/10.1038/s41598-023-32555-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-32555-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.