Abstract
While the breed of cattle can impact on the composition and structure of microbial communities in the rumen, breed-specific effects on rumen microbial communities have rarely been examined in sheep. In addition, rumen microbial composition can differ between ruminal fractions, and be associated with ruminant feed efficiency and methane emissions. In this study, 16S rRNA amplicon sequencing was used to investigate the effects of breed and ruminal fraction on bacterial and archaeal communities in sheep. Solid, liquid and epithelial rumen samples were obtained from a total of 36 lambs, across 4 different sheep breeds (Cheviot (n = 10), Connemara (n = 6), Lanark (n = 10) and Perth (n = 10)), undergoing detailed measurements of feed efficiency, who were offered a nut based cereal diet ad-libitum supplemented with grass silage. Our results demonstrate that the feed conversion ratio (FCR) was lowest for the Cheviot (most efficient), and highest for the Connemara breed (least efficient). In the solid fraction, bacterial community richness was lowest in the Cheviot breed, while Sharpea azabuensis was most abundant in the Perth breed. Lanark, Cheviot and Perth breeds exhibited a significantly higher abundance of epithelial associated Succiniclasticum compared to the Connemara breed. When comparing ruminal fractions, Campylobacter, Family XIII, Mogibacterium, and Lachnospiraceae UCG-008 were most abundant in the epithelial fraction. Our findings indicate that breed can impact the abundance of specific bacterial taxa in sheep while having little effect on the overall composition of the microbial community. This finding has implications for genetic selection breeding programs aimed at improving feed conversion efficiency of sheep. Furthermore, the variations in the distribution of bacterial species identified between ruminal fractions, notably between solid and epithelial fractions, reveals a rumen fraction bias, which has implications for sheep rumen sampling techniques.
Similar content being viewed by others
Introduction
Ruminant livestock contribute significantly to food security by converting human indigestible plant matter, into high quality sources of dairy and meat proteins, for human consumption1. A sustainable supply of animal derived protein over the next decades will be key to meeting the nutritional requirements of an estimated nine billion people by 20502. However, livestock production systems are also a major source of anthropogenic greenhouse gas emissions with enteric fermentation estimated to contribute to 35–40% of global methane emissions3. As a result, there is an urgent need to increase animal protein production to fulfil nutritional demand while simultaneously improving the livestock industry's environmental sustainability metrics. Increasing the feed conversion efficiency of livestock is proposed as a mitigation solution for the livestock industry, as more feed efficient ruminants emit less methane than their less efficient counterparts4,5,6. In addition, improvements to feed efficiency are likely to benefit farm profitability7 while reducing the quantity of global land dedicated to producing feed for the livestock industry8.
Mountain or hill sheep production is a significant agricultural enterprise that provides social and economic health in rural areas across the globe, while also protecting natural habitats and promoting biodiversity9. In Ireland and the UK, popular hill sheep breeds include the Scottish Blackface (SB) and the Cheviot. SB are mountain breeds which display adaptive tolerance to harsh environmental conditions and challenging terrains with low-energy vegetation10. The wide distribution of SB breeds across the UK and Ireland has led to evolutionary changes within the breed, influenced largely by environmental pressures between different habitats10. As a result, a range of different strains of SB breed exist today including the Lanark, Perth and Connemara breed types which all vary in body and wool composition. The Cheviot breed is also well-suited to highland farming, and while not as resilient as the SB11, they are slightly larger and produce lambs that mature quickly12.
Sheep, like all ruminants, rely on a complex and dynamic microbial ecosystem (anaerobic bacteria, archaea, fungi and protozoa) within their rumen to derive energy from feed13. The rumen is composed of three environmental niches, namely the solid-, liquid-, and epithelial-fractions14,15,16. The solid fraction, comprised of ingested feed, is primarily colonised by feed adherent microbes that breakdown fibrous matter14. The liquid fraction consists of the fluid within the rumen and provides an environment for free living microbes involved in the metabolism of soluble nutrients17. Finally, the epithelial fraction refers to the epithelial lining of the rumen, which harbours microbes active in tissue recycling18, oxygen scavenging19, and urea hydrolysis20 and is critically important for bioconversions and nutrient uptake as the cellular interface with the host animal. Research in both bovine and ovine models have reported differences in the microbial taxonomic profiles between ruminal fractions14,16 with the epithelial being mostly distinct from the solid and liquid fractions, whereas the solid and liquid fractions tend to be more similar17,21. To date, most studies investigating breed effects have been conducted using the rumen digesta samples with minimal exploration of the microbial community associated with the epithelial fraction.
Research has revealed links between the rumen microbiota, feed efficiency and methane emissions in both cattle and sheep, with differences in microbial diversity and abundances between divergent animal cohorts21,22,23,24. Understanding factors that influence the composition and diversity of the rumen microbiome is critical for improving strategies to enhance feed efficiency and reduce ruminant methane emissions. Recently, studies in cattle have shown that microbial taxonomic profiles differ between breeds25,26, suggesting that host genetics may regulate the composition of the rumen microbiome. However, such effects have not been explored in sheep.
It is unclear whether breed specific findings in cattle can be translated to sheep. Taxa-specific research is imperative given the importance of the global sheep industry from environmental, economic and social perspectives. In addition, while cattle studies have provided some indication that breed plays an important role in shaping the rumen microbiome, to date, such effects have not been investigated across all three ruminal fractions. Hence, the objectives of the current study are twofold. Firstly, to investigate the effect of breed on bacterial and archaeal populations in the solid, liquid and epithelial rumen fractions of sheep, and secondly to investigate the effect of the ruminal fraction on the microbial populations in each of the breeds, using 16S rRNA amplicon sequencing.
Results
Breed differences in animal feed conversion and economic trait performance
Throughout the feed intake measurement period, summary statistics shows animals on test had an average DMI of 1.11 kg/d (SD = 0.18), ADG of 0.27 kg/d (SD = 0.1), FCR of 4.04 kg of DMI/ Kg of ADG (SD = 0.1), start weight of 29.60 kg (SD = 3.7), final live weight of 46.00 kg (SD = 2.9), carcass weight of 20.20 kg (SD = 1.6), and a KO% of 44.1% (SD = 2.3). Average daily gain (P = 0.005), FCR (P = 0.035), CW (P < 0.04) and start weight (P < 0.036) were all significantly affected by breed. Summary statistics, along with comparisons amongst breeds for animal performance, feed intake and feed efficiency are displayed in Table 1. In summary, the Cheviot breed had the lowest FCR and the highest ADG, carcass weight, and start weight among all breeds, with differences in ADG and FCR being significant when compared to the Connemara breed and differences in carcass and start weight being significant when compared to the Lanark breed. In addition, the Cheviot breed had the fastest maturing lambs with 80% of lambs reaching maturity within the first 42 days (data not shown) and a mean LW of 47.1 kg (Table 1).
Overall microbial community structure
After data processing, filtering, and removal of chimeras and lowly sequenced samples a total of 5,411,353 reads remained, with an average of 91.3% of reads surviving. The average number of reads per sample was 64,420, which mapped to 2547 ASVs. After removal of taxa unassigned at the phylum level 2434 ASV’s remained. Analysis of the ASVs across all samples revealed that bacteria and archaea represented 95.4 and 4.6% of the microbial population, respectively. A total of 19 bacterial taxa were classified at the phylum level, with Firmicutes being the most abundant (45.8%), followed by Bacteroidetes (33.4%) and Proteobacteria (8.0%). There were 192 taxa classified at the genus level, with Prevotella_1 (13.1%) and Prevotella_7 (13.1%) being the most dominant followed by Succinivibrio (6.3%). Methanobrevibacter was shown to be the most abundant archaeal genus. (78.1%). In this study, no non-methanogenic archaeal taxa were identified.
Breed effects on bacterial and archaeal populations in the solid ruminal fraction
In the solid ruminal fraction, a total of 1706 bacterial ASVs agglomerated to 227 genera, 89 families, 51 orders, 27 classes and 16 phyla. Firmicutes (48.2%) Bacteroidetes (30.1%), Fibrobacterota (6.1%) were the three most abundant bacteria phyla (Fig. 1). Prevotella_7 (9.7%), Prevotella_1 (9.2%), unclassified Lachnospiraceae (7.6%), Fibrobacter (6.1%) and Ruminococcus_1 (5.6%) were the 5 most dominant bacteria genera (Fig. 2). A total of 27 archaeal ASVs were identified and agglomerated to 4 genera (Methanobrevibacter, Methanosphera, Methanimicrococcus and Candidatus methanomethylophilus), three families, three orders three classes and one phylum. Methanobrevibacter was the most dominant archaeal genus (72.9%).
Stack barchart representing the mean relative abundance of the 5 most dominant phyla across breeds (i.e. Cheviot, Connemara, Lanark, Perth) for solid liquid and epithelial ruminal fractions. Solid (Cheviot n = 8, Connemara n = 5, Lanark n = 9, Perth n = 7), liquid (Cheviot n = 9, Connemara n = 5, Lanark n = 9, Perth n = 5), epithelial (Cheviot n = 9, Connemara n = 3, Lanark n = 6, Perth n = 8).
Stack barchart representing the mean relative abundance of the 10 most dominant genera across breeds (i.e. Cheviot, Connemara, Lanark, Perth) for solid liquid and epithelial ruminal fractions. Solid (Cheviot n = 8, Connemara n = 5, Lanark n = 9, Perth n = 7), liquid (Cheviot n = 9, Connemara n = 5, Lanark n = 9, Perth n = 5), epithelial (Cheviot n = 9, Connemara n = 3, Lanark n = 6, Perth n = 8).
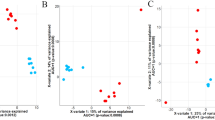
Alpha diversity analysis revealed that breed had an effect on solid associated bacterial and archaeal community richness and bacteria community PD (ANOVA, P < 0.05) (Table 2). Such differences were observed between the Cheviot and Lanark breeds, with the Cheviot exhibiting the least and the Lanark exhibiting the most rumen microbial diversity among the breeds. Based on weighted and unweighted UniFrac distances, beta diversity analysis showed no differences in overall community composition across the breeds (PERMANOVA, P > 0.05) for either bacterial or archaeal communities (Table 3).
The abundance of Sharpea at the genus level, Sharpea azabuensis at the species level and an unclassified ASV (ASV37) belonging to the family Lachnospiraceae were affected by breed (LRT, P.adj < 0.05) (Table 4). Sharpea (Wald, P.adj < 0.001; Log2FC = 4.37) and Sharpea azabuensis (Wald, P.adj < 0.001; Log2FC = 4.58) were higher in Perth compared to Cheviot. ASV37 (family Lachnospiraceae) was more abundant in Cheviot (Wald, P.adj < 0.01; Log2FC = 3.22) and Perth (Wald, P.adj < 0.001; Log2FC = 3.4) compared to Lanark (Table 4). Pairwise analysis between each of the breeds revealed a further 2 bacterial ASVs as differentially abundant. ASV48 classified to the genus Prevotella_9 was higher in Lanark compared to Perth (Wald, P.adj < 0.0001; Log2FC = 7.88) and Cheviot (Wald, P.adj < 0.01; Log2FC = 9.11), and ASV329 classified to the genus Pyramidobacter was higher in Lanark compared to Cheviot (Wald, P.adj < 0.05; Log2FC = 5.52). At the genus level P-2534-18B5_gut group (ASV17), belonging to phylum Bacteroidetes, was higher in the Perth (Wald, P.adj < 0.05; Log2FC = 5.78) and Lanark (Wald, P.adj < 0.05; Log2FC = 6.82) breeds compared to Cheviot, and Candidatus Saccharimonas was higher in the Lanark compared to the Cheviot (Wald, P.adj < 0.05; Log2FC = 5.66). Similarly, at the family level P-2534-18B5_gut group (ASV17) was higher in the Perth (Wald, P.adj < 0.01; Log2FC = 5.88) and Lanark (Wald, P.adj < 0.01; Log2FC = 6.80) breeds compared to Cheviot, and Saccharimonadaceae (ASV317) was higher in the Lanark (Wald, P.adj < 0.01; Log2FC = 5.48) and Connemara (Wald, P.adj < 0.05; Log2FC = 6.89) breeds compared to the Cheviot. At the order level Coriobacteriales was higher in the Lanark compared to the Perth (Wald, P.adj < 0.05; Log2FC = 1.17) (Table 5). One archaea ASV belonging to the genus Candidatus Methanomethylophilus (ASV337) was higher in the Perth (Wald, P.adj < 0.01; Log2FC = 3.12) and Lanark (Wald, P.adj < 0.05; Log2FC = 3.21) compared to Cheviot (Table 5).
Breed effects on bacterial and archaeal populations in the liquid ruminal fraction
For the liquid ruminal fraction, a total of 1790 bacteria ASVs agglomerated to 236 genera, 95 families, 57 orders, 29 classes and 17 phyla. Firmicutes (43.1%), Bacteroidetes (37.1%), and Proteobacteria (8.9%) were the most dominant phyla (Fig. 1). Prevotella 7 (12.2%), Prevotella 1 (11.7%), unclassified Lachnospiraceae (6.2%), Succinivibrio (5.8%) and Succiniclasticum (4.4%) were the five most dominant genera (Fig. 2). Twenty six archaea ASVs were available for analysis, which agglomerated to four genera (Methanobrevibacter, Methanosphera, unclassified Methanomethylophilaceae and Candidatus Methanomethylophilus), two families, two orders two classes and one phylum. Methanobrevibacter was the most dominant genus (78.9%).
Although there was no effect of breed on alpha diversity indices for bacteria communities (ANOVA, P > 0.05), breed did have an impact on the richness of archaeal communities. (ANOVA, P < 0.05) (Table 1). The Lanark breed had the highest level of archaeal community richness, whereas the Cheviot breed had the lowest level (Table 5). Based on weighted and unweighted UniFrac distances, the analysis of beta diversity showed no differences in overall community composition among breeds (PERMANOVA, P > 0.05) for either bacterial or archaeal communities (Table 3).
The likelihood ratio test detected no breed effect (LRT, P.adj > 0.05) on the abundance of bacterial or archaeal taxa across all taxonomic ranks. Pairwise analysis between each of the breeds revealed five taxa at the ASV level as differentially abundant. Two ASVs, ASV23 (Wald, P.adj < 0.01; Log2FC = 5.94) and ASV43 (Wald, P.adj < 0.05; Log2FC = 2.51) classified to the family Muribaculaceae were higher in Cheviot compared to Perth, ASV44 classified to the genus Acetitomaculum was higher in Cheviot (Wald, P.adj < 0.05; Log2FC = 10.26) compared to Connemara, ASV55 classified as Sharpea azabuensis was higher in Perth (Wald, P.adj < 0.01; Log2FC = 4.81) compared to Cheviot, and ASV20 classified to Lachnospiraceae NK3A20 group was higher in the Lanark (Wald, P.adj < 0.05; Log2FC = 3.06) compared to Connemara. At the genus level Sharpea (Wald, P.adj < 0.001; Log2FC = 5.05) was higher in the Perth compared to the Cheviot, ASV223 classified to order Rhodospirillales (Wald, P.adj < 0.01; Log2FC = 7.03) was higher in the Cheviot compared to the Perth, and ASV461 classified to Clostridiales_vadinBB60 group was higher in the Lanark (Wald, P.adj < 0.05; Log2FC = 5.86) compared to the Connemara. At the family level Muribaculaceae (Wald, P.adj < 0.05; Log2FC = 2.23) was higher in Cheviot compared to Perth, ASV223 classified to order Rhodospirillales was higher in the Cheviot compared to the Connemara (Wald, P.adj < 0.05; Log2FC = 7.11) and Perth (Wald, P.adj < 0.01; Log2FC = 6.81), ASV461 classified to Clostridiales_vadinBB60 group was higher in the Cheviot (Wald, P.adj < 0.05; Log2FC = 5.82) and Lanark (Wald, P.adj < 0.01; Log2FC = 6.09) compared to the Connemara, and ASV17 classified to P-2534-18B5_gut group was higher in the Perth (Wald, P.adj < 0.05; Log2FC = 4.75) and Lanark (Wald, P.adj < 0.05; Log2FC = 5.87) compared to the Cheviot. At the order level Rhodospirillales was higher in the Cheviot compared to the Connemara (Wald, P.adj < 0.05; Log2FC = 7.29) and Perth (Wald, P.adj < 0.01; Log2FC = 6.93), and Betaproteobacteriales was higher in the Cheviot compared to the Connemara (Wald, P.adj < 0.05; Log2FC = 3.03). At the class level the abundance of Alphaproteobacteria was higher in the Cheviot compared to the Perth (Wald, P.adj < 0.01; Log2FC = 7.11) and Connemara (Wald, P.adj < 0.05; Log2FC = 7.52). Finally at the phylum level the abundance of Proteobacteria was higher in the Perth compared to the Connemara (Wald, P.adj < 0.05; Log2FC = 2.62) (Table 5).
Breed effects on bacterial and archaeal populations in the epithelial ruminal fraction
In the epithelial ruminal fraction, a total of 1891 bacteria ASVs agglomerated to 231 genera, 89 families, 52 orders, 29 classes and 17 phyla. Firmicutes (46.3%), Bacteroidetes (33.4%), and Proteobacteria (10.1%) were the most dominant phyla (Fig. 1). Prevotella 1 (10.0%), Prevotella 7 (8.9%), Succinvibrio (5.9%), unclassified Lachnospiraceae (5.7%), and Ruminococcus 2 (5.0%) were the five most dominant genera (Fig. 2). Twenty eight archaeal ASVs were available for analysis, which agglomerated to five genera (Methanobrevibacter, Methanosphera, Methanimicrococcus, unclassified Methanomethylophilaceae and Candidatus Methanomethylophilus), three families, three orders, three classes and one phylum. Methanobrevibacter was the most dominant genus (82.0%).
Alpha diversity analysis revealed that while breed had no effect on epithelial associated archaeal community indices, it had a significant effect on bacteria community richness and inverse Simpson diversity (ANOVA, P < 0.05) (Table 2). Beta diversity analysis based on weighted and unweighted UniFrac distances, found no differences in community composition among the breeds (PERMANOVA, P > 0.05), for either bacterial or archaeal communities (Table 3).
The abundance of Family XIII at the family level and an unclassified ASV (ASV379) belonging to Family XIII at the ASV level were affected by breed (LRT, P.adj < 0.01) (Table 4). Family XIII was higher in Lanark compared to Cheviot (Wald, P.adj < 0.05; Log2FC = 1.41) and Perth (Wald, P.adj < 0.05; Log2FC = 1.13), and ASV379, belonging to Family XIII, was higher in the Lanark breed (Wald, P.adj < 0.05; Log2FC = 2.88) when compared to Perth breed (Table 5). Pairwise analysis revealed a further seven bacterial ASVs as differentially abundant. ASV37 classified to the Lachnospiraceae family was higher in the Perth (Wald, P.adj < 0.05; Log2FC = 2.72) compared to the Lanark, ASV123 classified to the genus Prevotella_1 was higher in the Connemara (Wald, P.adj < 0.01; Log2FC = 7.85) compared to the Lanark, ASV633 classified to the genus Ruminococcacese UCG-010 was higher in the Perth (Wald, P.adj < 0.05; Log2FC = 4.0) compared to the Lanark, ASV24 classified to the genus Succiniclasticum was lower in the Connemara compared to Cheviot (Wald, P.adj < 0.0001; Log2FC = 24.14), Lanark (Wald, P.adj < 0.0001; Log2FC = 23.67) and Perth (Wald, P.adj < 0.0001; Log2FC = 25.98) breeds, ASV74 classified to the genus Syntrophococcus was higher in the Lanark breed compared to Connemara (Wald, P.adj < 0.05; Log2FC = 5.42), ASV33 classified to the genus Ruminococcus_1 was higher in Perth (Wald, P.adj < 0.05; Log2FC = 4.81) and Connemara (Wald, P.adj < 0.01; Log2FC = 7.80) compared to Cheviot, and ASV118 also classified to the genus Ruminococcus_1 was higher in Perth (Wald, P.adj < 0.05; Log2FC = 5.52) compared to Lanark. At the genus level, Sharpea was higher in the Perth compared to the Cheviot (Wald, P.adj < 0.05; Log2FC = 3.13), ASV361 classified to Family XIII AD3011 group was higher in the Lanark (Wald, P.adj < 0.01; Log2FC = 2.28) when compared to the Cheviot, while ASV69 classified to Ruminococcaceae UCG-014 (Wald, P.adj < 0.05; Log2FC = 2.77) and ASV406 classified to Family XIII UCG-001 (Wald, P.adj < 0.05; Log2FC = 3.10) were both higher in the Cheviot when compared to the Lanark. At the family level Atopobiaceae was higher in the Lanark (Wald, P.adj < 0.05; Log2FC = 1.96) compared to Perth, and Synergistaceae was higher in the Lanark compared to the Cheviot (Wald, P.adj < 0.05; Log2FC = 2.19) (Table 5).
Effect of ruminal fraction on bacterial and archaeal populations across breeds
Bacterial and archaeal populations across ruminal fractions were investigated for Cheviot, Lanark and Perth breeds, and only included animals where all three ruminal fractions were available. Firmicutes was the most abundant phylum in the Cheviot (mean, solid = 51%, liquid = 41%, epithelial = 45%), Lanark (mean, solid = 44%, liquid = 39%, epithelial = 43%) and Perth (mean, solid = 49%, liquid = 46%, epithelial = 49%) breeds (Fig. 3). In the epithelial fraction Prevotella_7 was the most abundant genus in the Cheviot (10.6%) and Lanark (8.3%) breeds, while Prevotella_1 (10.0%) was the most dominant genus for the Perth breed. In the liquid fraction Prevotella_1 was the most dominant in the Cheviot (12.1%) breed, while Prevotella_7 was most dominant in Lanark (11.1%) and Perth (13.1%) breeds. In the solid ruminal fraction unclassified Lachnospiracheae, Prevotella_1 and Prevotella_7 were most abundant in the Cheviot (9.9%), Lanark (10.5%) and Perth (11.1%) breeds, respectively (Fig. 4).
Stack barchart representing the mean relative abundance of the 5 most dominant phyla across ruminal fractions (i.e. solid, liquid and epithelial) for Cheviot, Lanark and Perth breeds. Cheviot (solid n = 7, liquid n = 5, epithelial n = 5), Lanark (solid n = 7, liquid n = 5, epithelial n = 5), Perth (solid n = 7, liquid n = 5, epithelial n = 5).
Stack barchart representing the mean relative abundance of the 10 most dominant bacterial genera across ruminal fractions (i.e. solid, liquid and epithelial) for Cheviot, Lanark and Perth breeds. Cheviot (solid n = 7, liquid n = 5, epithelial n = 5), Lanark (solid n = 7, liquid n = 5, epithelial n = 5), Perth (solid n = 7, liquid n = 5, epithelial n = 5).
For the Cheviot breed, bacterial community alpha diversity measures were not affected by ruminal fraction (ANOVA, P > 0.1). For the Lanark breed, bacterial community Shannon diversity was affected by ruminal fraction (ANOVA, P < 0.05). For the Perth breed, bacterial community richness (observed ASV) and phylogenetic diversity (PD) were affected by ruminal fraction (ANOVA, P < 0.05), with the rumen epithelial fraction exhibiting greater diversity than solid and liquid ruminal fractions (Table 6). For all three breeds, archaeal community alpha diversity measures were not affected by ruminal fraction (ANOVA, P > 0.1). Beta diversity analysis showed that bacterial and archaeal community composition were also unaffected by ruminal fraction for all breeds analysed (PERMANOVA, P > 0.1) (Table 7).
Overall, ruminal fraction influenced 36 taxonomic groups across all ranks, representing 19 distinct ASVs, in the three breeds studied (LRT, P < 0.05). Ruminal fraction influenced the abundance of 18 taxa (11 distinct ASVs) in the Lanark breed, the most of any of breeds studied. ASV141, classified to the phylum Epsilonbacteraeota and the genus Campylobacter, was affected by ruminal fraction (LRT, P < 0.05) at all taxonomic ranks (i.e. phylum to ASV) and found to be significantly more abundant in the epithelial fraction when compared to the solid fraction. The abundance of ASV449, classified to the genus Desulfobulbus, was affected by ruminal fraction at the order, family and genus taxonomic ranks (LRT, P.adj < 0.05), found to be significantly more abundan in the epithelial ruminal fraction. At the genus level the abundance of Butyrivibrio 2, Fretibacterium, Howardella, and an unclassified ASV (ASV219) belonging to family Neisseriaceae were all affected by ruminal fraction (LRT, P < 0.05) and significantly higher in the epithelial fraction. Conversely, the abundance of Shutterella and two unclassified ASVs belonging to families Family XII UCG-001 and Eggerthellaceae were highest in the solid ruminal fraction (Wald, P.adj < 0.05). At the ASV level, the abundance of two unclassified ASVs; ASV210 and ASV239, belonging to genus Mogibacterium and family Family XIII were affected by ruminal fraction (LRT, P < 0.05) and highest in the epithelial ruminal fraction (Wald, P < 0.05). In the Cheviot breed, the abundance of 15 taxa (8 unique ASVs) were affected by ruminal fraction. ASV141 (Campylobacter) from taxonomic ranks phylum to genus and ASV449 (Desulfobulbus) from order to genus were differentially abundant and significantly more abundant in the epithelial ruminal fraction (Wald, P < 0.05). At the family level, Neisseriaceae and an unclassified ASV, ASV198, belonging to the order Coriobacteriales, were affected by ruminal fraction, with the epithelial and solid ruminal fractions, respectively, containing a higher proportion of these bacteria. At the genus level, the abundance of Mogibacterium, Butyrivibro 2 and two unclassified ASVs, ASV142 (F_Erysipelotrichaceae_UCG-004) and ASV263 (F_Burkholderiaceae), were affected by ruminal fraction (LRT, P < 0.05), with highest abundances observed in the epithelial ruminal fraction. In the Perth breed, the abundance of the bacterial phylum Tenericutes and an unclassified archaeal genus, ASV475, belonging to the family Methanomethylophilaceae were impacted by ruminal fraction (LRT, P0.05) (Table 8) Taken together, the majority of differences in microbial abundance were observed between the solid and epithelial ruminal fractions, as shown in Table 9, which summarises all the results of pairwise analysis between fractions.
Bacterial and archaeal genera associated with FCR and ADG
Spearman's correlation analysis was performed between the relative abundance of genera and animal production traits; FCR and ADG to find potential drivers of feed efficiency in the solid, liquid and epithelial fractions. After adjusting for repeated hypotheses testing, no genera were determined to be statistically significant. Therefore, putative drivers of FCR and ADG were considered to have a (P < 0.05). In the solid fraction, four bacterial genera showed significant negative correlations with FCR: Succinivibrionaceae (ρ = − 4.1), Lachnospira (ρ = − 3.9), Syntrophococcus (ρ = − 3.8) and an unclassified genus (ASV9) belonging to the order Gastranaerophilales (ρ = − 4.1). Ruminococcaceae UCG-013 (ρ = − 4.1) positively associated with ADG, while Lachnospiraceae NK3A20 group (ρ = − 3.8) was negatively correlated with ADG. In the liquid ruminal fraction the genus Acetitomaculum (ρ = − 3.8) an unclassified ASV belonging to the order Gastranaerophilales (ρ = − 4.4) and the archaeal genus Candidatus Methanomethylophilus (ρ = − 3.8) negatively correlated with FCR. Prevotella 9 (ρ = 3.8), Roseburia (ρ = 4.9), and 5 unclassified genera belonging to the families Ruminococcaceae-UCG-013 (ρ = 4.5), -UCG-002 (ρ = 4.4), -UCG-014 (ρ = 4.1), -UCG-010 (ρ = 4.1), and Lachnospiraceae (ρ = 3.9), and an unclassified genus belonging to order Mollicutes (ρ = 4.9) positively associated with ADG. In the epithelial fraction we observed no significant associations with FCR. Prevotella 9 (ρ = 4.8), 4 unclassified genera belonging to the families Ruminococcaceae-UCG-013 (ρ = 4.9), -UCG-009 (ρ = 4.7), -UCG-014 (ρ = 4.1) and Lachnospiraceae (ρ = 4.5), an unclassified genus belonging to order Mollicutes_RF39 (ρ = 4.6) and the archaeal genus Methanosphera (ρ = 4.6) positively associated with ADG. Mogibacterium (ρ = − 4.1) and 2 unclassified genera belonging to the families Prevotellaceae (ρ = − 4.3) and Christensenellaceae (ρ = 4.3) negatively associated with ADG (Table 10).
Discussion
Prior to the completion of this study, the effects of breed on the microbial composition of the rumen liquid, solid and epitheliuem fractions in hill sheep, were unknown. However, recent studies in cattle have demonstrated that microbial taxonomic profiles vary between breeds, where the abundance of particular microbial species are regulated by host genetics27. As the rumen the rumen is comprised of 3 interconnecting microbial ecosystems; solid-, liquid- and epithelial ruminal fractions, we investigated (1) the effect of sheep breed on bacterial and archaeal populations in all three ruminal fractions, and (2) the effect of ruminal fraction on those populations in three breeds of sheep (i.e. Cheviot, Lanark and Perth). Our results provide the first report that diversity and abundance of bacterial and archaeal taxa in the solid, liquid and epithelial rumen fractions of sheep are influenced by breed. Our results expand and reinforce previous research in cattle showing differences in bacteria populations between breeds and ruminal fractions.
In the current study, breed was found to influence important production traits related to host feed efficiency, including FCR and ADG. Cheviot lambs were found to have the lowest mean FCR, indicating that it was the most feed efficient breed. However, the difference was only significant when compared to the Connemara breed, which had the highest mean FCR. Additionally, the Cheviot breed also had the fastest maturing lambs in the study, with 80% of lambs reaching maturity (> 40 kg) within the first 42 days of the study, with a mean LW of 47.1 kg. Among the SB strains the Perth had the lowest FCR. No differences in FCR and ADG between the Cheviot, Lanark and Perth were found. Although this is the first study to compare FCR and ADG between these mountain/hill sheep breeds, the findings are in line with a previous study that found that metabolic differences between six British sheep breeds (i.e. SB, Welsh Mountain, Cheviot, Suffolk Down, Kent, and Hampshire Down) were mostly similar11. However, when subjected to environmental stresses such as wind and rain, differences were apparent, with the SB found to more stress-tolerant than the Cheviot11.
Metagenomic studies investigating host genetic effects on the rumen microbiota have to date been performed using original rumen digesta25,28, which comprises both the liquid and solid ruminal fractions. The purpose of this study was to investigate the influence of sheep breed on bacteria associated with each of the three fractions independently. Our findings demonstrate that breed contributed to significant variations in alpha diversity (i.e. observed ASV’s and PD) in the solid, but not in the liquid, ruminal fraction. The solid fraction the Cheviot breed harbored a bacterial community that was less rich and more phylogenetically related than those of the SB, which was significant when compared to the Lanark breed. Previously, lower rumen microbial alpha diversity and richness were linked to higher feed efficiency in cattle22. An efficient rumen microbiome is considered to be less diverse and more specialised in metabolizing feed and delivering energy to the host22, which could be a factor influencing the greater FCR observed for the Cheviot breed in the current study. Our beta diversity analysis showed that bacterial communities associated with the liquid and solid fractions were not affected by breed, suggesting a large overlap of community representatives among breeds. Taken together our findings on bacteria diversity and composition contrast with an analogous study conducted in cattle25. Li et al.25 reported no significant differences in alpha diversity among three breeds of cattle; while PCoA based on Bray Curtis distances revealed that the Kinsella Hybrid breed exhibited a distinct bacteria community composition to that of the Angus and Charlaois breeds used in the study25. Conversely, an earlier study in cattle found both alpha and beta diversities differing between Holstein and Jersey cows28. Variations in community composition and diversity may be attributed to differences of animal model, management practice, diet, environment, age or analytical approaches used. We consider that a combination of these factors might explain differences between the current study and those studies mentioned. Although no major differences in bacteria community composition were observed, the abundance of several taxonomic groups were affected by breed in the solid ruminal fraction: Sharpea at the genus level, and Sharpea azabuensis and an unclassified ASV belonging to the family Lachnospiraceae at the species level. The Perth breed exhibited highest abundance of Sharpea azabuensis which was significant in comparison to the Cheviot breed. Sharpea azabuensis is a strictly anaerobic gram-positive bacterium that can metabolise a variety of sugars including d-glucose, d-fructose, d-galactose and sucrose producing lactate as the primary end product29. Previous studies investigating the rumen microbiota of sheep divergent for methane emissions have reported an enrichment of Sharpea azabuensis in lower methane emitting cohorts30,31. As a result, we suspect that the greater abundance of Sharpea azabuensis in the Perth breed may be suggestive of lower methane production, however, due to the lack of methane emissions data recorded in this study, this should be considered with caution. Acetitomaculum was identified as a dominant bacterial genus in the liquid ruminal fraction, with a mean relative abundance of 3.5%. Its abundance was found to be negatively correlated with FCR, indicating a potential role in enhancing host feed efficiency. Indeed, the abundance of an unclassified ASV within the genus (ASV44) was found to be higher in feed efficient Cheviots, which was significant when compared to the Connemara breed. Acetitomaculum has one known species, A. ruminis, an acetogenic bacterium capable of heterotrophic and autotrophic growth32. It possible that its higher abundance may be contributing to Cheviot feed efficiency by shifting H2 away from methanogenesis and towards acetogenesis, reducing dietary energy loss33. Alternatively, Acetitomaculum may be contributing to the improved FCR in the Cheviots through metabolic pathways other than reductive acetogenesis due to the organism's ability to metabolise a wide range of substrates and its inability to compete with methanogens for H2, especially at low H2 concentrations32.
Bacteria associated with the rumen epithelium maintain close interactions with the host and have been shown to correlate with ruminal epithelial tissue gene expression34,35, suggesting that host genetics may influence this bacterial population more than those in the solid and liquid fractions. In the current study, sheep breed was found to significantly contribute to differences in the alpha diversity (i.e. observed ASVs and inverse Simpson), but not beta diversity of the epithelial-associated bacterial community. In the context of alpha diversity, the Cheviot breed harbored the fewest number of observable ASVs, while the Connemara harbored the most. Moreover, when compared to Connemara and Perth, the Cheviot and Lanark breeds had a significantly higher mean inverse Simpson index. This finding suggests that, while epithelial community richness was lowest for the Cheviot breed, the community was more uniformly distributed with respect to species abundance than those of the Connemara and Perth breeds. Firmicutes, Bacteroidetes, and Proteobacteria were the most predominant bacterial phyla, which is consistent with previous studies exploring epithelial bacterial communities in cattle36,37. ASV24 classified to the genus Succiniclasticum was shown to be more abundant in the epithelia of the Cheviot, Lanark, and Perth breeds when compared to the Connemara breed. Succiniclasticum is a gram-negative rod-shaped anaerobe that ferments succinate and converts it to propionate38, an important precursor of glucose in the rumen39. The higher abundance of Succiniclasticum in those breeds may have contributed to their enhanced FCR compared to the Connemara breed by supplying enough extra propionate to boost gluconeogenesis, which is important for animal growth and production40. The abundance of Ruminococcus 1 was significantly higher in the Perth and Connemara breeds relative to the Cheviot breed. Ruminococcus spp. are core members of the rumen microbiome41, and its association with the rumen epithelial could indicate that its abundance is under host genetic regulation. Indeed, previous research carried out by Li et al. showed Ruminococcus was heritable in cattle (h2 = 0.16 ± 0.08; mean ± SE), and variations in its abundance were associated with a single nucleotide polymorphism (SNP) in the RAPH1 gene27. The genus comprises some of the most proficient and best described cellulolytic degraders, including R. albus and R. flavefacians42. Consequently, it is probable that the Connemara and Perth breed are genetically selecting for a higher abundance of Ruminococcus, which may have allowed these SB breeds to evolve into successful mountain sheep able to thrive in poor grazing areas with low-energy vegetation.
Bacterial community profiles can differ across ruminal fractions in both bovine and ovine ruminants14,16. Therefore, we investigated the influence of ruminal fraction on bacterial populations in the Cheviot, Connemara, and Perth breeds individually. Our findings show that ruminal fraction had no effect of alpha diversity measures in the Cheviot breed. However, in the Lanark breed the liquid fraction exhibited a significantly lower Shannon diversity, while in the Perth breed the epithelial fraction exhibited a significantly higher community richness and PD when compared to the other fractions, respectively. It is widely reported in the literature that the bacterial community composition of the epithelial fraction is distinct from those communities associated with rumen content14,16,43. In contrast to prior studies16,18, our beta diversity analysis revealed no significant differences in community composition across ruminal fractions for any of the breeds studied. The reason for this finding is unclear, though it could be related to variations in dietary management between studies44,45,46. While no major compositional differences were seen in this study, ruminal fraction had a significant impact on the abundance of taxa commonly associated with the rumen epithelium, with the majority of differences occurring between the epithelial and solid fractions in all breeds. When considering the Lanark and Cheviot breeds, several taxa within the order Clostridiales (Family XII, Butyrivibrio 2, Mogibacterium and Lachnospiraceae UCG-008) were significantly more abundant in the epithelial ruminal fraction. Clostridiales are obligate anaerobes that have previously been discovered to interact with the rumen epithelium47,48 and reported to be key components of its core microbiota45. In addition to Clostridiales, Campylobacter was also found to be significantly abundant in the epithelial fraction of the Cheviot and Lanark breeds. Campylobacter is an asaccharolytic microaerophilic bacterium that has frequently been identified as associating with the rumen epithelium16,49,50, and its capacity to consume oxygen demonstrates its functional significance in maintaining the rumen's anaerobic environment51.
Archaea are the sole producers of methane within the rumen, which is an important homeostatic process that regulates the partial pressure of hydrogen52. However, methanogenesis is estimated to result in a 2–12% loss in feed efficiency to the host53, and is further supported by research that has revealed associations between ruminant methane emissions and host feed efficiency54. Moreover, the abundance of methanogenic archaea in the rumen has been linked to both methane emissions55 and feed efficiency56. To date, research in cattle has shown that archaea taxonomic abundances vary between breeds of cattle25 and some archaeal species have been found to be heritable27,57, signalling a potential host genetic effect on the community. Therefore, in the present study we explored the effect of sheep breed on archaeal populations associated with the solid, liquid and epithelial fractions. Alpha diversity analysis showed that breed had a significant effect on the richness of the solid and liquid associated communities, whereby the Cheviot breed exhibited the lowest community richness. Beta diversity analysis revealed no significant effect of breed on community composition, suggesting the presence of a shared core archaea community among breeds. Similarly, differential abundance analysis revealed no overall influence of breed on taxonomic abundances; however, pairwise comparison between breeds shows that the genus Candidatus Methanomethylophilus was significantly more abundant in Perth and Lanark breeds compared to the Cheviot breed in the solid ruminal fraction. Furthermore, results from our correlation analysis showed Methanosphaera and Candidatus Methanomethylophilus associated with improved ADG and feed efficiency in the epithelial and liquid ruminal fractions, respectively. Candidatus Methanomethylophilus is a H2-dependent methylotrophic methanogen, which derives its energy from the metabolism of methanol and methylaimines58,59 In our previous study with sheep, Candidatus Methanomethylophilus was identified in both the solid and liquid ruminal fractions, but significant correlation was not observed between its abundance and FCR21. In contrast, Li et al. (2019) found the abundance of Candidatus Methanomethylophilus significantly higher in H-RFI Charlois steers when compared to L-RFI counterparts, however, similar findings were not observed in the Angus or Hybrid Kinsella breeds used in that same study25. Furthermore, Methanosphaera and Candidatus Methanomethylophilus have previously been linked to lower methane emitting in sheep and cattle31,60, respectively. However, given the complexity of methane synthesis in the rumen, associating higher or lower methane production to individual taxonomic groups may be unrealistic61,62.
The results in the current study further support findings in the literature that breed/host genetics can influence the microbial community structure within the rumen. This could have applications for breeding programs, where microbiomes that are better at utilizing feed and producing less methane could potentially be selected for63,64. However, the heritability of rumen microbiome composition across generations needs further investigation in livestock ruminants, including with microbiome transplant experiments. Because of the functional redundancy of the rumen microbiome65, where phylogenetically distant microbes may have identical metabolic capabilities, taxonomic differences observed between breeds in the current study may not necessarily reflect functional divergence. Future research would benefit from coupling microbial community composition with rumen chemistry, in addition to multi-omics approaches (i.e. meta-genomics/transcriptomics), which would give a better indication of the rumen microbiota's varied metabolic capacity between sheep breeds.
Conclusions
In summary, this study demonstrated the breed of sheep has an effect on the bacterial and archaea taxonomic abundance within the rumen, which can have significant implications for improving feed efficiency and reducing methane emissions. However, further research is required to determine if the taxonomic differences observed signifies functional variation between the breeds. Furthermore, we observed differences in the distribution of bacterial taxa between ruminal fractions, which supports previous studies and highlights a rumen fraction bias of importance to rumen sampling strategies for rumen sampling strategies.
Methods
Animal model
The Teagasc Animal Ethics Committee authorised all treatments involving animals in this investigation, which was conducted under experimental licence (No:P19132/P028) from Ireland’s Health Product Regulatory Authority (HPRA) in compliance with ARRIVE guidelines and the European Union protection of animals used for scientific purposes regulations 2012 (S.I. No 543 of 2012).
Over a 3-month period, data was collected on 36 ram lambs enrolled in a feed efficiency measurement test. Lambs included in this study originated from four different breeds: Cheviot (n = 10), Connemara (n = 6), Lanark (n = 10) and Perth (n = 10). After weaning, lambs were individually penned on plastic slat-floored feeding pens (182 cm L × 122 cm W). Lambs were allowed tactile, olfactory, and visual contact with each other through the pen partitions. The mean body weight of animals at the beginning of the measurement period was 29.6 kg (SD = 3.7). Throughout the trial period, all lambs were offered a cereal-based nut ad libitum, with fresh concentrates supplied daily and refusals removed (i.e., troughs emptied and cleaned) weekly. Concentrates were weighed daily in the morning, and daily intake was estimated by subtracting the weekly total intake from the number of refusals and dividing by seven. Concentrates were supplemented with unrestricted access to perennial rye-grass silage (Lolium perenne) to maintain rumen health (100-g/d DM). Silage was offered fresh daily and refusals removed twice weekly during morning feeding. At no point were animals without access to concentrates or silage during the ad libitum feeding period. Silage intake was not measured as consumption was low. Table 11 contains the ingredients and chemical composition of concentrate and silage used in the study. At all times throughout the measurement period lambs had access to fresh drinking water. The feed intake measurement period ceased when lambs reached a target slaughter weight of > 40 kg. Lambs were slaughtered at the Kepak Ltd abattoir in Athleague in Co. Roscommon on three separate dates when lamb maturity was reached; 29th November 2017, 13th December 2017 and 17th January 2018. The abattoir was approximately 56 km (55 min) from the Teagasc research farm in Athenry, Co. Galway, Ireland. Prior to slaughter, feed and water (at the farm) were withheld for 2 h for all sheep in the study, since differences in time off feed may have affected rumen microbial community composition.
Phenotypic data collected throughout the trial period included the animals weight at the beginning of the trial period (Start weight); dry matter intake (DMI), described as the amount of feed (kg) the lambs consumed; average daily gain (ADG) was calculated by dividing the total weight gain over the trial period divided by the number of days animals were on trial before slaughter; feed conversion ratio (FCR) was calculated by dividing DMI by ADG. Live weight (LW) was the weight of lambs before slaughter. LW gain was the difference in weight at the beginning and end of the trial period. Carcass weight refers to the weight of the carcass after the offal has been removed following slaughter. KO% refers to the weight of the carcass as a percentage of the animal's live weight prior to slaughter.
Sample collection
Samples of ruminal fractions were collected immediately after slaughter. Rumen fluid and solid fractions were separated into 25 ml tubes, by filtering rumen digesta through four layers of sterile cheesecloth. To collect rumen epithelial samples, papillae were cut from dorsal, ventral, cariad and caudal regions of the rumen wall using sterilised scissors, approx. 1 cm2, and subsequently rinsed with cold sterile saline solution (0.9% w/v NaCl). Samples from all three ruminal fractions were frozen immediately in liquid nitrogen after separation and then stored at − 80 °C. A total of 90 samples were available for the current study, 28 epithelial samples (Cheviot n = 9, Connemara n = 3, Lanark n = 7, Perth n = 9), 30 liquid (Cheviot n = 9, Connemara n = 5, Lanark n = 9, Perth n = 7), and 32 solid ruminal samples (Cheviot n = 8, Connemara n = 6, Lanark n = 9, Perth n = 9).
Rumen microbial DNA extraction and library preparation
Under liquid nitrogen, each sample was homogenised to a fine frozen powder using a pestle and mortar. Extraction of microbial DNA from the samples was performed using the method described by Yu and Morrison66. Amplicon libraries were created from 25 ng of rumen microbial DNA using two rounds of PCR amplification as described in the Illumina Miseq 16S Sample Preparation Guide, with minor alterations to cycle length as described by McGovern et al.17. 515F/806R primers67, built with Nextera over hang adapters, and 2× KAPA Hifi HotStart ReadyMix DNA polymerase were used for the first round of PCR amplification, targeting the V4 hyper-variable region of the 16S rDNA (Roche Diagnositics, West Sussex, UK). The first round of PCR was performed at 95 °C for 3 min, then 20 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and 72 °C for 5 min. To enable the attachment of dual indices and Illumina sequencing adapters using the Nextera XT indexing kit, a second round of PCR was conducted at 95 °C for 3 min, followed by 8 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and 72 °C for 5 min (Illumina, San Diego, CA, USA). Following PCR rounds 1 and 2, the amplicons were purified using the Qiaquick PCR Purification Kit (Qiagen, Manchester, UK). To remove adaptor primers, amplicons were pooled together in identical concentrations and gel purified using the Qiagen Gel Extraction Kit (Qiagen, Manchester, UK). Using the QIAquick PCR purification kit, the amplicons were again purified to eliminate any agarose residues (Qiagen, Manchester, UK). Amplicon purity was measured using the Nanodrop 1000, followed by confirmation using the Qubit fluorometer and the KAPA SYBR FAST universal kit with Illumina Primer Premix (Roche Diagnositics, West Sussex, UK). Amplicon libraries were diluted and denatured according to the Illumina Miseq 16S Sample Preparation Guide, and sequencing was performed on the Illumina MiSeq using the 500 cycle version 2 MiSeq reagent kit (Illumina, San Diego, CA, USA).
Bioinformatics
Amplicon reads were quality assessed using FASTQC (version 0.11.5)68. Adapters and ambiguous basecalls were subsequently removed using Cutadapt (version 1.18)69. The amplicon reads were processed and analyzed using the Divisive Amplicon Denoising Algorithm 2 (DADA2) (DADA2)(version 1.18.0), as described in Callahan et al.70. The DADA2 tutorial available at https://benjjneb.github.io/dada2/tutorial.html (version 1.12) was followed for read filtering, dereplication, sample inference, chimera elimination, paired end read merging, and taxonomy categorization. Taxonomic classification was performed to the species level using the SILVA classification databases (version 132)71. The final output from DADA2 was an Amplicon Sequence Variant (ASV) table and a corresponding taxonomy table. A phylogenetic tree was created using the R package Phangorn72. Prior to downstream analysis, a Phyloseq object including the ASV table, taxonomy table, phylogenetic tree, and experimental metadata was created using the R/Bioconductor package Phyloseq (version 1.26)73. Six samples (2 solid, 2 liquid and 2 epithelial) were removed prior to downstream analysis as a result of having a substantially reduced number of reads (< 100 reads).
Compositional and statistical analysis
Animal production trait data were first verified for normality and homogeneity using the Shapiro Wilks and Levenes tests, respectively, and then compared among breeds using a two-way ANOVA followed by a post-hoc Tukey HSD. DMI records were missing from one animal in each of the Connemara, Perth and Lanark breeds, as a result those animals were omitted from DMI and FCR comparisons.
To profile dominant bacterial and archaea taxa, raw counts were converted to relative abundances and the mean and standard deviation relative abundance of dominant phyla and genera were reported. To examine the effect of breed on bacterial and archaeal populations the data was first stratified according to ruminal fraction (i.e. solid, liquid and epithelial) and then compared between breeds (i.e. Cheviot, Connemara, Lanark and Perth). Similarly, to examine the effect of ruminal fraction on microbial profiles data was stratified according to breed and compared between fractions. For fraction analysis only animals where all three ruminal fraction were available were considered. Due to low numbers of biological replicates the Connemara breed was excluded from ruminal fractions analysis. Prior to diversity analysis raw counts were normalised to even sampling depth using the scaling with ranked subsampling (SRS) method74 and rarefaction curves were generated to assess sequencing effort. Following assessment of rarefaction curves, sample 2083-Solid, which had a sequencing depth of 13,922, was removed due to a loss in community diversity when rarefying. Shannon, inverse Simpson, Faiths phylogenetic diversity (PD), and observed ASVs (richness) diversity indices were used to generate within-sample (alpha) diversity metrics. Alpha diversity data were checked for normality and homogeneity using Shapiro Wilks test and Levenes test prior to statistical analysis. A two-way ANOVA was used to test the null hypothesis that no difference on mean alpha diversity measures existed between groups. For beta diversity analysis, dissimilarities in community composition were measured using both weighted and unweighted UniFrac distances and visualised using principle coordinate analysis (PCoA). Differences in community composition was tested with PERMANOVA and conducted with 9999 permutations using the Adonis function from the R/Bioconductor package Vegan (version 2.5-5)75. Differential abundance (DA) analysis was conducted using both the likelihood ratio test (LRT) and the Wald’s test from the DESeq2 package76. Only taxa with a relative abundance larger than 0.01 percent and a prevalence greater than 50 percent were considered for DA analysis. An a priori q-value threshold was set at 0.05. The date of sample collection was included as a covariate to adjust for variations in abundance associated with different slaughter dates. Lastly, Spearman correlation coefficients (P < 0.05) were done to assess the correlation between bacterial and archaeal genera and animal production traits; FCR and ADG, in order to identify potential feed efficiency drivers. Only genera with a relative abundance larger than 0.01 percent and a prevalence greater than 50 percent were considered for correlation analysis.
Ethics declaration
Irelands Health Products Regulatory Authority reviewed and authorised the animal study in compliance with the European Union (EU) protection of animals used for scientific purposes regulations 2012 (S.I No 543 of 2012).
Data availability
Data used for this study were deposited into NCBI Sequence Read Archieve (SRA), published under accession number PRJNA781265.
References
Oltjen, J. W. & Beckett, J. L. Role of ruminant livestock in sustainable agricultural systems. J. Anim. Sci. 74, 1406–1409 (1996).
Henchion, M., Hayes, M., Mullen, A. M., Fenelon, M. & Tiwari, B. Future protein supply and demand: Strategies and factors influencing a sustainable equilibrium. Foods 6, 53 (2017).
Dopelt, K., Radon, P. & Davidovitch, N. Environmental effects of the livestock industry: The relationship between knowledge, attitudes, and behavior among students in Israel. Int. J. Environ. Res. Public Health 16, 1359 (2019).
Hegarty, R. S., Goopy, J. P., Herd, R. M. & McCorkell, B. Cattle selected for lower residual feed intake have reduced daily methane production1,2. J. Anim. Sci. 85, 1479–1486 (2007).
Fitzsimons, C., Kenny, D. A., Deighton, M. H., Fahey, A. G. & McGee, M. Methane emissions, body composition, and rumen fermentation traits of beef heifers differing in residual feed intake. J. Anim. Sci. 91, 5789–5800 (2013).
Beauchemin, K. A., Ungerfeld, E. M., Eckard, R. J. & Wang, M. Review: Fifty years of research on rumen methanogenesis: lessons learned and future challenges for mitigation. Animal 14, 2–16 (2020).
Kenny, D. A., Fitzsimons, C., Waters, S. M. & McGee, M. Invited review: Improving feed efficiency of beef cattle—The current state of the art and future challenges. Animal 12, 1815–1826 (2018).
van Zanten, H. H. E., Mollenhorst, H., Klootwijk, C. W., van Middelaar, C. E. & de Boer, I. J. M. Global food supply: Land use efficiency of livestock systems. Int. J. Life Cycle Assess. 21, 747–758 (2016).
Byrne, D., Keena, C., Maguire, F., Sheridan, H. & Gorman, M. Farming the uplands-where to from here? In Teagasc Hill Sheep Conference (2017).
Carlyle, W. J. The changing distribution of breeds of sheep in Scotland, 1795–1965. Agric. Hist. Rev. 27, 19–29 (1979).
Blaxter, K. L., Clapperton, J. L. & Wainman, F. W. The extent of differences between six British breeds of sheep in their metabolism, feed intake and utilization, and resistance to climatic stress. Br. J. Nutr. 20, 283–294 (1966).
Kirton, A. et al. A comparison between 15 ram breeds for export lamb production 1. Liveweights, body components, carcass measurements, and composition. New Zealand J. Agric. Res. 38, 347–360 (1995).
Huws, S. A. et al. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: Past, present, and future. Front. Microbiol. 9, 2161 (2018).
Li, Z., Mu, C., Xu, Y., Shen, J. & Zhu, W. Changes in the solid-, liquid-, and epithelium-associated bacterial communities in the rumen of Hu lambs in response to dietary urea supplementation. Front. Microbiol. 11, 244 (2020).
Ji, S. et al. Comparison of rumen bacteria distribution in original rumen digesta, rumen liquid and solid fractions in lactating Holstein cows. J. Anim. Sci. Biotechnol. 8, 16 (2017).
Ren, Q. et al. Bacterial communities in the solid, liquid, dorsal, and ventral epithelium fractions of yak (Bos grunniens) rumen. MicrobiologyOpen 9, e963 (2020).
McGovern, E. et al. 16S rRNA sequencing reveals relationship between potent cellulolytic genera and feed efficiency in the rumen of bulls. Front. Microbiol. 9, 1842–1842 (2018).
Dinsdale, D., Cheng, K. J., Wallace, R. J. & Goodlad, R. A. Digestion of epithelial tissue of the rumen wall by adherent bacteria in infused and conventionally fed sheep. Appl. Environ. Microbiol. 39, 1059–1066 (1980).
Cheng, K. J., McCowan, R. P. & Costerton, J. W. Adherent epithelial bacteria in ruminants and their roles in digestive tract function. Am. J. Clin. Nutr. 32, 139–148 (1979).
Cheng, K. J. & Wallace, R. J. The mechanism of passage of endogenous urea through the rumen wall and the role of ureolytic epithelial bacteria in the urea flux. Br. J. Nutr. 42, 553–557 (1979).
McLoughlin, S. et al. Rumen microbiome composition is altered in sheep divergent in feed efficiency. Front. Microbiol. 11, 1981 (2020).
Shabat, S. K. B. et al. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10, 2958–2972 (2016).
Myer, P. R., Smith, T. P. L., Wells, J. E., Kuehn, L. A. & Freetly, H. C. Rumen microbiome from steers differing in feed efficiency. PLoS ONE 10, e0129174 (2015).
Ellison, M. J. et al. Diet and feed efficiency status affect rumen microbial profiles of sheep. Small Rumin. Res. 156, 12–19 (2017).
Li, F., Hitch, T. C. A., Chen, Y., Creevey, C. J. & Guan, L. L. Comparative metagenomic and metatranscriptomic analyses reveal the breed effect on the rumen microbiome and its associations with feed efficiency in beef cattle. Microbiome 7, 6–6 (2019).
Noel, S. J. et al. Rumen and fecal microbial community structure of Holstein and Jersey dairy cows as affected by breed, diet, and residual feed intake. Animals 9, 498 (2019).
Li, F. et al. Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome 7, 92 (2019).
Paz, H. A., Anderson, C. L., Muller, M. J., Kononoff, P. J. & Fernando, S. C. Rumen bacterial community composition in Holstein and Jersey cows is different under same dietary condition and is not affected by sampling method. Front. Microbiol. 7, 1206–1206 (2016).
Kumar, S. et al. Sharpea and Kandleria are lactic acid producing rumen bacteria that do not change their fermentation products when co-cultured with a methanogen. Anaerobe 54, 31–38 (2018).
Kamke, J. et al. Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea-enriched microbiome characterised by lactic acid formation and utilisation. Microbiome 4, 56 (2016).
Kittelmann, S. et al. Two different bacterial community types are linked with the low-methane emission trait in sheep. PLoS ONE 9, e103171 (2014).
Le Van, T. D. et al. Assessment of reductive acetogenesis with indigenous ruminal bacterium populations and Acetitomaculum ruminis. Appl. Environ. Microbiol. 64, 3429–3436 (1998).
Karekar, S., Stefanini, R. & Ahring, B. Homo-acetogens: Their metabolism and competitive relationship with hydrogenotrophic methanogens. Microorganisms 10, 397 (2022).
Liu, K. et al. Ruminal microbiota–host interaction and its effect on nutrient metabolism. Anim. Nutr. 7, 49–55 (2021).
Chen, Y., Oba, M. & Guan, L. L. Variation of bacterial communities and expression of Toll-like receptor genes in the rumen of steers differing in susceptibility to subacute ruminal acidosis. Vet. Microbiol. 159, 451–459 (2012).
Wetzels, S. U. et al. Epimural bacterial community structure in the rumen of Holstein cows with different responses to a long-term subacute ruminal acidosis diet challenge. J. Dairy Sci. 100, 1829–1844 (2017).
Anderson, C. J., Koester, L. R. & Schmitz-Esser, S. Rumen epithelial communities share a core bacterial microbiota: A meta-analysis of 16S rRNA gene Illumina MiSeq sequencing datasets. Front. Microbiol. 12, 625400 (2021).
van Gylswyk, N. O. Succiniclasticum ruminis gen. nov., sp. nov., a ruminal bacterium converting succinate to propionate as the sole energy-yielding mechanism. Int. J. Syst. Evol. Microbiol. 45, 297–300 (1995).
Elliot, J. M. Propionate metabolism and vitamin B12. In Digestive Physiology and Metabolism in Ruminants: Proceedings of the 5th International Symposium on Ruminant Physiology, held at Clermont — Ferrand, on 3rd–7th September, 1979 (eds Ruckebusch, Y. & Thivend, P.) 485–503 (Springer Netherlands, 1980).
Young, J. W. Gluconeogenesis in cattle: Significance and methodology1. J. Dairy Sci. 60, 1–15 (1977).
Wirth, R. et al. The planktonic core microbiome and core functions in the cattle rumen by next generation sequencing. Front. Microbiol. 9, 2285 (2018).
Yeoman, C. J. et al. In vivo competitions between Fibrobacter succinogenes, Ruminococcus flavefaciens, and Ruminoccus albus in a gnotobiotic sheep model revealed by multi-omic analyses. MBio 12, e03533-20 (2021).
Cammack, K. M., Austin, K. J., Lamberson, W. R., Conant, G. C. & Cunningham, H. C. Ruminant nutrition symposium: Tiny but mighty: The role of the rumen microbes in livestock production. J. Anim. Sci. 96, 752–770 (2018).
Zhang, R., Ye, H., Liu, J. & Mao, S. High-grain diets altered rumen fermentation and epithelial bacterial community and resulted in rumen epithelial injuries of goats. Appl. Microbiol. Biotechnol. 101, 6981–6992 (2017).
Petri, R. M. et al. Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Appl. Environ. Microbiol. 79, 3744–3755 (2013).
Zhang, J. et al. Effect of dietary forage to concentrate ratios on dynamic profile changes and interactions of ruminal microbiota and metabolites in Holstein heifers. Front. Microbiol. 8, 2206 (2017).
Chen, Y., Penner, G. B., Li, M., Oba, M. & Guan, L. L. Changes in bacterial diversity associated with epithelial tissue in the beef cow rumen during the transition to a high-grain diet. Appl. Environ. Microbiol. 77, 5770–5781 (2011).
Tan, R. S. G., Zhou, M., Li, F. & Guan, L. L. Identifying active rumen epithelial associated bacteria and archaea in beef cattle divergent in feed efficiency using total RNA-seq. Curr. Res. Microb. Sci. 2, 100064 (2021).
Pacífico, C. et al. Unveiling the bovine epimural microbiota composition and putative function. Microorganisms 9, 342 (2021).
Schären, M. et al. Alterations in the rumen liquid-, particle- and epithelium-associated microbiota of dairy cows during the transition from a silage- and concentrate-based ration to pasture in spring. Front. Microbiol. 8, 744–744 (2017).
Mann, E., Wetzels, S. U., Wagner, M., Zebeli, Q. & Schmitz-Esser, S. Metatranscriptome sequencing reveals insights into the gene expression and functional potential of rumen wall bacteria. Front. Microbiol. 9, 43 (2018).
Moss, A. R., Jouany, J.-P. & Newbold, J. Methane production by ruminants: Its contribution to global warming. Ann. Zootech. 49, 231–253 (2000).
Johnson, K. A. & Johnson, D. E. Methane emissions from cattle. J. Anim. Sci. 73, 2483–2492 (1995).
Alemu, A. W., Vyas, D., Manafiazar, G., Basarab, J. A. & Beauchemin, K. A. Enteric methane emissions from low- and high-residual feed intake beef heifers measured using GreenFeed and respiration chamber techniques. J. Anim. Sci. 95, 3727–3737 (2017).
Wallace, R. J. et al. The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 16, 1–14 (2015).
Zhou, M. & Hernandez-Sanabria, E. Characterization of variation in rumen methanogenic communities under different dietary and host feed efficiency conditions, as determined by PCR-denaturing gradient gel electrophoresis analysis. Appl. Environ. Microbiol. 76, 3776–3786 (2010).
Goodrich, J. K. et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19, 731–743 (2016).
Borrel, G. et al. Genome sequence of “Candidatus Methanomethylophilus alvus” Mx1201, a methanogenic archaeon from the human gut belonging to a seventh order of methanogens. J. Bacteriol. 194, 6944–6945 (2012).
Noel, S. J., Højberg, O., Urich, T. & Poulsen, M. Draft genome sequence of “Candidatus Methanomethylophilus” sp. 1R26, enriched from bovine rumen, a methanogenic archaeon belonging to the Methanomassiliicoccales order. Genome Announc. 4, e01734-15 (2016).
Martínez-Álvaro, M. et al. Identification of complex rumen microbiome interaction within diverse functional niches as mechanisms affecting the variation of methane emissions in bovine. Front. Microbiol. 11, 659 (2020).
Greening, C. et al. Diverse hydrogen production and consumption pathways influence methane production in ruminants. ISME J. 13, 2617–2632 (2019).
GhanbariMaman, L. et al. Co-abundance analysis reveals hidden players associated with high methane yield phenotype in sheep rumen microbiome. Sci. Rep. 10, 4995 (2020).
Gonzalez-Recio, O., Zubiria, I., García-Rodríguez, A., Hurtado, A. & Atxaerandio, R. Short communication: Signs of host genetic regulation in the microbiome composition in 2 dairy breeds: Holstein and Brown Swiss. J. Dairy Sci. 101, 2285–2292 (2018).
Difford, G. F. et al. Host genetics and the rumen microbiome jointly associate with methane emissions in dairy cows. PLoS Genet. 14, e1007580 (2018).
Weimer, P. J. Redundancy, resilience, and host specificity of the ruminal microbiota: Implications for engineering improved ruminal fermentations. Front. Microbiol. 6, 296 (2015).
Yu, Z. & Morrison, M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36, 808–812 (2004).
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 108, 4516–4522 (2011).
Andrews, S. FastQC: a quality control tool for high throughput sequence data. 2010. (2017).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Callahan, B. Silva taxonomic training data formatted for DADA2. Silva Version 132 (2018).
Schliep, K. P. phangorn: Phylogenetic analysis in R. Bioinformatics 27, 592–593 (2011).
McMurdie, P. J. & Holmes, S. Phyloseq: A bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. In Biocomputing 2012 235–246 (World Scientific, 2012).
Beule, L. & Karlovsky, P. Improved normalization of species count data in ecology by scaling with ranked subsampling (SRS): Application to microbial communities. PeerJ 8, e9593 (2020).
Dixon, P. VEGAN, A package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550–550 (2014).
Acknowledgements
The authors would like to acknowledge financial support from DAFM (RAMLAM–11/SF/310) and MASTER (EU Horizon 2020 funded project, grant agreement No 818368). SM is funded by the Teagasc Walsh Scholarship programme (Ref: 2015218). CS acknowledges funding support from Science Foundation Ireland.
Author information
Authors and Affiliations
Contributions
F.C., N.C. and M.D. designed and conceptualised the sheep production experiments. S.W., C.S. and S.M. designed and conceptualised microbiome study. Y.M., C.C.S. and P.S. performed DNA extraction, 16S rRNA library preparation and sequencing. S.M. carried out bioinformatic analysis, detailed interpretation of the data, and drafted the manuscript with support from S.W. and C.S. First draft of manuscript was revised and finalised by S.W. and C.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
McLoughlin, S., Spillane, C., Campion, F.P. et al. Breed and ruminal fraction effects on bacterial and archaeal community composition in sheep. Sci Rep 13, 3336 (2023). https://doi.org/10.1038/s41598-023-28909-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-28909-1
This article is cited by
-
Rumen fermentation and microbiota in Shami goats fed on condensed tannins or herbal mixture
BMC Veterinary Research (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.