Abstract
The parasitoid wasp, Ixodiphagus hookeri (Hymenoptera: Encyrtidae), is the natural enemy of a wide range of hard and soft tick species. While these encyrtid wasps are supposed to be distributed worldwide, only a few studies report on their actual distribution around the globe. Within a shotgun sequencing-based metagenome analysis, the occurrence of I. hookeri was screened at multiple Ixodes ricinus (Acari: Ixodidae) tick sampling points in Hungary to contribute to the assessment of the distribution patterns of the parasitoid wasps in Central Europe. To our knowledge, the first report of the species in Hungary and the description of the southernmost I. hookeri associated geoposition in Central Europe took place within our study. I. hookeri infested I. ricinus nymphs were detected at five sampling points in Hungary. The results show that the exact distribution range of I. hookeri is still barely studied. At the same time, unprecedented public health issues being brought about by climate change might require steps toward the exploitation of the tick biocontrol potential and as an ecological bioindicator role of the parasitoid wasp in the future.
Similar content being viewed by others
Introduction
The emergence of vector-borne diseases that can affect human and animal populations is strongly influenced by climate change, urbanization, and globalization1. Among the most significant arthropod vectors (e.g. ticks, fleas, black flies, mosquitoes, or sand flies), ticks transmit a markedly broad spectrum of pathogenic microorganisms, including various protozoa, rickettsiae, spirochaetes, and viruses2. Since Hungary is situated in the southern part of Central Europe, climate change may facilitate the expansion of certain tick species to this landlocked country from the neighboring Mediterranean region. Currently, 27 hard tick species (Ixodidae) have been described in Hungary3. As part of the VectorNet project, the European Centre of Disease Prevention and Control (ECDC) currently monitors seven tick species, including Ixodes ricinus, that commonly transfer diseases to humans and animals4. Naturally, further tick species may also serve as vectors for microorganisms of human or veterinary medical significance5. Out of the seven tick species monitored by ECDC, all except for Ornithodoros spp. are hard ticks present in Hungary3.
Several studies report the incidence trends of tick-borne encephalitis (TBE) and Lyme borreliosis (LB), the most prevalent tick-borne infection in Europe6,7. In the case of LB, which has higher country-wise incidence rates than TBE, decade-long trends of incidence rates are not consistent along the countries around the world, increasing, and decreasing tendencies both appear. On the other hand, reports of the geographic distribution of LB show a clear expansion, especially towards higher altitudes and latitudes6,7. Although global trends of the incidence rates of these TBDs are not consistent6,7, changes in the distribution range of European I. ricinus populations are. Enhanced surveillance and diagnostic measures raise awareness of the changing geographical distribution, density, and activity of the I. ricinus, the primary vector of TBE and LB in Europe8. As a consequence of climate change, I. ricinus expanded its distribution to areas of higher altitude and latitude apart from its prior range, and its northerly shift within the European continent has also been documented9,10,11.
As climate change, accompanied by various sociodemographic alterations, brings unprecedented challenges related to vector-borne diseases12,13, the need for the development of control methods against tick populations is a public highlight. Several methods have been introduced to address this issue. These control methods often rest on either conventional chemical acaricides or on further alternatives, such as biological control methods assisted by the natural enemies of ticks14,15,16,17,18. A line of biological control methods against ticks could be the Encyrtidae family members which are small-sized, parasitoid, or hyperparasitoid wasps distributed all around the globe. Due to their efficacy and target specificity, numerous wasps from this family are used as biological pest control, while several additional encyrtid species are documented as promising candidates for this role19,20,21. Ixodiphagus spp., including I. hookeri22, are encyrtid wasps attacking a wide range of tick species that have received relatively much attention as a specific and effective, natural alternative for biological hard tick control23,24. Interestingly, I. hookeri appears to have alternating preferences for the tick species and developmental stage of its hosts at geographically distant locations25,26. In European settings, Ixodiphagus wasps are described to parasitize the larvae and nymphs of hard ticks with a clear predilection for unfed nymphs24. If oviposition occurs in larvae, transstadial transmission through the molting of the ticks to nymphs can also occur27. Wasp eggs start their embryonic development in engorging or engorged nymphs. Wasp larvae feed on tick tissues and emerge as fully grown adults causing the death of the host before it can reach the adult stage23,24.
Ixodiphagus wasps have been associated with several hard and soft tick genera belonging to the families of Ixodidae and Argasidae, including Ornithodoros, Amblyomma, Dermacentor, Haemaphysalis, Hyalomma, Ixodes and Rhipicephalus23. Studies conducted in Europe revealed that I. ricinus appears to be the preferred species of the European I. hookeri, while another common tick species, Dermacentor reticulatus is supposed not to be chosen as a host by the European representatives of the parasitoid wasps24.
While Ixodiphagus spp. have been detected in many countries and in a diverse range of hard and soft tick species, parasitoid wasps have been less studied in Hungary despite their potential to reduce tick populations and tick-borne disease cases. In the present study, our aim was to confirm the presence I. hookeri in a diverse set of locations in Hungary using a modern, sensitive metagenomic approach28,29. Due to their high density in Hungary, high public health significance as TBD transmitters, and potential to host Ixodiphagus wasps, I. ricinus ticks were decided to be assessed for the parasitoids. Based on our approach, genomic information of the European populations of I. hookeri may also be obtained, which can serve as a reference for further studies.
Materials and methods
Between March and August of 2019, in two I. ricinus metagenome surveys, questing ticks, were collected from 21 geopositions in Hungary by flagging and dragging. One of the surveys was performed as a country-wide climatically designed sampling (17 sites)30. Further samples were collected in 4 sites popular for outings and dog walking. The closest settlements of sampling points were: Kissziget (a), Sárvár (b), Mosonmagyaróvár (c), Sáska (d), Darány (e), Somogybabod (f), Pénzesgyőr (g), Pécsvárad (h), Vérteskozma (i), Németkér (j), Törökbálint (k), Normafa (l), Nagy-Hideg-Hegy (m), 10th district of Budapest (n), Pusztavacs (o), Kékes (p), Lillafüred (q), Aggtelek (r), Háromhuta (s), Nyíregyháza (t), Nyíradony (u). The collected ticks were frozen at − 18 °C. In the laboratory, the ticks were classified taxonomically using standard morphological keys31, and 10 nymphs and 10 adult females of I. ricinus per sampling sites (Fig. 2) were selected randomly. Before DNA extraction, the ticks were washed twice with 99.8% alcohol.
The blackPREP Tick DNA/RNA Kit (Analytik Jena GmbH) was used for the DNA isolation. Isolated total metagenome DNA was used for library preparation. In vitro fragment libraries were prepared using the NEBNext Ultra II DNA Library Prep Kit for Illumina. Paired-end fragment reads were generated on an Illumina NextSeq sequencer using TG NextSeq 500/550 High Output Kit v2 (300 cycles). Primary data analysis (base-calling) was carried out with Bbcl2fastq software (v2.17.1.14, Illumina).
On the raw sequencing data, quality-based filtering and trimming were performed by TrimGalore (v.0.6.6, https://github.com/FelixKrueger/TrimGalore), setting 20 as a quality threshold, retaining reads longer than 50 bp only. Using the remained reads, a de novo assembly was performed by MEGAHIT (v1.2.9)32 using default settings. The resulting contigs were taxonomically classified using Kraken2 (k = 35)33 with the NCBI non-redundant nucleotide database34. Contigs were predicted as I. hookeri by taxon classification and were checked with BLAST35 on the partial sequence of I. hookeri 28S ribosomal RNA gene (MH077537.1) as a reference. Multiple sequence alignment was done by MAFFT (v7.490)36. All data management procedures, analyses, and plottings37,38 were performed in the R environment (v4.2.1)39.
Results
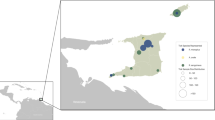
Of the 21 adult female samples (10 individuals per sample) examined, we did not find any contigs with reasonable evidence for I. hookeri origin. In five of the 21 nymph samples (10 individual nymphs per sample), namely sample b, c, d, g, n, contigs deriving from I. hookeri were found. The sequence identity of the contigs deposited in GenBank to the I. hookeri 28S rRNA gene was 378/386 (97.9%), 556/559 (99.4%), 439/447 (98.2%), 445/453 (98.2%), and 300/308 (97.4%) for samples b (accession id: OQ316579), c (OQ316577), d (OQ316581), g (OQ316578), and n (OQ312115), respectively. The multiple sequence alignments of the contigs with the partial reference sequence of I. hookeri 28S rRNA gene (MH077537.1) are shown in Fig. 1.The figure shows that the sequence of the generated contigs varies from the reference sequence at 8 positions (A434T, A491C, A499G, A666G, A670G, T677G, G678T, C688G). In the annotation of the altered positions, the first letter refers to the base in the reference sequence, the following numbers specify the genomic position of the polymorphism, and the last letter indicates the base detected in our samples. By position A434T, transversions were identified in sample b, c, d, and g. All our samples included A491C and A499G mutations. A666G, A670G, T677G, G678T, and C688G polymorphisms occurred in sample b, d, g, and n. In the easternmost sample (n), we found one position (C565T) that differs from all other Hungarian samples and the reference sequence as well. The geoposition of the samples is presented in Fig. 2.
Multiple sequence alignments. The contigs were predicted to have I. hookeri origins based on the partial reference sequence of I. hookeri 28S ribosomal RNA gene (MH077537.1). Letters on a blue background indicate sequence identity from less than all the samples, and letters on a brown background indicate the parts where all 5 samples and the reference sequence are identical. Positions with yellow backgrounds indicate polymorphisms. The geopositions of the Hungarian samples are presented in Fig. 2.
Geopositions of the metagenomically analyzed samples in Hungary. The red dots represent the sampling points we have found I. hookeri sequences, the blue ones where we have not. The inset map shows the study region, Hungary in Europe, colored yellow. Neighboring countries are presented by ISO 3 character codes (AUT: Austria, HRV: Croatia, ROU: Romania, SRB: Serbia, SVK: Slovakia, SVN: Slovenia, UKR: Ukraine).
Discussion
The findings that no reads deriving from I. hookeri were detected in adult I. ricinus samples collected between the end of March and the middle of May, while nymphs were associated with this species, promote former theories of the life cycle of the parasitoid wasps23,24. I. hookeri eggs may have been laid in the larvae or in nymphs before winter or, less probably, during spring, as the wasps were formerly associated with the possibility of surviving winter conditions40,41.
To our knowledge, this is the first report on evidence of the presence of Ixodiphagus wasps, namely I. hookeri in Hungary. This finding expands the localities associated with I. hookeri within Europe. All except one sampling points that were proven to host I. hookeri are located in western Hungary. The cluster of the four western Hungarian sampling points lays close to Austria’s and Slovakia’s borders. While the presence of I. hookeri has not been published in Austria; to our best knowledge, Slovakian reports of the occurrence of the wasps exist. I. hookeri has previously been identified at three locations within Slovakia; near Šoporña, associated to Haemaphysalis concinna42, close to the capital of Slovakia, Bratislava, in I. ricinus43,44 and in the Slovak Karst, isolated from both I. ricinus and H. concinna41. According to our hypothesis, the western Hungarian cluster of sampling points b, c, d, and g (Sárvár, Mosonmagyaróvár, Sáska, and Pénzesgyőr, respectively) may be associated with˝ the wasp populations described by Bratislava43,44 and by Šoporña42 in Slovakia. Despite being situated close to areas where I. hookeri is present, no I. hookeri DNA was detected from ticks in sampling points m, p, q, r, and s (Nagy-Hideg-Hegy, Kékes, Lillafüred, Aggtelek, and Háromhuta, respectively). Shifting slightly to the east, sampling point n (Budapest) represented the closest occurrence of the wasps to Slovakia. Considering the physical proximity between sampling point r and the Slovak Karst, where the report of Buczek and colleagues was released, the possibility of receiving false negative results is raised. The basis of receiving false negative results will be described further on.
Other European countries where the presence of I. hookeri has been reported include the Czech Republic (detected in former Czechoslovakia)45, Finland46, France47,48, the Georgia49, Germany24,26,50, Italy51, the Netherlands27,52, the United Kingdom53, Russia (detected in the Ussuri forest, in the Asian part of the former Soviet Union)54 and Ukraine (detected in the former Soviet Union)55. To our knowledge, sampling point d (Sáska) in Hungary represents the southernmost detection point of I. hookeri within Central Europe. The detection of I. hookeri in Hungary may serve as a novel hint regarding the potential distribution I. hookeri at the Balkan Peninsula, where the species appears to be little studied.
As mentioned above, wasp-negative sampling points can be wasp-invaded. Even though next-generation sequencing (NGS) based metagenomic approach appears to be just as or even more sensitive as polymerase chain reaction (PCR) based target detection techniques28,29,56, certain limitations can be addressed. Within the pool of reads deriving from the shotgun-sequenced metagenome that contains genome fragments from every organism present in the sample, lower relative abundance rates of an individual species serve with relatively lower read counts from its genome. In other words, shotgun sequencing preserves the original relative read abundance rates of the various organisms of the samples and may represent fewer reads of certain species by non-targeted runs57,58. Moreover, the I. hookeri reference sequence, that the metagenomic read sets were aligned to only represented a smaller fragment, namely the unique 28S ribosomal RNA gene of the full I. hookeri genome. Thus only I. hookeri reads deriving from this part of the genome could have been aligned, that further increases the chance of false-negative sampling points for the wasps. Besides the above-mentioned reasons, due to the low European tick parasitization rates of I. hookeri, the 10 nymphs collected at the sampling points may miss the wasps by chance alone.
Nevertheless, NGS-based approaches have a prospering future within the studies of parasitoids of public health significance, such as I. hookeri. According to Collatz and colleagues, large geographical distance and climatic differences (e.g. presence in Africa, Asia, Europe, and North America)24,25,59,60 may even underlie divergence and distinct taxonomic categorization of I. hookeri to different species, subspecies or at least strains24. Concurrently, publications on I. hookeri indicate a certain extent of behavioral and host preferences at different continents24,25,61. To assess the basic variation in behavioral traits of I. hookeri or to identify specific characteristics of subgroups that can be better utilized by the biological control methods, the study of the I. hookeri genome or at least specific genome regions, such as 28S rRNA or 16S rRNA genes, may become inevitable, similarly to other weighty insect groups52,62,63.
The improvement of our knowledge of I. hookeri with either traditional or genomic methods could facilitate the assessment of its potential as a means of biological control, while limitations and doubts about the wasps’ biocontrol potential could be addressed with more research. Attempted mass releases of the parasitoid wasps in the U.S and in the former Soviet Union between 1920 and 194040,64,65 were unsuccessful as far as causing noticeable reduction of tick populations. One reason for this may be that I. hookeri requires high tick host densities and superabundant tick populations to reach its ideal abundance61,66. Inadequate numbers of parasitoids released compared to the geographical areas may have also undermined these trials23. On the other hand, the parasitoid wasps have been transported to the sites of attempted mass releases from great distances, sometimes even from different continents (e.g. from France to the U.S.)40,64 without any considerations regarding their host preferences, climatic adaptations, or behavioral attributes, that have, since then turned out to be rather specific to their geographic locations of origin24,25,61. In a global perspective, climate-associated occurrence rate alterations23,25 or differences in other characteristics of I. hookeri, such as the duration of its development, may also be underlain by host-related factors. Synchronization with the maturation of the tick host and, indirectly, with the main activity period of the vertebrate hosts of the ticks throughout the year may play a role in the life cycle of the wasps24.
Furthermore, we do not know how great the tick populations would be without the endemic I. hookeri populations and how much the parasitoid wasps contribute to maintaining the equilibrium of the communities in which they are included67. Nonetheless, the hypothesis regarding sufficiently high tick host densities and superabundant parasitoid host populations is in line with findings regarding the bioindication potential of certain insect species, including parasitoid wasps68. If so, this potential may also be worth further observation.
Conclusively, assessment of existing populations and further examinations on entomologic and genomic traits along with ecological roles could help understand and exploit the Ixodiphagus wasps’ potential as a biological tick control method or as a potential bioindicator species.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Chala, B. & Hamde, F. Emerging and re-emerging vector-borne infectious diseases and the challenges for control: A review. Front. Public Health https://doi.org/10.3389/fpubh.2021.715759 (2021).
Jongejan, F. & Uilenberg, G. The global importance of ticks. Parasitology 129, S3–S14 (2004).
Hornok, S., Kováts, D., Horváth, G., Kontschán, J. & Farkas, R. Checklist of the hard tick (Acari: Ixodidae) fauna of Hungary with emphasis on host-associations and the emergence of Rhipicephalus sanguineus. Exp. Appl. Acarol. 80, 311–328 (2020).
ECDC. Surveillance and disease data—Tick maps. https://www.ecdc.europa.eu/en/diseasevectors/surveillance-and-disease-data/tick-maps (2022). Accessed: 2022–09–02.
Brites-Neto, J., Duarte, K. M. R. & Martins, T. F. Tick-borne infections in human and animal population worldwide. Vet. World 8, 301 (2015).
Hubálek, Z. Epidemiology of Lyme borreliosis. Lyme Borreliosis 37, 31–50 (2009).
Rizzoli, A. et al. Lyme borreliosis in Europe. Eurosurveillance 16, 19906 (2011).
Marques, A. R., Strle, F. & Wormser, G. P. Comparison of Lyme disease in the United States and Europe. Emerg. Infect. Dis. 27, 2017 (2021).
Jaenson, T. G., Jaenson, D. G., Eisen, L., Petersson, E. & Lindgren, E. Changes in the geographical distribution and abundance of the tick Ixodes ricinus during the past 30 years in Sweden. Parasit. Vectors 5, 1–15 (2012).
Medlock, J. M. et al. Driving forces for changes in geographical distribution of Ixodes ricinus ticks in Europe. Parasit. Vectors 6, 1–11 (2013).
Semenza, J. C. & Suk, J. E. Vector-borne diseases and climate change: a European perspective. FEMS Microbiol. Lett. https://doi.org/10.1093/femsle/fnx244 (2018).
Sutherst, R. W. Global change and human vulnerability to vector-borne diseases. Clin. Microbiol. Rev. 17, 136–173 (2004).
Tabachnick, W. Challenges in predicting climate and environmental effects on vector-borne disease episystems in a changing world. J. Exp. Biol. 213, 946–954 (2010).
Sonenshine, D. E., Kocan, K. M. & de la Fuente, J. Tick control: Further thoughts on a research agenda. Trends Parasitol. 22, 550–551 (2006).
Willadsen, P. Tick control: Thoughts on a research agenda. Vet. Parasitol. 138, 161–168 (2006).
Goolsby, J. A. et al. Rationale for classical biological control of cattle fever ticks and proposed methods for field collection of natural enemies. Subtrop. Agric. Environ. 66, 7–15 (2016).
Singh, N. et al. Effect of immersion time on efficacy of entomopathogenic nematodes against engorged females of cattle fever tick, Rhipicephalus (= Boophilus) microplus. Southwest. Entomol. 43, 19–28 (2018).
Černý, J. et al. Management options for Ixodes ricinus-associated pathogens: A review of prevention strategies. Int. J. Environ. Res. Public Health 17, 1830 (2020).
Kapranas, A. et al. Encyrtid parasitoids of soft scale insects: Biology, behavior, and their use in biological control. Annu. Rev. Entomol. 60, 195–211 (2015).
Chirinos, D. T. & Kondo, T. Description and biological studies of a new species of Metaphycus Mercet, 1917 (Hymenoptera: Encyrtidae), a parasitoid of Capulinia linarosae Kondo & Gullan. Int. J. Insect Sci. 11, 1179543319857962 (2019).
Polaszek, A., Noyes, J. S., Russell, S. & Ramadan, M. M. Metaphycus macadamiae (Hymenoptera: Encyrtidae)–a biological control agent of macadamia felted coccid Acanthococcus ironsidei (Hemiptera: Eriococcidae) in Hawaii. PLoS ONE 15, e0230944 (2020).
Howard, L. Another chalcidoid parasite of a tick. Can. Entomol. 40, 239–241 (1908).
Hu, R., Hyland, K. & Oliver, J. A review on the use of Ixodiphagus wasps (Hymenoptera: Encyrtidae) as natural enemies for the control of ticks (Acari: Ixodidae). Syst. Appl. Acarol. 3, 19–28 (1998).
Collatz, J. et al. A hidden beneficial: Biology of the tick-wasp Ixodiphagus hookeri in Germany. J. Appl. Entomol. 135, 351–358 (2011).
Takasu, K. & Nakamura, S. Life history of the tick parasitoid Ixodiphagus hookeri (Hymenoptera: Encyrtidae) in Kenya. Biol. Control. 46, 114–121 (2008).
Collatz, J. et al. Being a parasitoid of parasites: host finding in the tick wasp Ixodiphagus hookeri by odours from mammals. Entomol. Experimentalis et Applicata 134, 131–137 (2010).
Krawczyk, A. I. et al. Tripartite interactions among Ixodiphagus hookeri, Ixodes ricinus and deer: Differential interference with transmission cycles of tick-borne pathogens. Pathogens 9, 339 (2020).
Plaire, D., Puaud, S., Marsolier-Kergoat, M.-C. & Elalouf, J.-M. Comparative analysis of the sensitivity of metagenomic sequencing and PCR to detect a biowarfare simulant (Bacillus atrophaeus) in soil samples. PLoS ONE 12, e0177112 (2017).
Wang, C.-X. et al. Comparison of broad-range polymerase chain reaction and metagenomic next-generation sequencing for the diagnosis of prosthetic joint infection. Int. J. Infect. Dis. 95, 8–12 (2020).
Tóth, A. G. et al. Ixodes ricinus tick bacteriome alterations based on a climatically representative survey in Hungary. bioRxiv (2022).
Estrada-Peña, A., Mihalca, A. D. & Petney, T. N. Ticks of Europe and North Africa: A Guide to Species Identification (Springer, 2018).
Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 20, 1–13 (2019).
Pruitt, K. D., Tatusova, T. & Maglott, D. R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33, D501–D504 (2005).
NCBI Resource Coordinators. Database resources of the national center for biotechnology information. Nucleic Acids Res. 44, D7 (2016).
Katoh, K. & Standley, D. M. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Tennekes, M. tmap: Thematic maps in R. J. Stat. Softw. 84, 1–39 (2018).
Bodenhofer, U., Bonatesta, E., Horejš-Kainrath, C. & Hochreiter, S. msa: An R package for multiple sequence alignment. Bioinformatics 31, 3997–3999 (2015).
R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria (2022).
Alfeev, N. & Klimas, Y. On the possibility of developing ichneumon flies, Hunterellus hookeri in climatic conditions of the USSR. Sovet. Vet. 15, 55 (1938).
Buczek, A., Buczek, W., Bartosik, K., Kulisz, J. & Stanko, M. Ixodiphagus hookeri wasps (Hymenoptera: Encyrtidae) in two sympatric tick species Ixodes ricinus and Haemaphysalis concinna (Ixodida: Ixodidae) in the Slovak Karst (Slovakia): Ecological and biological considerations. Sci. Rep. 11, 1–10 (2021).
Slovák, M. Finding of the endoparasitoid Ixodiphagus hookeri (Hymenoptera, Encyrtidae) in Haemaphysalis concinna ticks in Slovakia. Biol. Bratislava 58, 890–894 (2003).
Rehacek, J. & Kocianova, E. Attempt to infect Hunterellus hookeri Howard (Hymenoptera, Encyrtidae), an endoparasite of ticks, with Coxiella burnetti. Acta Virol. 36, 492–492 (1992).
Bohacsova, M., Mediannikov, O., Kazimirova, M., Raoult, D. & Sekeyova, Z. Arsenophonus nasoniae and Rickettsiae infection of Ixodes ricinus due to parasitic wasp Ixodiphagus hookeri. PLoS ONE 11, e0149950 (2016).
Boucek, Z. & Verny, V. A parasite of ticks, the chalcid Hunterellus hookeri in Czechoslovakia. Zool. Listy 3, 109–111 (1954).
Sormunen, J. J., Sippola, E., Kaunisto, K. M., Vesterinen, E. J. & Sääksjärvi, I. E. First evidence of Ixodiphagus hookeri (Hymenoptera: Encyrtidae) parasitization in Finnish castor bean ticks (Ixodes ricinus). Exp. Appl. Acarol. 79, 395–404 (2019).
Doby, J. & van Laere, G. Hunterellus hookeri howard, 1907, Hymenoptère Chalcididae parasite de la tique Ixodes ricinus dans l’ouest et le centre de la France. Bull. de la Société française de parasitologie 11, 265–270 (1993).
Plantard, O. et al. Detection of Wolbachia in the tick Ixodes ricinus is due to the presence of the hymenoptera endoparasitoid Ixodiphagus hookeri. PLoS ONE 7, e30692 (2012).
Japoshvili, G. New records of Encyrtids (Hymenoptera: Chalcidoidea: Encyrtidae) from Georgia, with description of seven new species. J. Asia-Pacific Entomol. 20, 866–877 (2017).
Walter, G. Beitrag zur Biologie der Schlupfwespe Hunterellus hookeri Howard (Hymenoptera: Encyrtidae) in Norddeutschland. Beitr. Naturkunde Niedersachsens 33, 129–133 (1980).
Ramos, R. A. N. et al. Occurrence of Ixodiphagus hookeri (Hymenoptera: Encyrtidae) in Ixodes ricinus (Acari: Ixodidae) in Southern Italy. Ticks Tick-borne Dis. 6, 234–236 (2015).
Tijsse-Klasen, E., Braks, M., Scholte, E.-J. & Sprong, H. Parasites of vectors—Ixodiphagus hookeri and its Wolbachia symbionts in ticks in the Netherlands. Parasit. Vectors 4, 1–7 (2011).
Luu, L. et al. Bacterial pathogens and symbionts harboured by Ixodes ricinus ticks parasitising red squirrels in the United Kingdom. Pathogens 10, 458 (2021).
Pervomaisky, G. S. On the infestation of Ixodes persulcatus by Hunterellus hookeri How. (Hymenoptera). Zool. Zhurnal 22, 211–213 (1943).
Klyushkina, E. A parasite of the ixodid ticks, Hunterellus hookeri. How in the Crimea. Zool. Zh. 37, 1561–1563 (1958).
Gorman, M., Xu, R., Prakoso, D., Salvador, L. C. & Rajeev, S. Leptospira enrichment culture followed by ONT metagenomic sequencing allows better detection of Leptospira presence and diversity in water and soil samples. PLOS Neglected Trop. Dis. 16, e0010589 (2022).
Ranjan, R., Rani, A., Metwally, A., McGee, H. S. & Perkins, D. L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 469, 967–977 (2016).
Laudadio, I. et al. Quantitative assessment of shotgun metagenomics and 16S rDNA amplicon sequencing in the study of human gut microbiome. OMICS 22, 248–254 (2018).
Munaf, H. et al. The first record of Hunterellus hookeri parasitizing Rhipicephalus sanguineus in Indonesia. Southeast Asian J. Trop. Medicine Public Heal. 7, 492 (1976).
Stafford, K. C. III., Denicola, A. J. & Kilpatrick, H. J. Reduced abundance of Ixodes scapularis (Acari: Ixodidae) and the tick parasitoid Ixodiphagus hookeri (Hymenoptera: Encyrtidae) with reduction of white-tailed deer. J. Med. Entomol. 40, 642–652 (2003).
Stafford, K. C. Jr., Denicola, A. J. & Magnarelli, L. A. Presence of Ixodiphagus hookeri (Hymenoptera: Encyrtidae) in two Connecticut populations of Ixodes scapularis (Acari: Ixodidae). J. Med. Entomol. 33, 183–188 (1996).
Gillespie, J., Johnston, J., Cannone, J. & Gutell, R. Characteristics of the nuclear (18S, 5.8 S, 28S and 5S) and mitochondrial (12S and 16S) rRNA genes of Apis mellifera (Insecta: Hymenoptera): Structure, organization, and retrotransposable elements. Insect Mol. Biol. 15, 657–686 (2006).
Zhao, Y., Zhang, W.-Y., Wang, R.-L. & Niu, D.-L. Divergent domains of 28S ribosomal RNA gene: DNA barcodes for molecular classification and identification of mites. Parasit. Vectors 13, 1–12 (2020).
Larrousse, F., King, A. G. & Wolbach, S. The overwintering in Massachusetts of Ixodiphagus caucurtei. Science 67, 351–353 (1928).
Smith, C. N. et al. Studies of parasites of the American dog tick. J. Econ. Entomol. https://doi.org/10.1093/jee/36.4.569 (1943).
Hu, R., Hyland, K. E. & Mather, T. N. Occurrence and distribution in Rhode Island of Hunterellus hookeri (Hymenoptera: Encyrtidae), a wasp parasitoid of Ixodes dammini. J. Med. Entomol. 30, 277–280 (1993).
Scatolini, D. & Penteado-Dias, A. A fauna de Braconidae (hymenoptera) como bioindicadora do grau de preservação de duas localidades do Estado do Paraná. Revista Brasileira de Ecol. 1, 84–87 (1997).
Anderson, A. et al. The potential of parasitoid Hymenoptera as bioindicators of arthropod diversity in agricultural grasslands. J. Appl. Ecol. 48, 382–390 (2011).
Funding
The European Union’s Horizon 2020 research and innovation program under Grant Agreement 874735 (VEO) supported the research.
Author information
Authors and Affiliations
Contributions
N.S. takes responsibility for the data’s integrity and the data analysis’s accuracy. N.S. and A.G.T. conceived the concept of the study. E.K. and M.G. performed sample collection and procedures. N.S. participated in the bioinformatic and statistical analysis. A.G.T. and N.S. participated in the drafting of the manuscript. A.G.T., N.S. and R.F. carried out the manuscript’s critical revision for important intellectual content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tóth, A.G., Farkas, R., Gyurkovszky, M. et al. First detection of Ixodiphagus hookeri (Hymenoptera: Encyrtidae) in Ixodes ricinus ticks (Acari: Ixodidae) from multiple locations in Hungary. Sci Rep 13, 1624 (2023). https://doi.org/10.1038/s41598-023-28969-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-28969-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.