Abstract
Stingless bees are a diverse group with a relevant role in pollinating native species. Its diet is rich in carbohydrates and proteins, by collecting pollen and nectar supplies the development of its offspring. Fermentation of these products is associated with microorganisms in the colony. However, the composition of microorganisms that comprise this microbiome and its fundamental role in colony development is still unclear. To characterize the colonizing microorganisms of larval food in the brood cells of stingless bees Frieseomelitta varia, Melipona quadrifasciata, Melipona scutellaris, and Tetragonisca angustula, we have utilized molecular and culture-based techniques. Bacteria of the phyla Firmicutes, Proteobacteria, Actinobacteria, and fungi of the phyla Ascomycota, Basidiomycota, Mucoromycota, and Mortierellomycota were found. Diversity analysis showed that F. varia had a greater diversity of bacteria in its microbiota, and T. angustula had a greater diversity of fungi. The isolation technique allowed the identification of 189 bacteria and 75 fungi. In summary, this research showed bacteria and fungi associated with the species F. varia, M. quadrifasciata, M. scutellaris, and T. angustula, which may play an essential role in the survival of these organisms. Besides that, a biobank with bacteria and fungus isolates from LF of Brazilian stingless bees was created, which can be used for different studies and the prospection of biotechnology compounds.
Similar content being viewed by others
Introduction
Stingless bees (Apidae, Meliponini) represent the most diverse eusocial bees, with more than 550 species distributed in tropical and subtropical regions1,2,3. Stingless bees are economically explored in Brazil's north and northeast regions due to the production of honey, wax, geopropolis, pollen, and the commercialization of colonies. However, the main socio-economic contribution is the pollination of native species4,5,6,7,8.
Brazilian stingless bee species present differences in behavior and caste differentiation; for instance, some stingless bees like Frieseomelitta varia and Tetragonisca angustula produce a royal cell to which the nurser worker will add a large quantity of LF (there is no different food such as royal jelly present in honeybee). Specifically, in the Melipona genus, there is no construction of royal cells and no production of exceptional food for the queen. The brood cells have the same size (volume) and receive the same quantity of equal food. Melipona has the “two loci/two alleles model” for cast differentiation, with queen development up to 25% under ideal food supply conditions9,10.
Despite the diverse mores of caste differentiation among stingless bee species, this process is regulated by juvenile hormone (JH) titter9,11. Although this hormone is produced by the corpora allata glands of stingless bees, the JH precursors can be obtained from metabolites of microorganisms associated with stingless bees. Recently, interesting works showed that Scaptotrigona depilis stingless bees have a mutualist relationship with fungi of the genera Monascus and Zygosaccharomyces present in larval food (LF), influencing the development of these bees12,13,14. Besides those relationships well established, the frequency with Candida apicola and Starmerella meliponinorum yeasts have been found in pollen and honey of Melipona quadrifasciata, and Tetragonisca angustula suggests a mutualistic relationship between them15.
These bees are the primary pollinators in Brazil, and the hive structure is entirely different from other social bees. Despite the significant importance of stingless bees for pollination, little is known about the microbiological community inside the hive16,17. Nectar and pollen collected by foragers are stored in sealed cerumen pots, fermented by microorganisms to supply the necessary nutrients for the development of larvae and the survival of adults17. Through these processes, the nectar—transformed into honey by bees and fermented by microorganisms—will be the primary source of carbohydrates for these bees. In addition, fermented pollen is the source of proteins, lipids, and other nutrients18,19,20,21. LF results from a mix of fermented pollen, honey, and glandular secretions of nurse bees and is richly associated with microbial communities16,22,23,24,25.
Microorganisms in the hive contribute to the development of the bee's immune system, aid in food digestion, and defend the hives against pathogens26,27,28. Bacteria of the genera Bacillus and Streptomyces were found in beehives of Melipona and Trigona. These organisms produce antimicrobial substances against fungi and Paenibacillus larvae besides presenting fermentative potential29,30,31,32.
However, they present the structure of cerumen pots to fermented stock food in the colony (pollen and honey pots) and the production of LF33. Despite the recent advances in microbiota research associated with stingless bee colonies, the composition of the LF microbiome of these bees still needs to be determined. The role of the microorganisms in the feeding and the manutention of bee colonies stays an open field for investigation.
Due to the importance of microorganisms to stingless bees, knowing the microbial community present in beehives becomes necessary. In this work, to better understand and create a microorganism collection from stingless bees, we describe the microbiome present in LF of four species of Brazilian stingless (Frieseomelitta varia, Melipona quadrifasciata, Melipona scutellaris, and Tetragonisca angustula) bee reared in the urban area of the Cerrado (Brazilian Savanna) biome (one of the most important and large biomes in Brazil despite little-studied34,35). The LF microbial—cultivated and non-cultivated—communities were molecularly characterized by Next Generation DNA Sequencing (NGS) of rRNA regions—V3/V4 of 16S and ITS1—and characterized by culturing-based techniques. Besides that, we created a biobank with 189 bacteria and 75 fungus isolates from the LF of Brazilian stingless bees, which can be used for different studies and the prospection of biotechnology compounds.
Results
Raw and filtered data from NGS
A total of 1,002,306 amplicons with good quality were obtained after filtering, merged (F and R), and chimera removing from 1,315,720 raw reads of the region V3/V4 of 16S rRNA gene from microorganism present in LF of the stingless bees. Reads were clustered into 129 ASVs for the bacterial community. For the ITS1 region, a total of 624,309 amplicons with good quality were obtained after filtering, merged, and chimera removing from 4,420,973 raw reads. The reads were clustered into 300 ASVs fungal communities (Table 1).
Bacterial community sequencing
The bacterial composition analysis classified one hundred and twenty-nine ASVs (See Supplementary Table S1). No ASVs were found simultaneously in the LF of the four bee species. Sixty-three exclusive ASVs were detected in F. varia LF, 23 in M. quadrifasciata, 25 in M. scutellaris, and 16 in T. angustula (Fig. 1A).
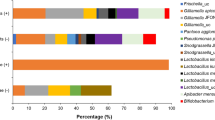
The most abundant phyla observed in the LF of the four bee species were Firmicutes and Proteobacteria. The phylum Actinobacteria was not represented in the species M. scutellaris and T. angustula (Table 2). The ASVs were divided into 26 genera; the most abundant genus in all bees was Lactobacillus (Fig. 1B). Fifteen exclusive bacteria genera showed in F. varia, five in M. quadrifasciata, and only the genus Anthococcus in M. scutellaris (See Supplementary Table S1). No exclusive genus was found in the LF of T. angustula; the genus Micrococcus is expected in the LF of F. varia and M. quadrifasciata. Only five ASVs were classified at a species level, including Bombella intestine, Corynebacterium lipophiloflavum, Serratia symbiotica from LF of F. varia, and Acinetobacter junii and Variovorax paradoxes from LF of M. quadrifasciata.
Fungal community sequencing
Three hundred ASVs were classified in fungal composition analysis from LF (See Supplementary Table S2). F. varia had 39 exclusive ASVs, M. quadrifasciata 68, M. scutellaris 45 and T. angustula 128. However, no common ASVs were found for the four stingless bee species (Fig. 2A).
Fungi of the phyla Ascomycota, Basidiomycota, Mucoromycota, and Mortierellomycota were identified. The phyla Ascomycota and Basidiomycota were the most abundant in LF of stingless bee species of this work. The only phylum in LF for the four species of stingless bees was Ascomycota (Table 3).
Stingless bees of the genus Melipona shared three exclusive ASVs, with ASV2 classified at the fungi kingdom, ASV44 as the genus Didymella, and ASV23 as Saccharomyces cerevisiae. The family emphasizing M. quadrifasciata and F. varia were Claridosporiaceae, and for T. angutula, Erysiphaceae (Fig. 2B) stood out. Forty-eight genera of fungi were identified, 11 were yeast genera, and 37 were filamentous fungi. In F. varia was found six genera of fungi, four species of yeasts, and two species of filamentous fungi. Different from the species M. quadrifasciata, M. scutellaris, and T. angustula, where filamentous fungal genera were most representative.
Fifty-eight fungal species were identified. The most abundant species in LF of F. varia were Cladosporium exasperatum and Saccharomyces cerevisiae. In M. quadrifasciata were Cladosporium delicatulum and Penicillium citrinum. In M. scutellaris, Cladosporium delicatulum, Bipolaris shoemakeri and T. angutula, Erysiphe quercicola and Saccharomyces cerevisiae (Fig. 2C). The F. varia LF showed three exclusive species, M. quadrifasciata, 21, M. scutellaris, five and T. angustula, 11.
Richness and diversity indexes
The richness and diversity indexes from bacterial community sequencing were higher in the samples of F. varia LF regarding the number of bacterial species. The LF of T. angustula showed the lowest diversity, while M. quadrifasciata presented the lowest species diversity indexes. Melipona scutellaris and T. angustula showed a similar Shannon and Simpson diversity index (Fig. 3A).
For Fungal Community Sequencing, the higher richness of fungi species was found in T. angustula and the lowest in F. varia. M. quadrifasciata and T. angustula LF had the highest Simpson and Shannon indices, and M. scutellaris had the lowest indices (Fig. 3B).
The Principal Coordinate Analysis (PCoA) of the bacteria community suggests different microbial patterns for M. quadrifasciata and M. scutellaris compared with F. varia and T. angustula LF (Fig. 4A). Patterns of F. varia and T. angustula showed similar compositions and were grouped separately from Melipona stingless bees, suggesting they present a similar species composition. Likewise, the PCoA analysis of the fungal community suggests different microbial among samples (Fig. 4B).
Cultivable fraction of the microbiome
A total of 189 bacteria were isolated in culture medium, 41 from LF of F. varia, 41 of M. quadrifasciata, 71 of M. scutellaris and 36 of T. angustula. Seventy-five fungi specimens were isolated from LF stingless bees, ten from F. varia, 18 from M. quadrifasciata, 29 from M. scutellaris, and 18 from T. angustula. The isolated microorganisms constructed of the Collection of Microorganisms Isolated from Stingless Bee of the Laboratory of Genetics, Institute of Biotechnology at Federal University of Uberlandia (CoMISBee, Table 4). It was possible to identify some of the bacteria by the Matrix Associated Laser Desorption-Ionization—Time of Flight (MALDI-TOF) method, 42 bacteria isolated from the LF of the four stingless bees were grouped into 12 genera (Fig. 5A). The isolated fungi have not yet been identified.
Stingless bees of the genus Melipona had the highest number of organisms cultivated and identified for MALDI. For M. scutellaris were identified 12 bacteria specimens, and eight for M. quadrifasciata, F. varia, and T. angustula had only two bacteria identified (Fig. 5B). The only genera observed by both NGS and culture-based methods were Enterococcus and Serratia, isolated from F. varia, and Pseudomonas, isolated from M. quadrifasciata.
Discussion
This work detected bacteria, fungi, and yeasts associated with LF of four species of Brazilian stingless bee (F. varia, M. quadrifasciata, M. scutellaris, and T. angustula) present in an urban Meliponary. These microorganisms are essential for colony maintenance, pollen maturation, and honey fermentation of from stingless bees16,22,23,24,25. Due to the lack of data about the microbiome in stingless bee, our goal was to produce the first descriptive data about microbiome present in the LF of these species. As the beehives were reared in the same place and samples were collected simultaneously, the data were suitable to be compared. Differences were found in the LF microbiota of the four stingless bees, showing high variability and richness composition in species25.
Sequencing the V3/V4 region of the 16S gene and region ITS1 and inference of ASV allowed assessing the microbial diversity in the LF of the four stingless bee species. The sequencing of the V3/V4 region of the 16S permitted the classification of 116 ASVs into 27 different genera and only five ASVs at the species level. This region was used due to its great taxonomic coverage to genus level36,37. This work is the first report of the bacterial species Acinetobacter junii, Bombella intestine, Corynebacterium lipophiloflavum, Serratia symbiotica, and Variovorax paradoxus in stingless bees. The sequencing of 16S regions enabled identifying the microbial community in the LF of stingless bees and the discovery of new species related to stingless bees38.
The bacterial community analysis showed that the most abundant bacteria in the LF of the stingless bees investigated were those of the Lactobacillus and the family Acetobacteraceae. This abundance has also been previously reported in the honey stomach/anterior intestine of Melipona39,40. Therefore, itmay explain their presence in food, as the nurser bee transports fermented honey and pollen by ingestion, storage in the honey stomach, and subsequent regurgitation into the brood cells4,41,42,43 .
In this study, 42 bacteria isolated by the culture-dependent method have been identified. No species of the genus Lactobacillus were found in these isolates, although we used MRS agar (selective for Lactobacillus—LB 172,234). The absence of these microorganisms in isolation shows that the database used for identification may not have covered the specimens present in CoMISBee, since bacteria of the genera Bacillus and Lactobacillus were found using NGS but were not identified and not isolated.
Bacteria of the genus Lactobacillus play an essential role in pollen and nectar processing, honey storage, and protection of these bees24,43,44. The genus Bacillus, although somewhat related to nectar and pollen processing, produces enzymes such as amylase, esters, lipases, proteases, aminopeptidases, phosphatases, and glycosidases23,45. Such enzymes had been described as present in the LF of M. scutellaris22. Also, a significant enrichment for metabolites related to lactose degradation and galactose metabolism showed in the LF of M. scutellaris46. The presence of this genus in the colonies of M. quadrifasciata may be related to the fermentation of pollen and honey and may also protect against pathogens by producing antimicrobial molecules24,47,48,49.
In this investigation, Paenibacillus sp. was isolated in M. scutellaris, but not identified in the NGS method. Species of this genus are significant producers of antimicrobials and various enzymes50. Paenibacillus polymyxa, found in the LF of the bee M. scutellaris, can produce antimicrobial compounds protecting these bees against pathogens51. However, although Paenibacillus harbors species beneficial to bees, some species of this genus are disease-causing honeybees50,52.
The discovery of bacteria of the genera Lysinibacillus and Serratia in the colonies of this work may be a warning of the health of these bees. There is only one report of pathogens associated with stingless bees, the bacterium Lysinibacillus sphaericus disease-causing in Tetragonula carbonaria53,54. Moreover, Serratia marcescens, an opportunistic disease-causing species in Apis mellifera54,55 , was found in this investigation in the LF of F. varia and M. scutellaris through the isolation technique. Specific studies on the impact of these microorganisms on the health conditions of these colonies and the identification of the species Lysinobacillus sp using other methodologies are necessary.
Interestingly, the bacteria Lysinobacillus sp. and Serratia marcescens were not found in DNA sequencing. It might show that other microorganisms present in the LF prevent the growth of S. marcescens and keep it so low that the extraction and sequencing technique could not assess this specie. It is essential to highlight that all colonies used for collecting LF were healthy.
The isolation of bacteria in a culture medium is essential for accessing these microorganisms and allowing their biotechnological use. Nonetheless, the fact that it does not identify among the isolated species some bacteria essential for the fermentation of LF reinforces the importance of molecular techniques to elucidate better the microbiome's diversity. Furthermore, this fact emphasizes the importance of using both methodologies to know of the microbiota of stingless bees38.
In addition to intimate relationship with bacteria, stingless bees have symbiont relationships with fungi3,12,14. The diversity of fungi in the LF of stingless bees was visualized with the sequencing of the ITS1 region. The sequencing of the ITS1 region was effective in showing the fungal composition56,57. Distinct species of fungi and yeasts were found in the LF of F. varia, M. quadrifasciata, M. scutellaris, and T. angustula. This work reinforces that filamentous fungi and yeasts are closely related to stingless bees 12,13,14.
The diversity of yeast species found in the LF of stingless bees varied among the bee species studied. Saccharomyces, Starmerella, and Candida, are the most common genera in this research. These genera were found in pollen and honey of F. varia, M. quadrifasciata, and T. angustula stingless bees58,59,60,61,62. This group of yeasts is related to pollen and honey fermentation3,15,60,63. The yeast Saccharomyces cerevisiae was observed in the LF of all bee species in this study. Detecting these organisms in stingless bees paves the way to finding new strains with significant biotechnological potential58.The species Saccharomyces cerevisiae, Starmerella meliponinorum, and Rhodotorula mucilaginosa were detected in T. angustula, and our work, suggesting that these yeasts may be related to the hives of these stingless bee species.
The highest diversity of yeast species was observed in the LF of M. quadrifasciata, including the genera Diutina, Meyerozyma, Rhodotorula, and Starmera, found only in the LF of this stingless bee. The genus Bannoa found only in M. scutellaris, the first report of the species Bannoa ogasawarensis related to a stingless bee. This specie has been reported to colonize dead leaves of plants64. The osmophilic yeast Zygosaccharomyces mellis, found only in T. angustula LF, was reported in the honey of the T. angustula65. The Zygosaccharomyces genus was described to be involved in food deterioration in the hive, but it can also establish mutualistic relationships with other organisms3,66.
This work observed filament fungi in the LF of all stingless bee species. Cladosporium was the only genus common among the four species67,68,69. The Cladosporium was previously reported in stingless bee of the Melipona genus69. The most representative filamentous fungi in the stingless bees species are described as phytopathogens70,71. However, they are used as a biological control for various pathogens. Epicoccum dendrobii produces biomolecules capable of inhibiting phytopathogens72. Twenty-one species of Penicillium and 16 of Talaromyces were observed in M. scutellaris75. The genus Penicillium were detected in the LF of F. varia and M. quadrisfaciata and Talaromyces in M. quadrifasciata.
Stingless bees may use spores produced by fungi as a protein source in times of pollen scarcity since the nutritional value of the spores is lower than that of the pollen21,74,75. T. angustula presented the most remarkable diversity of filamentous fungi and is considered a specialist species in colonizing diverse niches. Therefore, it can use fungi as a protein source for to maintain the colonies60.
The diversity of bacteria and fungi in stingless bees follows the diversity indexes evaluated. On the one hand, bee species with a more extraordinary richness of bacteria present a lower richness of fungal species, as occurred in F. varia. On the other hand, the inverse relationship between bacterial and fungal species richness is genuine, as occurred in T. angustula. The yeast diversity found in the LF in our work is like the yeast diversity observed in the honey of the species F. varia, M. quadrifasciata, and T. angustula61, indicating the diversity of yeasts is related to the honey of these species.
In addition, fungi and bacteria are organisms known for their ability to produce metabolites with antimicrobial activity. Bacteria of the genera Bacillus, Lactobacillus, Micrococcus, Pseudomonas, Providencia, Serratia, and Vagococcus, found in this work, can produce compounds that suppress the growth of other bacterial genera76,77,78,79. Fungi of the genera Cladosporium, Aspergillus, or Penicillium can reduce the number of bacteria in LF by secreting antimicrobial molecules capable of inhibiting bacterial strains67,68,80,81. This fact may explain the difference in the composition of fungi, yeasts, and bacteria in the LF of the different bee species.
Our findings on the microbiota of LF reinforce the importance of vertical transmission of the microbiota from social contact (trophallaxis) between individuals in colonies82. Furthermore, they suggest that part of the microbiota associated with LF can pass between bees in the nectar collection process and during the transfer of honey and matured pollen to the brood cells40,43.
This work opens a new perspective in the knowledge of Brazilian stingless bees' microbiomes, which still needs to be explored. In addition, the microorganisms found here can be an essential source for discovering and prospecting novel bioactive compounds. Recently, our group showed that some of these isolated bacteria (genera Providencia, Serratia, and Vagococcus) have a biotechnological potential, showing antimicrobial activity against multiresistant hospital bacteria76.
This research described the microbiota associated with the species F. varia, M. quadrifasciata, M. scutellaris, and T. angustula and discussed their possible roles in maintaining and protecting of the colonies. This work is the first investigation about the microbiota of the LF of stingless bees. It opens the pathway for elucidating the role of bacteria and fungi in the survival of these organisms. Besides that, the first biobank with 189 bacteria and 75 fungus isolates from the LF of Brazilian stingless bees was also created. It can be used for different studies, such as the screening of bioactive compounds with the potential for treating a wide range of diseases.
Methods
Sample collection
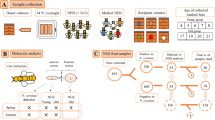
The LF samples were collected from only one beehive of each of the four stingless bees’ species (Frieseomelitta varia, Melipona quadrifasciata, Melipona scutellaris, and Tetragonisca angustula) and processed to obtain microorganism isolated by the culture technique (Fig. 6) or for DNA extraction. The beehives were kept in an urban meliponary in Uberlândia city, Minas Gerais, Brazil. All samples were collected in the springer season, October 2018.
Methodology for cultivating larval food from four stingless bee species with different dilutions and culture mediums. Brain Heart Infusion (BHI), Nutrients (NUT), Tryptone soy agar (TSA), Man, Rogosa and Sharpe (MRS), Mac Conkey (MCK), Mannitol Salgado (MAS), Yeast Malt Agar (ISP), Potato Dextrose Agar 2% (PDA2) and 20% (PDA20) glucose, PDA acidified with tartaric acid (PDAa), PDA NaCl 5% (PDA5s), Oatmeal Agar (OAT), Yeast Extract-Peptone-Dextrose (YPD), DRBC (Dichloran Rose-Bengal Chloramphenicol) e Sabouraud (SAB). Images of representative method from ‘Smart Servier Medical Art’ (https://smart.servier.com/).
Larval food collection
Brood cells were collected from beehives and stored in sterile Petri dishes. In the laminar flow cabinet, the brood cell was cleaned with sterilized distilled water for 1 min and rinsed three times with 70% ethanol. Then, the brood cells were opened with a sterile pipette tip, the eggs were removed, and the LF was collected (the LF from the cell with larvae was discarded). At least 1.0 mL of LF was collected from the brood cells of each colony.
Molecular characterization of the microbiome
DNA extraction
One hundred microliters of LF collected were used for DNA extraction, following the manufacturer’s instructions for bacterial and fungal DNA extractions using the DNeasy® Blood & Tissue (Qiagen) kit. The DNA quantity was measured using NanoDrop 2000 Spectrophotometer, and quality was analyzed by agarose gel 1% electrophoresis stained with ethidium bromide.
Amplicon sequencing and data analysis
For the construction of libraries, 30 ng of DNA was used as input to amplify the V3/V4 region of the 16S rRNA gene with primers 341F (CCTACGGGRSGCAGCAG) and 806R (GGACTACHVGGGTWTCTAAT)36,37, or ITS1 region with the primers IST1(GAACCWGCGGARGGATCA) and ITS2 (GCTGCGTTCTTCATCGATGC)56,83. The amplicons were sequenced in the paired-end reads (2 × 250 bp) of 500 cycles on the Illumina MiSeq platform. The raw data was processed using the package DADA2 in R version 4.0.284 to generate the Amplicon Sequence Variants.
The reads of the V3/V4 region of the 16S rRNA gene were analyzed according to the “DADA2 Pipeline Tutorial” (https://benjjneb.github.io/dada2/tutorial.html). The workflow has been changed in the trinLeft parameter for trimming the primers and using the values 17 for forward, 20 for reverse reads, and truncLen; the forward reads were truncated to 290 and reverse reads to 200 sizes. Taxonomy was assigned with Silva 138 database85.
The amplicons of the ITS1 region were analyzed according to “DADA2 ITS Pipeline Tutorial (1.8)” (https://benjjneb.github.io/dada2/ITS_workflow.html). The Cutadapt86 was used to remove the primers and the minLen parameter to adjust the sequencing quality. The workflow has been changed in parameters trimLeft (default 0), truncLen (default 0), and minLen, removing reads with lengths less than 50. The Amplicon Sequence Variants (ASV) were classified using UNITE ITS database using pre-trained.
The alfa and beta diversity analyses were performed with phyloseq, ggplot2, and vegan packages in R87 using the “DADA2 Pipeline Tutorial”.
Microbiome analysis by cultivation-based techniques and construction of biobank.
Microorganisms’ isolation
Three hundred microliter of LF were collected from brood cells of each colony and used to prepare of three different solutions. First, one hundred microliters of the sample were resuspended in 10 ml of solution 1% NaCl, 100 μL of LF in 10 ml of peptone water, and 100 of LF μL in 10 ml of 15GF broth26, as shown in Fig. 125. Then, the second and third solutions were incubated for 24 h at 32ºC ± 0,5 (Incoterm® Dual Sensor Digital Thermo-Hygrometer) because of is the average internal hive temperature.
All these three solutions were named “mother solution”. From the mother solutions, the serial solutions were made to the order of 10–4, and 100 μL of each was plated on the surface of different culture mediums and incubated in a bacteriological incubator at 32ºC ± 0,5 and checked for 48 h for bacterial growth and seven days for fungal growth.
After the incubation, each colony was isolated on BHI agar for bacteria and PDA agar for fungi. Colonies were characterized by visible morphological differences and gram staining (Newprov). The isolated microorganisms were preserved in LB broth (Luria Bertani) plus 20% glycerol and kept in an ultra-freezer at −80ºC.
Identification by MALDI-TOF of cultivable bacteria
The bacteria were plated on BHI for taxonomic identification and incubated at 37ºC ± 1 for 24 h. An isolated colony of each strain was collected from the agar using an inoculation loop and inactivated with absolute ethanol. Bacterial preparation was performed following the manufacturer's protocol88. Spectra were acquired using a Flex Control Microflex LT mass spectrometer (Bruker Daltonics) with a 60 Hz nitrogen laser. The spectra of each sample were processed using MALDI Biotyper software version 3 (Bruker Daltonics) and assembled to generate a Master Spectral Library (MSP) for strains using the BioTyper MSP breeding standard88,89.
Data availability
The files have been deposited in the SRA (Bioproject: PRJNA860336).
References
Michener, CD. The bees of the world. vol. 1 (JHU Press, 2000).
Melo, G. Stingless Bees (Meliponini). Encyclopedia of Social Insects. (Springer, 2020).
de Paula, G. T., Menezes, C., Pupo, M. T. & Rosa, C. A. Stingless bees and microbial interactions. Curr. Opin. Insect Sci. vol. 44 41–47 Preprint at https://doi.org/10.1016/j.cois.2020.11.006 (2021).
Kerr, W. E., Carvalho, G. A. & Nascimento, V. A. Abelha uruçu: biologia, manejo e conservacao. Manejo da Vida Silvestre (1996).
Villas-Bôas, Jerônimo. Manual tecnológico: mel de abelhas sem ferrão (2012).
Contrera, F. A. L., Menezes, C. & Venturieri, G. C. Revista Brasileira de Zootecnia New horizons on stingless beekeeping (Apidae, Meliponini). (2011).
Alves, R. D. O., Sodré, G. D. S., Souza, B. D. A., Carvalho, C. D. & Fonseca, A. A. O. Desumidificação: uma alternativa para a conservação do mel de abelhas sem ferrão. Mensagem Doce 91, 2–8 (2007).
Dutra, R. P. et al. Antileishmanial activity and chemical composition from Brazilian geopropolis produced by stingless bee Melipona fasciculata. Rev. Bras 29, 287–293 (2019).
Kerr, W. E. Genetic determination of castes in the genus melipona. Genetics 35, 143–152 (1950).
de Carvalho, W. J. et al. Characterization of antennal sensilla, larvae morphology and olfactory genes of Melipona scutellaris stingless bee. PLoS ONE 12, e0174857 (2017).
Kerr, W. E. Sex determination in honey bees (Apinae and Meliponinae) and its consequences. Braz. J. Genet. 20, 601–612 (1997).
Paludo, C. R. et al. Stingless Bee Larvae require fungal steroid to pupate. Sci. Rep. 2018 8:1 8, 1–10 (2018).
Paludo, C. R. et al. Microbial community modulates growth of symbiotic fungus required for stingless bee metamorphosis. PLoS One 14, (2019).
Menezes, C. et al. A Brazilian social bee must cultivate fungus to survive. Curr. Biol. 25, 2851–2855 (2015).
Daniel, H. M. et al. Starmerella neotropicalis f. a., sp. nov., a yeast species found in bees and pollen. Int. J. Syst. Evol. Microbiol. 63, 3896–3903 (2013).
Gilliam, M., Buchmann, S. L., Lorenz, B. J. & Roubik, D. W. Microbiology of the larval provisions of the stingless bee, Trigona hypogea, an obligate necrophage. Biotropica 17, 28–31 (1985).
Engel, P. & Moran, N. A. The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. vol. 37 699–735 Preprint at https://doi.org/10.1111/1574-6976.12025 (2013).
Hassan, K. M. et al. Profiling pH and moisture content of stingless bee honey in closed and opened cerumen honey pots. J Phys Conf Ser 1892, 012032 (2021).
Engel, M. S. & Dingemans-Bakels, F. Nectar and pollen resources for stingless bees (Meliponinae, Hymenoptera) in Surinam (South America). Apiologie 11, 341–350 (1980).
Rebelo, K. S., Ferreira, A. G. & Carvalho-Zilse, G. A. Physicochemical characteristics of pollen collected by Amazonian stingless bees. Ciência Rural 46, 927–932 (2016).
Leonhardt, S. D., Dworschak, K., Eltz, T. & Bï Uthgen, N. Foraging loads of stingless bees and utilisation of stored nectar for pollen harvesting*. Apidologie 38, 125–135 (2007).
Menezes, C., Paludo, C. R. & Pupo, M. T. A review of the artificial diets used as pot-pollen substitutes. in Pot-Pollen in Stingless Bee Melittology (eds. Vit, P., Pedro, S. R. M. & Roubik, D. W.) 253–262 (Springer International Publishing, 2018). doi:https://doi.org/10.1007/978-3-319-61839-5.
Gilliam, M., Roubik, D. W. & Lorenz, B. J. Microorganisms associated with pollen, honey, and brood provisions in the nest of a stingless bee, Melipona fasciata. Apidologie 21, 89–97 (1990).
Mohammad, S. M., Mahmud-Ab-Rashid, N. K. & Zawawi, N. Probiotic properties of bacteria isolated from bee bread of stingless bee Heterotrigona itama. J Apic Res https://doi.org/10.1080/00218839.2020.1801152 (2020).
Ueira-Vieira, C. et al. Pollen diversity and pollen ingestion in an Amazonian stingless bee, Melipona seminigra (Hymenoptera, Apidae). J Apic Res 52, 173–178 (2013).
Martin, F. P. J. et al. Probiotic modulation of symbiotic gut microbial–host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol 4, 157 (2008).
Mazmanian, S. K., Cui, H. L., Tzianabos, A. O. & Kasper, D. L. An Immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005).
Voulgari-Kokota, A., McFrederick, Q. S., Steffan-Dewenter, I. & Keller, A. Drivers, Diversity, and Functions of the Solitary-Bee Microbiota. Trends Microbiol. vol. 27 1034–1044 Preprint at https://doi.org/10.1016/j.tim.2019.07.011 (2019).
Kroiss, J. et al. Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol 6, 261–263 (2010).
Promnuan, Y., Kudo, T. & Chantawannakul, P. Actinomycetes isolated from beehives in Thailand. World J Microbiol Biotechnol 25, 1685–1689 (2009).
Menezes, C., Vollet-Neto, A., Contrera, F. A. F. L., Venturieri, G. C. & Imperatriz-Fonseca, V. L. The Role of Useful Microorganisms to Stingless Bees and Stingless Beekeeping. in Pot-Honey 153–171 (Springer New York, 2013). doi:https://doi.org/10.1007/978-1-4614-4960-7_10.
Menegatti, C. et al. Meliponamycins: antimicrobials from stingless bee-associated Streptomyces sp. J Nat Prod 83, 610–616 (2020).
Prato, M. Influência da quantidade de alimento sobre a produção de sexuados e a determinação de castas em três espécies de abelhas sem ferrão. (Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto - USP, 2015).
Myers, N., Mittermeier, R. A., Mittermeier, C. G., da Fonseca, G. A. B. & Kent, J. Biodiversity hotspots for conservation priorities. Nature | vol. 403 (2000).
Strassburg, B. B. N. et al. Moment of truth for the Cerrado hotspot. Nat Ecol Evol 1, 0099 (2017).
de Sousa, L. P. Bacterial communities of indoor surface of stingless bee nests. PLoS ONE 16, e0252933 (2021).
Takahashi, S., Tomita, J., Nishioka, K., Hisada, T. & Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 9, e105592 (2014).
Liu, S. et al. Opportunities and challenges of using metagenomic data to bring uncultured microbes into cultivation. Microbiome 2022 10:1 10, 1–14 (2022).
Cerqueira, A. E. S. et al. Extinction of anciently associated gut bacterial symbionts in a clade of stingless bees. ISME J. 15, 2813–2816 (2021).
Marçal, L. N. Comunidades bacterianas associadas a colônias de abelhas amazônicas sem ferrão da espécie Melipona seminigra: diversidade e potencial enzimático. (2017).
Nogueira-Neto, P. Vida e criação de abelhas indígenas sem ferrão. (1997).
Vit, P., Roubik, D. W. & Pedro, S. R. M. Pot-Honey: A legacy of stingless bees. . (Springer, 2012).
Vásquez, A. & Olofsson, T. C. The lactic acid bacteria involved in the production of bee pollen and bee bread. J Apic Res 48, 189–195 (2009).
Kešnerová, L. et al. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol 15, e2003467 (2017).
Wang, M., Zhao, W. Z., Xu, H., Wang, Z. W. & He, S. Y. Bacillus in the guts of honey bees (Apis mellifera; Hymenoptera: Apidae) mediate changes in amylase values. Eur J Entomol 112, 619–624 (2015).
Borges, L. D. F. et al. 10-hydroxy-2E-decenoic acid (10HDA) does not promote caste differentiation in Melipona scutellaris stingless bees. Sci Rep 11, 9882 (2021).
Yoshiyama, M. & Kimura, K. Bacteria in the gut of Japanese honeybee, Apis cerana japonica, and their antagonistic effect against Paenibacillus larvae, the causal agent of American foulbrood. J Invertebr Pathol 102, 91–96 (2009).
Machado, J. O. Simbiose entre as abelhas sociais brasileiras (Meliponinae, Apidae) e uma espécie de bactéria. Cienc Cult 23, 625–633 (1971).
Paludo, C. R. et al. Whole-Genome Sequence of Bacillus sp. SDLI1, Isolated from the Social Bee Scaptotrigona depilis. Genome Announc 4, (2016).
Nicholas Grady, E., MacDonald, J., Liu, L., Richman, A. & Yuan, Z.-C. Current knowledge and perspectives of Paenibacillus: a review. Microb Cell Fact 15, 203 (2016).
Menegatti, C. et al. Paenibacillus polymyxa associated with the stingless bee melipona scutellaris produces antimicrobial compounds against entomopathogens. J Chem Ecol 44, 1158–1169 (2018).
Genersch, E. American Foulbrood in honeybees and its causative agent. Paenibacillus larvae. https://doi.org/10.1016/j.jip.2009.06.015 (2009).
Shanks, J. L., Haigh, A. M., Riegler, M. & Spooner-Hart, R. N. First confirmed report of a bacterial brood disease in stingless bees. J Invertebr Pathol 144, 7–10 (2017).
Fünfhaus, A., Ebeling, J. & Genersch, E. Bacterial pathogens of bees. Curr Opin Insect Sci 26, 89–96 (2018).
Raymann, K., Coon, K. L., Shaffer, Z., Salisbury, S. & Moran, N. A. Pathogenicity of serratia marcescens strains in honey bees. mBio 9, (2018).
Schmidt, P.-A. et al. Illumina metabarcoding of a soil fungal community. Soil Biol Biochem 65, 128–132 (2013).
Bazzicalupo, A. L., Bálint, M. & Schmitt, I. Comparison of ITS1 and ITS2 rDNA in 454 sequencing of hyperdiverse fungal communities. Fungal Ecol 6, 102–109 (2013).
Silva, M. S. et al. Selection of yeasts from bee products for alcoholic beverage production. Braz. J. Microbiol. 51, 323–334 (2020).
Teixeira, A. C. P. et al. Starmerella meliponinorum sp. nov., a novel ascomycetous yeast species associated with stingless bees. Int J Syst Evol Microbiol 53, 339–343 (2003).
da Costa Neto, D. J. & Morais, P. B. de. The vectoring of Starmerella species and other yeasts by stingless bees in a Neotropical savanna. Fungal Ecol 47, (2020).
Echeverrigaray, S. et al. Yeast biodiversity in honey produced by stingless bees raised in the highlands of southern Brazil. Int J Food Microbiol 347, 109200 (2021).
Rosa, C. A. et al. Yeast communities associated with stingless bees. FEMS Yeast Res 4, 271–275 (2003).
Santos, A. R. O. et al. Starmerella camargoi f.A., sp. nov., starmerella ilheusensis f.a., sp. nov., Starmerella litoralis f.a., sp. nov., starmerella opuntiae f.a., sp. nov., Starmerella roubikii f.a., sp. nov. and Starmerella vitae f.a., sp. nov., isolated from flowers and bees, and transfer of related candida species to the genus Starmerella as new combinations. Int. J. Syst. Evol. Microbiol. 68, 1333–1343 (2018).
Parra, P. P. & Aime, M. C. New species of Bannoa described from the tropics and the first report of the genus in South America. Mycologia 111, 953–964 (2019).
Matos, T. T. S. et al. Kluyveromyces osmophilus is not a synonym of Zygosaccharomyces mellis; reinstatement as Zygosaccharomyces osmophilus comb. nov. Int. J. Syst. Evol. Microbiol. 70, 3374–3378 (2020).
Paludo, C. R. et al. Stingless bee larvae require fungal steroid to pupate. Sci Rep 8, (2018).
Sugijanto, N. E. & Dorra, B. L. Antimicrobial activity of Cladosporium oxysporum endophytic fungus extract isolated from Aglaia odorata Lour. Indonesian J. Med. 01, 108–115 (2016).
Ding, L., Qin, S., Li, F., Chi, X. & Laatsch, H. Isolation, Antimicrobial activity, and metabolites of Fungus Cladosporium sp. associated with red alga porphyra yezoensis. Curr Microbiol 56, 229–235 (2008).
Souza, J. R. S. et al. Occurrence of filamentous fungi associated with stingless bees Melipona in meliponaries at the metropolitan region of Manaus, Amazonas. Rev. da Biol. 18, 1–5 (2018).
Modro, A. F. H., Silva, I. C., Message, D. & Luz, C. F. P. Saprophytic fungus collection by africanized bees in Brazil. Neotrop Entomol 38, 434–436 (2009).
Suwannarach, N., Kumla, J., Khuna, S., Cheewangkoon, R. & Lumyong, S. First report of cape gooseberry scab caused by Cladosporium exasperatum in Thailand. Plant Dis https://doi.org/10.1094/PDIS-12-21 (2022).
Taguiam, J. D., Evallo, E. & Balendres, M. A. Epicoccum species: ubiquitous plant pathogens and effective biological control agents. Eur J Plant Pathol 159, 713–725 (2021).
Barbosa, R. N. et al. New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie Van Leeuwenhoek 111, 1883–1912 (2018).
Eltz, T., Brühl, C. A. & Görke, C. Collection of mold (Rhizopus sp.) spores in lieu of pollen by the stingless bee Trigona collina. Insectes Soc 49, 28–30 (2002).
Oliveira, M. L. & Morato, E. F. Stingless bees (Hymenoptera, Meliponini) feeding on stinkhorn spores (Fungi, Phallales): robbery or dispersal?. Rev Bras Zool 17, 881–884 (2000).
Santos, A. C. C. et al. Antimicrobial activity of supernatants produced by bacteria isolated from Brazilian stingless bee’s larval food. BMC Microbiol 22, (2022).
Clements, T., Ndlovu, T. & Khan, W. Broad-spectrum antimicrobial activity of secondary metabolites produced by Serratia marcescens strains. Microbiol Res 229, 126329 (2019).
Feliatra, F., Batubara, U. M., Nurulita, Y., Lukistyowati, I. & Setiaji, J. The potentials of secondary metabolites from Bacillus cereus SN7 and Vagococcus fluvialis CT21 against fish pathogenic bacteria. Microb Pathog 158, 105062 (2021).
Kerr, J. R. Bacterial inhibition of fungal growth and pathogenicity. Microb Ecol Health Dis 11, 129–142 (1999).
SILVA, E. M. S. et al. Metabolites from endophytic Aspergillus fumigatus and their in vitro effect against the causal agent of tuberculosis. Acta Amazon 48, 63–69 (2018).
Petit, P., Lucas, E. M. F., Abreu, L. M., Pfenning, L. H. & Takahashi, J. A. Novel antimicrobial secondary metabolites from a Penicillium sp. isolated from Brazilian cerrado soil. Electron. J. Biotechnol. 12, (2009).
Kwong, W. K. et al. Evolutionary biology dynamic microbiome evolution in social bees.
White, T. J., Bruns, T., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. in PCR Protocols 315–322 (Elsevier, 1990). doi:https://doi.org/10.1016/B978-0-12-372180-8.50042-1.
Callahan, B. J. et al. Dada2: high-resolution sample inference from illumina amplicon data. 13, (2016).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41, D590–D596 (2012).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17, 10 (2011).
Regina, A. L. A. et al. A watershed impacted by anthropogenic activities: Microbial community alterations and reservoir of antimicrobial resistance genes. Sci. Total Environ. 793, 148552 (2021).
dos Santos, R. G. et al. Exploring the MALDI Biotyper for the Identification of Corynebacterium pseudotuberculosis biovar Ovis and Equi. J Am Soc Mass Spectrom 33, 2055–2062 (2022).
Singhal, N., Kumar, M., Kanaujia, P. K. & Virdi, J. S. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol 6, 791 (2015).
Acknowledgements
We thank to Rede Genoma de Minas Gerais and FAPEMIG for making this research possible.
Funding
This project was funded by the Research Support Foundation of the State of Minas Gerais (FAPEMIG APQ-02766-17, APQ-00269-22 and, RED-00132-16), and National Council of Scientific and Technological Development (CNPq) and FAPEMIG for INCT -TeraNano, CNPq grant number: 403193/2022-2 and CBB-APQ-03613-17. The ACCS and other graduate students receive financial support for Coordination for the Improvement of Higher Education Personnel (CAPES) and CNPQ. Biological material of the F. varia, M. quadrifasciata, M. scutellaris, and T. angustula was obtained under Brazilian laws. The species does not fall under the IUCN Red List categories as a threatened species. The specie M. scutellaris is endangered on the official list of species of the Brazilian fauna of the ICMBio.
Author information
Authors and Affiliations
Contributions
C.U.V., R.C.C.D. and A.C.C.S. conceived and supervised the project. A.C.C.S. and L.D.F.B. collected the larval food and performed analyzes. V.A.C.A. and N.D.C.R. performed the MALDI-TOF analyzes for assembly. G.R.F., A.R.S. and A.C.C.S. performed the statistical analyses. C.U.V., A.M.B. and A.C.C.S. wrote the draft. All authors discussed the results and commented on the manuscript. All authors approved its final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Santos, A.C.C., Borges, L.D.F., Rocha, N.D.C. et al. Bacteria, yeasts, and fungi associated with larval food of Brazilian native stingless bees. Sci Rep 13, 5147 (2023). https://doi.org/10.1038/s41598-023-32298-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-32298-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.