Abstract
In a moss sample collected in Ribeiro Frio, Madeira, Paramacrobiotus gadabouti sp. nov. was found and described using the integrative taxonomy approach. The new species is described based on morphological and morphometric data from both phase-contrast light microscopy (PCM), as well as scanning electron microscopy (SEM). Moreover, four DNA markers, three nuclear (18S rRNA, 28S rRNA, ITS-2) and one mitochondrial (COI) markers, were used to elucidate the phylogenetic position of the new species within the family Macrobiotidae. The new species has a microplacoid that placed it within Parmacrobiotus richtersi group and exhibit richtersi-type eggs having processes terminated with cap-like structures. Paramacrobiotus gadabouti sp. nov. is most similar to Pam. alekseevi, Pam. filipi and Pam. garynahi, but differs from them mainly in details of egg morphology and morphometrics. Unlike other species from this group, which were confirmed as bisexual and showed limited distribution, Paramacrobiotus gadabouti sp. nov. is yet another parthenogenetic species with a wide distribution, demonstrating that at least some tardigrades confirm to the hypothesis of 'everything is everywhere'.
Similar content being viewed by others
Introduction
The Phylum Tardigrada currently consists of over 1400 species1,2,3,4 that inhabit terrestrial and aquatic (freshwater and marine) environments throughout the world5,6,7. Knowledge of terrestrial tardigrades of Madeira, Portugal is rather poor as to date, only 33 species (22 Eutardigrada and 11 Heterotardigrada taxa) have been reported from this region8,9,10.
The genus Paramacrobiotus Guidetti et al.11, currently comprises 43 formally named species4. It was formally erected in 2009 based on morphological and genetic analyses11. Morphologically two distinct species groups are present in the genus, one exhibiting a microplacoid within the pharynx, i.e. richtersi group and the second one without microplacoid, i.e. areolatus group. This phenotypic difference led Kaczmarek et al.12 to propose these two groups to constitute separate subgenera for which specific names were clarified by Marley et al.13. However, their erection was subseqently questioned independently based on two phylogenetic analyses14,15. Within the genus Paramacrobiotus bisexual and unisexual species/populations have been observed and reported in the past (e.g. in populations of Pam. richtersi (Murray, 1911)16 from Italy (bisexual and unisexual); according to modern taxonomy they probably constitute distinct species), Pam. areolatus (Murray, 1907)17 from Italy (bisexual) and Svalbard (unisexual), Pam. tonolli (Ramazzotti, 1956)18 (bisexual) from USA, Pam. fairbanksi Schill, Förster, Dandekar and Wolf, 201019 (unisexual) from Antarctic, Italy, Poland, Spain and USA, Pam. kenianus Schill, Förster, Dandekar and Wolf, 201019 (unisexual) from Kenya and Pam. palaui (unisexual) Schill, Förster, Dandekar and Wolf, 201019 from Micronesia, Pam. depressus Guidetti, Cesari, Bertolani, Altiero and Rebecchi, 201914 (bisexual) from Italy, Pam. celsus Guidetti, Cesari, Bertolani, Altiero and Rebecchi, 201914 (bisexual) from Italy, Pam. spatialis Guidetti, Cesari, Bertolani, Altiero and Rebecchi, 201914 (bisexual) from Italy, Pam. arduus Guidetti, Cesari, Bertolani, Altiero and Rebecchi, 201914 (bisexual) from Italy, Pam. experimentalis Kaczmarek, Mioduchowska, Poprawa and Roszkowska, 202020 (bisexual) from Madagascar15,19,20,21,22,23,24,25,26,27,28,29. Importantly, Guidetti et al.14 also concluded that unisexual species like Pam. fairbanksi have a wider geographical range compared to bisexual Paramacrobiotus taxa. Subsequetly, Stec et al.15 corroborated this hypothesis additionally suggesting that the wide distribution of some taxa of the genus may be caused by human-mediated dispersion, since most of these populations were found in populated areas with trade and touristes.
In the present paper, we provide a description of a new parthenogenetic and widespread Paramacrobiotus species based on its population discovered in Madeira. The study was framed within an integrative taxonomy with detailed morphological and genetic analyses. We also conducted species molecular delimitation analyses based on all COI sequences of the genus Paramacrobiotus available in GenBank. Finally, we reconstructed the multilocus phylogeny of superclade II of the family Macrobiotidae (sensu Stec et al.29) to elucidate the phylogenetic position of the new species.
Material and methods
Sample processing
The moss sample was collected in Ribeiro Frio, Madeira (32°44′36.7"N, 16°54′28.0"W) in September 2019. The sample was packed in paper envelope, dried at a temperature of ca. 25 °C and delivered to the laboratory at the Faculty of Biology, Adam Mickiewicz University in Poznań, Poland. Tardigrades were extracted from the samples and studied following the protocol of Stec et al.30.
Tardigrade culture
Specimens of a new species have been cultured continuously since February 2022. The cultures were maintained in plastic Petri dishes containing a mixture of ddH2O and “Żywiec Zdrój” spring water (3:1). To aid tardigrades locomotion, each Petri dish bottom was scratched with fine sandpaper. The culture was maintained in an environmental chamber (model POL ST1 BASIC) at 18 °C and fed once per week on rotifers (2 ml of culture of Lecane inermis (Bryce 1892))31. Once per week, the medium was exchanged using a sterile plastic pipette to avoid contamination. To establish the type of reproduction in the new species, 50 eggs were collected and incubated in a glass cube and inspected daily. Upon hatching, each juvenile was transferred to a single well of 24 well plates with scratched bottom. The isloated individuals were then observed and fed every week.
Genotyping
Prior to DNA extraction, each tardigrade specimen was examined in vivo under PCM (400 × magnification). In order to obtain voucher specimens, genomic DNA was extracted from individual animals following a Chelex 100 resin (BioRad) extraction method32 with modifications according to Stec et al.33.
Two conservative nuclear ribosomal subunit genes were sequenced, i.e. 18S rRNA, 28S rRNA as well as nuclear ITS-2 (internal transcribed spacer 2) and mitochondrial COI (cytochrome C oxidase subunit I) barcode sequences. Fragments of the nuclear genes were amplified using the following primers: 18S_Tar_Ff1 (5ʹ-AGGCGAAACCGCGAATGGCTC-3ʹ) and 18S_Tar_Rr1 (5ʹ-GCCGCAGGCTCCACTCCTGG-3ʹ; Stec et al.34) for the 18S rRNA gene fragment; 28SF0001 (5ʹ-ACCCvCynAATTTAAGCATAT-3ʹ) and 28SR0990 (5ʹ-CCTTGGTCCGTGTTTCAAGAC-3ʹ; Mironov et al.35) for the 28S rRNA gene fragment; ITS–3 (5′- GCATCGATGAAGAACGCAGC.-3′) and ITS-4 (5′- TCCTCCGCTTATTGATATGC-3′; White et al.36) for the ITS-2 gene fragment. In turn, the COI molecular marker was amplified using universal primers: HCO2198 (5ʹ-TAAACTTCAGGGTGACCAAAAAATCA-3ʹ) and LCO1490 (5ʹ-GGTCAACAAATCATAAAGATATTGG–3ʹ; Folmer et al.37). All PCR reactions were performed in 20 μl volume containing 0.8 × JumpStart Taq ReadyMix (1 U of JumpStart Taq DNA polymerase, 4 mM Tris–HCl (pH 8.3), 20 mM KCl, 0.6 mM MgCl2, 0.08 mM of dNTP; Sigma-Aldrich), 0.4 μM of proper forward and reverse primers and ca. 1 ng of DNA. The PCR cycling profiles to amplify the 28S rRNA, ITS-2 and COI sequences were performed according to the protocols described in Kaczmarek et al.20. In turn, 18S rRNA sequences were amplified according to the protocol described in Stec et al.34. The reactions were performed in a BiometraTProfessional thermocycler. The PCR products were cleaned up by exonuclease I (20 U/μl, Thermo Scientific) and alkaline phosphatase FastAP (1 U/μl, Thermo Scientific). The Sanger sequencing method was carried out in both directions using the BigDyeTM terminator cycle sequencing and ABI Prism 3130xl genetic analyser (Life Technologies). In case ITS-2 gene fragment poor sequencing results have been obtained. Finally, this molecular marker was not applied in the analysis.
Phylogenetic analysis and molecular species delimitation
Phylogenetic analyses were performed in order to establish phyletic position of the new species and reconstruct the relationships within Macrobiotidae clade II (sensu Stec et al.29). The data set was compiled from taxa/specimens for which DNA sequences of at least two (out of all four commonly used 18S rRNA, 28S rRNA, ITS-2, COI) molecular markers are available and suitable for concatenation (Table 1). The DNA sequences of Macrobiotus rybaki Vecchi & Stec, 202138 and Sisusbiotus spectabilis Thulin, 192839, and Mesobiotus datanlanicus Stec, 201940 were used as the outgroup. The sequences were aligned using the AUTO method (for COI and ITS2) and the Q-INS-I method (for ribosomal markers: 18S rRNA and 28S rRNA) of MAFFT version 741,42 and manually checked against non-conservative alignments in BioEdit. Then, the aligned sequences were trimmed to: 994 (18S rRNA), 811 (28S rRNA), 487 (ITS-2), 658 (COI) bp and concatenated using SequenceMatrix43. Before partitioning, the concatenated alignment was divided into six data blocks constituting three separate blocks of ribosomal markers and three separate blocks of three codon positions in COI data set. Using PartitionFinder44 under the Akaike Information Criterion (AIC), the best scheme of partitioning and substitution models were chosen for Bayesian phylogenetic analysis. Before running phylogenetic analysis, we also performed a substitution saturation test with DAMBE for two variable DNA fragments that were used in our analyses, namly COI and ITS245,46. Bayesian inference (BI) marginal posterior probabilities were calculated for the concatenated (18S rRNA+28S rRNA+ITS-2+COI) data set using MrBayes v3.247. Random starting trees were used and the analysis was run for ten million generations, sampling the Markov chain every 1000 generations. An average standard deviation of split frequencies of < 0.01 was used as a guide to ensure the two independent analyses had converged. The program Tracer v1.648 was then used to ensure Markov chains had reached stationarity, and to determine the correct ‘burn-in’ for the analysis which was the first 10% of generations. The effective sample size (ESS) values were greater than 200 and the consensus tree was obtained after summarising the resulting topologies and discarding the ‘burn-in’. Maximum-likelihood (ML) tree was computed using RAxML v8.0.1949. Strength of support for internal nodes of ML construction was measured using 1,000 rapid bootstrap replicates. The consensus tree was viewed and visualised by FigTree v.1.4.3 available from http://tree.bio.ed.ac.uk/software/figtree. The best evolutionary models of sequence evolution selected for BI and ML analyses, as well as the results of saturation test are given in supplementary materials (SM.01). Networks of haplotypes of the new species were prepared using PopARTver.1.7 (http://popart.otago.ac.nz) with the implementation of Median-Joining method50 For this purpose, we used all COI and ITS-2 sequences od speciemens of the new species that were present in our phylogenetic analyses (N = 5 for ITS-2 and N = 8 for COI).
Using the COI data set comprising all Paramacrobiotus sequences of this marker avilabe in GenBank (80 sequences), we performed two genetic species delimitation analyses. According to the recommendation by Fontaneto et al.63 one of them was a tree-based method, the Poisson Tree Processes (bPTP64), whereas the second one was a distance-based method, the Assemble Species by Automatic Partitioning (ASAP65). For the bPTP analysis, we computed a ML tree using RAxML v8.0.1949 also with prior search of the best model and partition scheme using PartionFinder266 (SM.01). The calculations were conducted on the bPTP webserver (http://species.h-its.org/ptp), with 500,000 MCMC generations, thinning the set to 100, burning at 10% and performing a search for ML and Bayesian solutions. For ASAP analysis we used the COI alignment as input data. The analyses were run on the respective server (https://bioinfo.mnhn.fr/abi/public/asap/asapweb.html) with default settings. All COI sequences used for the analyses and their outputs are given within the supplementary materials (SM.02).
Microscopy and imaging
In total 33 animals, 5 exuvium, 2 simplex and 24 eggs were mounted on microscope slides in the Hoyer’s medium, and then examined under Olympus BX41 Phase-contrast light Microscope (PCM) associated with Olympus SC50 digital camera (Olympus Corporation, Shinjuku-ku, Japan). Thirty animals and 10 eggs were prepared for Scanning Electron Microscope (SEM) observation according to the protocol in Roszkowska et al.67 and examined under high vacuum in Hitachi S3000N SEM (Hitachi, Japan). All figures were assembled in Corel Photo-Paint 2017. For deep structures that could not be fully focused on a single photograph, a series of 2–10 images were taken every ca. 0.5 μm and then manually assembled into a single deep-focus image in Corel Photo-Paint 2017.
Morphometrics and morphological nomenclature
All measurements are given in micrometers [μm]. Structures were measured only if their orientation was suitable. Body length was measured from the anterior extremity to the end of the body, excluding the hind legs. The types of bucco-pharyngeal apparatuses and claws were classified according to Pilato and Binda68. All measurements and terminology of adults and eggs of Paramacrobiotus were prepared according to Kaczmarek and Michalczyk69 and Kaczmarek et al.12. Terminology describing the oral cavity armature (OCA) follows Michalczyk and Kaczmarek70. The macroplacoid length sequence was indicated according to Kaczmarek et al.71. Morphological states of the cuticular bars on legs follow Kiosya et al.72. The pt ratio is the ratio of the length of a given structure to the length of the buccal tube expressed as a percentage73. Morphometric data were handled using the “Parachela” ver. 1.8 template available from the Tardigrada Register74. Row morphometric data for the new species are given in Supplementry Materials (SM.03). Tardigrade taxonomy follows Bertolani et al.75 and Stec et al.29. Genus abbreviations follow Perry et al.76.
Comparative material
For comparison with the new species, holotype and/or paratypes of Pam. derkai (Degma, Michalczyk and Kaczmarek, 2008)77, Pam. experimentalis, Pam. fairbanksi Schill, Förster, Dandekar and Wolf, 201019, Pam. filipi Dudziak, Stec and Michalczyk, 202057, Pam. garynahi (Kaczmarek, Michalczyk and Diduszko, 2005)78, Pam. huziori (Michalczyk and Kaczmarek, 2006)79, Pam. intii Kaczmarek, Cytan, Zawierucha, Diduszko and Michalczyk, 201471, Pam. lachowskae Stec, Roszkowska, Kaczmarek and Michalczyk, 201858, Pam. magdalenae (Michalczyk and Kaczmarek, 2006)80, Pam. sklodowskae (Michalczyk, Kaczmarek and Węglarska, 200681) and Pam. spinosus Kaczmarek, Gawlak, Bartels, Nelson and Roszkowska, 201712 were examined. Moreover, for species identification, the key in Kaczmarek et al.12 and original descriptions were also used (i.e.15,78,82).
Results
Phylogeny and species delimitation
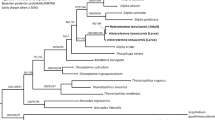
Both phylogenetic analyzes resulted with trees of similar topology and mostly well-supported nodes in which Paramacrobiotus and Tenuibiots are monophyletic genera, whereas Minibiotus was recovered paraphyletic (Fig. 1). Monophyly was not confirmed for Pam. richtersi and Pam. areolatus morpho-groups since representatives of the latter form a paraphyletic group caused by Pam. lachowskae which cluster together with the former morpho-group (Fig. 1). The sequences of the new species obtained in this study clastered together with Paramacrobiotus aff. richtersi populations from France, Portugal, Australia and Tunisia previously reported by Stec et al.15, forming a monophyletic clade staying in sister relationship with Paramacrobiotus aff. richtersi population from Hungary. Haplotype networks showed higher haplotype diversity in case of COI than in ITS-2 marker, with same COI haplotype shered sometimes with populations from very distinct localities (Fig. 2). Molecular species delimitation analyzes recovered 22 and 29 putative species for ASAP and bPTP methods, respectively, with all valid nominal taxa delineated coherently as distinct entities (SM.02). Importantly, 9 ASAP and 12 bPTP entities were delimited from COI sequences without assignment to any nominal Paramacrobiotus species. Single locus delimitations confirmed the results from multilocus phylogeny, recognizing the newly studied population and Paramacrobiotus aff. richtersi populations from France, Portugal, Australia and Tunisia as a single species (Fig. 1; SM.02) which is formally described below.
Maximum likelihood (ML) phylogeny constructed from concatenated sequences of the family Macrobiotidae (18S rRNA + 28S rRNA + ITS-2 + COI; Table 1). Numbers above branches indicate bootstrap support values, while Bayesian posterior probabilities (pp) are given below branches. Bootstrap < 50 and pp < 0.90 are not shown. Taxa of the Pam. richtersi and Pam. areolatus complex are indicated by blue and red branches, respectively. The outgroup is indicated in gray font. The scale bar represents substitutions per position.
Haplotype Median Joining networks for mitochondrial (COI) and nuclear (ITS-2) markers of P. gadabouti sp. nov: (A)—COI; (B)—ITS-2. Haplotypes are represented by coloured circles. The size of circles is proportional to the number sequences/specimens of each particular haplotype. Sequence/specimen names correspond with names presented in phylogenetic tree in Fig. 1. Numbers in brackets indicate the numbers of mutations between the haplotypes.
Taxonomic Account
-
Phylum: Tardigrada (Doyère, 1840)83
-
Class: Eutardigrada (Richters, 1926)84
-
Order: Macrobiotoidea (Thulin, 1928)39
-
Family: Macrobiotidae (Thulin, 1928)39
-
Genus: Paramacrobiotus (Guidetti et al., 2009)11
Paramacrobiotus gadabouti sp. nov. Kayastha, Stec, Mioduchowska and Kaczmarek.
(Figs. 2, 3, 45, 6 and 7; Tables 2 and 3).
Paramacrobiotus gadabouti sp. nov.: (A)—claws II (paratype, PCM); (B)—claws IV (paratype, PCM); (C)—claws II (paratype, SEM); (D)—claws IV (paratype, SEM). Filled unindented arrowhead represents smooth lunulae, empty unindented arrowhead represents granulation, empty indented arrowheads represent single continuous bar and filled indented arrowheads represent doubled muscle attachments. Scale bars in µm.
Paramacrobiotus gadabouti sp. nov.: (A)—bucco-pharyngeal apparatus (dorso–ventral projection) general view (paratype); (B)—placoid morphology in dorsal view (paratype); (C)—ventral placoids (paratype); (D)—oral cavity armature (paratype, PCM) seen from the dorsal side; (E)—oral cavity armature (paratype, PCM) seen from the ventral side. Empty indented arrowhead represents third macroplacoid with sub-terminal constriction, filled indented arrowhead represents third macroplacoid with central constriction in ventral side, filled arrow represents first band of teeth, empty arrow represents second band of teeth, filled unindented arrowhead represents third band of teeth from dorsal side and empty unindented arrowhead represents third band of teeth from ventral side. Scale bars in µm.
Type Material. Holotype (slide M50/4 (+ 6 paratypes (3 animals + 2 exuvium + 1 simplex) on the same slide)) and 101 paratypes (29 animals + 3 exuvium + 1 simplex + 24 eggs; slides: M50/*, where the asterisk can be substituted by any of the following numbers: 1–3, 5–20), 4 exoskeleton after DNA extraction (M50.1/S, M50.2/S, M50.3/S and 50.4/S) and 30 animals + 10 eggs on one SEM stub.
Description (measurements and statistics in Table 2). Animals: Body colour transparent/white, eyes absent in living specimens (Fig. 3A–B). Except for granulation on legs I–IV (Fig. 4A–D), cuticle is smooth, i.e. without gibbosities, papillae, pores, spines or sculpturing. The leg granulation is present on the external surface of legs I–III and on lateral and dorsal surfaces of the hind legs (Fig. 4A–D). Claws of the hufelandi type, stout (Fig. 4A–D). Primary branches with distinct accessory points. Smooth lunulae present under all claws (Fig. 4A–D, filled unindented arrowhead). Single continuous cuticular bar constricted in the middle and paired muscle attachments below claws I–III present (Fig. 4A–D, empty indented arrowhead and filled indented arrowhead).
Bucco-pharyngeal apparatus of the Macrobiotus type (Fig. 5A–C), with ventral lamina and ten peribuccal lamellae (Fig. 6A). Mouth antero-ventral. The OCA is composed of three bands of teeth (similar on dorsal and ventral sides) (Figs. 5D–E and 6A–C). The first band of teeth consists of small cones (granules in PCM) situated at the anterior portion of the oral cavity, and just behind the base of the peribuccal lamellae (4–5 rows) (Figs. 5D, 6B, filled arrow). The second band of teeth positioned in the rear of the oral cavity between the ring fold and the third band of teeth (Figs. 5D, 6B, empty arrow) and composed of larger cones (small ridges parallel to the main axis of the buccal tube in PCM), arranged in one row that runs around the oral cavity wall (Figs. 5D–E and 6B, filled unindented arrowhead). The third band of teeth positioned just before the buccal tube opening and composed of dorsal and ventral portion (Figs. 5D–E and 6B–C). The dorsal portion of the third band comprises three, distinctly separated, long and thin ridges (Fig. 5D and 6C). Similarlty, the ventral portion is composed of three distinct teeth with two ventro-lateral onces in shape of ridges and one medio-ventral tooth being often divided into 2–3 smaller granular teeth (Fig. 5E). Additional teeth absent (Figs. 5D–E and 6A–C). Pharyngeal bulb spherical, with triangular apophyses and three rod-shaped macroplacoids. Macroplacoid length sequence 2 < 1 < 3 (Fig. 5A–C). The first macroplacoid without constrictions, but distinctly narrower anteriorly. The second macroplacoid of uniform width and without constrictions. The third macroplacoid with a sub-terminal constriction (Fig. 5A–B; empty unindented arrowhead). Microplacoid present, triangular in shape (Fig. 5A–B).
Paramacrobiotus gadabouti sp. nov.: (A)—mouth with ten peribuccal lamellae (paratype, SEM); (B)—oral cavity armature with first and second band of teeth (paratype, SEM); (C)—oral cavity armature with third band of teeth (paratype, SEM) from dorsal side. Filled arrow represents first band of teeth, empty arrow represents second band of teeth and filled unindented arrowhead represents third band of teeth. Scale bars in µm.
Eggs: Laid freely, white, spherical exhibiting ornamentations of the richtersi type (Fig. 7A–B). Processes in the shape of rounded or truncated cones (Fig. 7A–F). Top endings of the processes with cap like structures (well visible in PCM in the process midsection and always well visible in SEM) (Fig. 7D–F). The surface of cap-like structrues is mostly rough with small granules and wrinkles that can be visible on its surface but only in SEM (Fig. 7D, F). Labyrinthine layer between process walls visible under PCM as a clear reticular pattern (Fig. 7C). Reticular pattern composed of regular and elongated mesh with straight or slightly sinuous margins. Egg shells areolated with a single ring of 10–12 areolae around each process (Fig. 7C–D). Internal surface of areolae clearly sculptured in PCM and pores that are visible only in SEM (Fig. 7C–D).
Reproduction: In the experimental setting with isolated individuals of the new species, eggs laying was observed in all matured animals. These eggs hatched into juveniles. Thus, we conclude the reproduction in Pam gadabouti sp. nov. to be parthenogenetic.
Type Locality: Portugal, Madeira Island, 32°44′36.7"N, 16°54′28.0"W, 647 m asl, Ribeiro Frio, moss from rock and rock wall, 23 Sptember 2019, coll. Łukasz Sługocki, Ricardo Araújo and J. J. Gonçalves Silva.
Additional Localities: (1) Portugal, Madeira Island, 32°49′06″N, 16°59′19″W, 299 m asl, Ponta Delgada, moss from rock, 21 February 2018, coll. Łukasz Michalczyk; (2) Australia, Western Australia State, 31°57′16″S, 115°50′40″E, 56 m asl, Perth, Kings Park, moss from tree, 22 March 2015, coll. Łukasz Michalczyk; (3) France, Île-de-France Region, 48°51′35.5″N, 2°23′40″E, 80 m asl, Paris, Père Lachaise Cemetery, moss from grave, 23 May 2016, coll. Witold Morek; (4) Tunisia, Beni M'tir, Jendouba Governorate, 36°73′92″N, 8°72′99″E, 516 m asl, moss from soil in a forest, 12 June 2015, coll. Jamila Ben Marnissi. All these additional localities have been previously reported in Stec et al.15.
Etymology: The name ‘gadabouti’ refers to the new species ubiquity; from Eng. ‘gadabout’: someone who restlessly moves from place to place seeking amusement or the companionship of others.
Type depositories: Holotype (M50/4 (+ 6 paratypes (3 animals + 2 exuvium + 1 simplex) on the same slide)) and 97 paratypes (slides: M50/*, where the asterisk can be substituted by any of the following numbers: 1–3, 5–20, 1/S, 2/S, 3/S and 4/S) were deposited at the Department of Animal Taxonomy and Ecology, Institute of Environmental Biology, Adam Mickiewicz University in Poznań, Uniwersytetu Poznańskiego 6, 61–614 Poznań, Poland and four paratypes (slides: M50/7 and M50/13 (3 animals and 1 egg)) were deposited at Institute of Systematics and Evolution of Animals, Polish Academy of Sciences, Sławkowska 17, 31–016, Kraków, Poland.
Discussion
Differential diagnosis of the new species
Paramacrobiotus gadabouti sp. nov. by having a microplacoid in the pharynx and eggs ornamentation of the richtersi type with egg processes ended with cap-like structures is similar to Pam. alekseevi82 (Tumanov 2005), Pam. filipi57 Dudziak, Stec and Michalczyk 2020 and Pam. garynahi78 (Kaczmarek, Michalczyk and Diduszko 2005). The new species differs specifically from:
-
1
Paramacrobiotus alekseevi, known only from type locality in Thailand82, by: the presence of pores inside egg areoles, a higher pt value of second macroplacoid (15.4–19.8 in the new species vs 9.8–14.5 in Pam. alekseevi) and a longer microplacoid (3.3–5.1 μm [pt = 6.7–9.3] in the new species vs 1.9–3.0 μm [pt = 4.0–6.2] in Pam. alekseevi). Remarks: The comparison was made based on the Pam. alekseevi original descripition77 as well as the amended description by Stec et al.71.
-
2
Paramacrobiotus filipi, known only from type locality in Malaysian part of Borneo57, by: the absence of dorsal cuticle granulation, a longer second macroplacoid (7.0–11.3 μm [pt = 15.4–19.8] in the new species vs 2.4–6.2 μm [pt = 8.0–13.8] in Pam. filipi), a higher pt value of macroplacoid row (60.7–69.8 in the new species vs 44.4–58.6 in Pam. filipi), a longer placoid row (34.9–53.6 μm [pt = 77.7–87.4] in the new species vs 17.4–34.5 μm [pt = 52.9–73.6] in Pam. filipi) and a larger full egg diameter (104.8–125.3 μm in the new species vs 99.0–104.5 μm in Pam. filipi).
-
3
Paramacrobiotus garynahi, known only from type locality in Baikal Region (Russia)78, by: medioventral tooth in the third band of teeth in the oral cavity divided, eggs chorion ornamentation of the richtersi type, i.e. areoles with pores inside (areolatus type with areoles wrinkled inside in Pam. garynahi), a higher pt value of macroplacoid and placoid rows (60.7–69.8 and 77.7–87.4, respectively, in the new species vs 44.4–56.9 and 55.3–70.3, respectively, in Pam. garynahi) and a smaller eggs bare and full diameter (64.3–91.7 μm and 104.8–125.3 μm, respectively, in new thespecies vs 96.0–132.0 μm and 142.0–180.0 μm, respectively, in Pam. garynahi).
Diversity and distribution of Paramacrobiotus taxa
Studies on the genus Paramacrobiotus become easier due to several revisions and redescriptions of Paramacrobiotus species which have been recently published (e.g.12,14,15). The barrier for tardigrade diversity studies is currently being broken down especially by an integrative approach implemented into taxonomic and faunistic research. The tight link between genetic data and the exact specimen/species name and its morphology provided by authors of integrative studies is and will be crucial to understand species diversity in the genus Paramacrobiotus. Similarly, to Stec et al.15, our molecular analyzes showed 9–10 taxa without a certain assignment to any nominal Paramacrobiotus species. They may constitute already known species that were described in the past based on morphology only and for which genetic data are lacking or they constitute new for science species avaiting their formal descriptions. Although the results indicate considerable diversity that is still hidden within the genus, it should be also noted that in our study more putative species were delimited by tree-based methods compared with distance-based methods. However, this finding is in line with recent studies on tardigrades, but also studies on other animal groups85,86,87. Based on the research which examined numerous Paramacrobiotus populations14,15, we can notice that many species in this genus (especially in the Pam. richtersi group) are extremely similar to each other often exhibiting a considerable intraspecific variation in egg chorion morphology. This makes many of these taxa as sutiable candidates to be considered as cryptic or pseudocryptic species14,15. Therefore, it seems very likely that future taxonomic studies on the genus Paramacrobiotus would be able to formally name many newly discovered evolutionary lineages only by rigorous tests of distinct species hypotheses with integrative methods.
Over the years, species of Paramacrobiotus have been recorded in various geographic regions. Nominal species of the genus have been found in six continents (Table 4). Additionally, there are many unconfirmed taxa from Pam. richtersi and Pam. areolatus group which are known from numerous localities around the world (see e.g.14,15,26,88,89,90,91,92). Importantly, verification of these records is now extremely hard and, in many cases, not possible because of the lack of genetic data and original material. The majority of the Paramacrobiotus species are known only from their type localities or from very restricted geographic ranges. However, some of them are reported from slightly wider geographical areas, like: Pam. danielae from Ecuador and Peru, and Pam. sklodowskae from Cyprus and Tunisia. There are also much wider distributed species, like, for example Pam. centesimus, known from Brazil and Ecuador, Pam. gerlachae from Costa Rica and the Seychelles, Pam. tonolli known from Canada and many states in USA or Pam. vanescens reported from the Democratic Republic of the Congo, the Republic of Guinea-Bissau, the Republic of Zambia and Tanzania15,88,89,90,91. However, the most widely distributed species in the genus Paramacrobiotus, which should be considered as truly cosmopolitan, is the parthenogenetic Pam. fairbanksi reported alredy from Antarctica, Italy, Poland, Spain and USA (Alaska)28. Furthermore, parthenogenetic Pam. gadabouti sp. nov. described here has been confirmed in our study to be present in Australia, France, Portugal and Tunisia (see Figs. 1 and 2). This, all together with Pam. fairbanksi, corroborate that at least some tardigrade species conform to “everything is everywhere” hypothesis. In contrast, other species from the Pam. richtersi group which are bisexual, in most cases the range seems to be limited and restricted e.g. Pam. experimentalis reported only from Madagascar, Pam. metropolitanus from Japan, Pam. celsus, Pam. depressus and Pam. spatialis reported only from Italian locations, but type species of the genus Pam. richtersi is reported from Ireland and Finland14,15,20,56,59. Importantly when comparing haplotype networks presented for Paramacrobiotus taxa in Guidetti et al.14 and haplotype network provided in our study (Fig. 2) one may see that the divergence between haplotypes in bisexual species (Pam. richtersi, Pam. celsus, Pam. arduus, Pam. depressus and Pam. spatialis) seems to be higher than divergence between haplotypes in parthenoegenetic species (Pam. fairbanksi, Pam. gadabouti). Unfortunately, it is premature to conclude if this result could be considered an actual biological pattern or if it simply reflects biases in the analysed data sets, that might be potentially caused by not very large number of sequences analysed per each studied species/population.
Paramacrobiotus gadabouti sp. nov. is the fourth tardigrade species known from more than one zoogeographic realm and the third known from both the Palaearctic and the Australasian realms. The first two being Echiniscus testudo83 (Doyère 1840) and Milnesium inceptum113 Morek, Suzuki, Schill, Georgiev, Yankova, Marley and Michalczyk 2019. This is all in line with the recent study on global distribution of the Minesium populations which demonstrated that most of the species have limited distribution; however, some others can be considered cosmopolitan114. These examples also confirm the hypothesis presented by Guidetti et al.14 that parthenogenetic tardigrades should have a wider distribution due to the adventage in inhabiting new places caused by asexual reproduction. On the other hand, it must be noted that most records of these four disuscussed parthenogenetic species (Ech. testudo, Mil. inceptum, Pam. fairbanksi, Pam. gadabouti sp. nov.) come from highly populated and often touristic places. Therefore, it is also likely that their wide distribution range was additionally enhanced by human-mediated dispersion15 or other vectors such as wind, mammals, birds and animals as evidence has been brought to light regarding the dispersal of tardigrades via these various other vectors115,116,117.
Data availability
The datasets generated and/or analysed during the current study are available in the GenBank repository, ACCESSION NUMBER OP394113–OP394114, OP394209–OP394212. The data of all sequences are available for public access.
References
Guidetti, R. & Bertolani, R. B. Tardigrade taxonomy: An updated check list of the taxa and a list of characters for their identification. Zootaxa 845, 1–46. https://doi.org/10.11646/zootaxa.845.1.1 (2005).
Degma, P. & Guidetti, R. Notes to the current checklist of Tardigrada. Zootaxa 1579, 41–53. https://doi.org/10.11646/zootaxa.1579.1.2 (2007)
Vicente, F. & Bertolani, R. Considerations on the taxonomy of the phylum Tardigrada. Zootaxa 3626, 245–248. https://doi.org/10.11646/zootaxa.3626.2.2 (2013).
Degma, P. & Guidetti, R. Actual checklist of Tardigrada species. (Version 41: Edition: 16-05-2022). (2009–2022).
Ramazzotti, G. & Maucci, W. Il phylum Tardigrada. III edizione riveduta e aggiornata. Mem. Ist. Ital. Idrobiol. 41, 1–1012 (1983).
Beasley, C. W. The phylum Tardigrada. in English Translation P. 3rd edn (eds Ramazzotti, G. & Maucci, W.) 1–1014 (Abilene, USA, 1995).
Nelson, D. R., Guidetti, R., Rebecchi, L., Kaczmarek, Ł. & McInnes, S. Phylum Tardigrada. in Thorp and Covich’s Freshwater Invertebrates 505–522 (Elsevier, 2020). https://doi.org/10.1016/B978-0-12-804225-0.00015-0.
Da Cunha, A. X. & do Nascimento-Ribeiro, F. A fauna de Tardígrados da Ilha da Madeira. Mem. Estud. Mus. Zool. Univ. Coimbra 1–24 (1962).
Fontoura, P., Pilato, G. & Lisi, O. Tardigrada from Santo Antão Island (Archipelago of Cape Verde, West Africa) with the description of a new species. Zootaxa 2838, 30–40. https://doi.org/10.11646/zootaxa.2838.1.2 (2011).
Gąsiorek, P., Vončina, K. & Michalczyk, Ł. Echiniscus testudo (Doyère, 1840) in New Zealand: Anthropogenic dispersal or evidence for the ‘Everything is Everywhere’ hypothesis?. N. Z. J. Zool. 46, 174–181. https://doi.org/10.1080/03014223.2018.1503607 (2019).
Guidetti, R., Schill, R. O., Bertolani, R., Dandekar, T. & Wolf, M. New molecular data for tardigrade phylogeny, with the erection of Paramacrobiotus gen. nov. J. Zool. Syst. Evol. 47, 315–321. https://doi.org/10.1111/j.1439-0469.2009.00526.x (2009).
Kaczmarek, Ł, Gawlak, M., Bartels, P. J., Nelson, D. R. & Roszkowska, M. Revision of the genus Paramacrobiotus Guidetti et al., 2009 with the description of a new species, re-descriptions and a key. Ann. Zool. 67, 627–656. https://doi.org/10.3161/00034541ANZ2017.67.4.001 (2017).
Marley, N. J. et al. A clarification for the subgenera of Paramacrobiotus Guidetti, Schill, Bertolani, Dandekar and Wolf, 2009, with respect to the International Code of Zoological Nomenclature. Zootaxa 4407, 130–134. https://doi.org/10.11646/zootaxa.4407.1.9 (2018).
Guidetti, R., Cesari, M., Bertolani, R., Altiero, T. & Rebecchi, L. High diversity in species, reproductive modes and distribution within the Paramacrobiotus richtersi complex (Eutardigrada, Macrobiotidae). Zool. Lett. 5, 1–28. https://doi.org/10.1186/s40851-018-0113-z (2019).
Stec, D., Krzywański, Ł, Zawierucha, K. & Michalczyk, Ł. Untangling systematics of the Paramacrobiotus areolatus species complex by an integrative redescription of the nominal species for the group, with multilocus phylogeny and species delineation in the genus Paramacrobiotus. Zool. J. Linn. Soc. 188, 694–716. https://doi.org/10.1093/zoolinnean/zlz163 (2020).
Murray, J. Scottish Tardigrada, a review of our present knowledge. Ann. Scot. Nat. Hist. 78, 88–95 (1911).
Murray, J. XXV.—Arctic Tardigrada, collected by Wm. S. Bruce. Trans. R. Soc. Edinb. 45, 669–681 (1907).
Ramazzotti, G. Tre nouve specie di Tardigradi ed altre specie poco comuni. Atti Soc. Nat. Milano 10, 284–291 (1956).
Schill, R. O., Förster, F., Dandekar, T. & Wolf, M. Using compensatory base change analysis of internal transcribed spacer 2 secondary structures to identify three new species in Paramacrobiotus (Tardigrada). Org. Divers. Evol. 10, 287–296. https://doi.org/10.1007/s13127-010-0025-z (2010).
Kaczmarek, Ł et al. Integrative description of bisexual Paramacrobiotus experimentalis sp. Nov. (Macrobiotidae) from republic of Madagascar (Africa) with microbiome analysis. Mol. Phylogenet. Evol. 145, 106730. https://doi.org/10.1016/j.ympev.2019.106730 (2020).
Bertolani, R. Partenogenesi geografica triploide in un Tardigrado (Macrobiotus richtersi). Rend. Acc. Naz. Lincei. Ser. 8, 487–489 (1971).
Bertolani, R. Sex ratio and geographic parthenogenesis in Macrobioutus (Tardigrada). Experientia 28, 94–95. https://doi.org/10.1007/BF01928285 (1972).
Bertolani, R. L. partenogenesi nei Tardigradi. Boll. Zool. 39, 577–581. https://doi.org/10.1080/11250007209431414 (1972).
Bertolani, R. Cytology and Reproductive Mechanisms in Tardigrades. I. 93–114 (East Tennesse State University Press, Johnson City, 1982).
Lemloh, M., Brümmer, F. & Schill, R. O. Life-history traits of the bisexual tardigrades Paramacrobiotus tonollii and Macrobiotus sapiens. J. Zool. Syst. Evol. Res. 49, 58–61. https://doi.org/10.1111/j.1439-0469.2010.00599.x (2011).
Guil, N. & Giribet, G. A comprehensive molecular phylogeny of tardigrades-adding genes and taxa to a poorly resolved phylum-level phylogeny. Cladistics 28, 21–49. https://doi.org/10.1111/j.1096-0031.2011.00364.x (2012).
Kosztyła, P. et al. Experimental taxonomy confirms the environmental stability of morphometric traits in a taxonomically challenging group of microinvertebrates. Zool. J. Linn. Soc. 178, 765–775. https://doi.org/10.1111/zoj.12409 (2016).
Kaczmarek, Ł et al. New records of Antarctic Tardigrada with comments on iterpopulation variability of the Paramacrobiotus fairbanksi Schill, Förster, Dandekar and Wolf, 2010. Diversity 12, 108. https://doi.org/10.3390/d12030108 (2020).
Stec, D., Vecchi, M., Calhim, S. & Michalczyk, Ł. New multilocus phylogeny reorganises the family Macrobiotidae (Eutardigrada) and unveils complex morphological evolution of the Macrobiotus hufelandi group. Mol. Phylogenet. Evol. 160, 106987. https://doi.org/10.1016/j.ympev.2020.106987 (2021).
Stec, D., Smolak, R., Kaczmarek, Ł & Michalczyk, Ł. An integrative description of Macrobiotus paulinae sp. Nov. (Tardigrada: Eutardigrada: Macrobiotidae: hufelandi group) from Kenya. Zootaxa 4052, 501–526. https://doi.org/10.11646/zootaxa.4052.5.1 (2015).
Bryce, D. On some moss-dwelling Cathypnadae; with descriptions of five new species. Sci. Gossip Lond. 28, 271–275 (1892).
Casquet, J., Thebaud, C. & Gillespie, R. G. Chelex without boiling, a rapid and easy technique to obtain stable amplifiable DNA from small amounts of ethanol-stored spiders. Mol. Ecol. Resour. 12(1), 136–141. https://doi.org/10.1111/j.1755-0998.2011.03073.x (2012).
Stec, D., Kristensen, R. M. & Michalczyk, Ł. An integrative description of Minibiotus ioculator sp. nov. from the Republic of South Africa with notes on Minibiotus pentannulatus Londoño et al., 2017 (Tardigrada: Macrobiotidae). Zool. Anz. 286, 117–134. https://doi.org/10.1016/j.jcz.2020.03.007 (2020).
Stec, D., Zawierucha, K. & Michalczyk, Ł. An integrative description of Ramazzottius subanomalus (Biserov, 1985 (Tardigrada) from Poland. Zootaxa 4300, 403–420. https://doi.org/10.11646/zootaxa.4300.3.4 (2017).
Mironov, S. V., Dabert, J. & Dabert, M. A new feather mite species of the genus Proctophyllodes Robin, 1877 (Astigmata: Proctophyllodidae) from the Long-tailed Tit Aegithalos caudatus (Passeriformes: Aegithalidae)—Morphological description with DNA barcode data. Zootaxa 3253, 54–61. https://doi.org/10.11646/zootaxa.3253.1.2 (2012).
White, T. J., Bruns, T., Lee, S. & Taylor, J. PCR Protocols: A Guide to Methods and Application 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1 (Academic Press, 1990).
Folmer, O., Black, M., Hoeh, W., Lutz, R. & Vrijenhoek, R. Phylogenetic uncertainty. Mol. Mar. Biol. Biotechnol. 3, 294–299 (1994).
Vecchi, M. & Stec, D. Integrative descriptions of two new Macrobiotus species (Tardigrada, Eutardigrada, Macrobiotidae) from Mississippi (USA) and Crete (Greece). ZSE 97, 281–306. https://doi.org/10.3897/zse.97.65280 (2021).
Thulin, G. Über die phylogenie und das system der. Hereditas 11, 207–266. https://doi.org/10.1111/j.1601-5223.1928.tb02488.x (1928).
Stec, D. Mesobiotus datanlanicus sp. nov., a new tardigrade species (Macrobiotidae: Mesobiotus harmsworthi group) from Lâm Đồng Province in Vietnam. Zootaxa 4679, 164–180. https://doi.org/10.11646/zootaxa.4679.1.10 (2019).
Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. NAR 30, 3059–3066. https://doi.org/10.1093/nar/gkf436 (2002).
Katoh, K. & Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298. https://doi.org/10.1093/bib/bbn013 (2008).
Vaidya, G., Lohman, D. J. & Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. https://doi.org/10.1111/j.1096-0031.2010.00329.x (2011).
Lanfear, R., Calcott, B., Ho, S. Y. & Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29(6), 1695–1701. https://doi.org/10.1093/molbev/mss020 (2012).
Xia, X., Xie, Z., Salemi, M., Chen, L. & Wang, Y. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 26, 1–7. https://doi.org/10.1016/S1055-7903(02)00326-3 (2003).
Xia, X. & Lemey, P. Assessing substitution saturation with DAMBE. In The Phylogenetic Handbook (eds Lemey, P. et al.) 615–630. https://doi.org/10.1017/CBO9780511819049.022 (Cambridge University Press, 2009).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. https://doi.org/10.1093/bioinformatics/btg180 (2003).
Rambaut, A., Suchard, M. A., Xie, D. & Drummond, A. J. Tracer v1. 6. 2014. (2015).
Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. https://doi.org/10.1093/bioinformatics/btu033 (2014).
Bandelt, H., Forster, P. & Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036 (1999).
Ehrenberg, C. G. Beitrag zur Bestimmung des Stationären Mikroskopischen Lebens in bis 20,000 Fuss Alpenhöhe. (1859).
Guil, N. & Guidetti, R. A new species of Tardigrada (Eutardigrada: Macrobiotidae) from Iberian Peninsula and Canary Islands (Spain). Zootaxa 889, 1–11. https://doi.org/10.11646/zootaxa.889.1.1 (2005).
Plate, L. H. Beiträge zur Naturgeschichte der Tardigraden. Zool. Jahrb. Abteilung Anat. Ontog. Tiere 3, 487–550. https://doi.org/10.5962/bhl.part.1265 (1889).
Kaczmarek, Ł, Kayastha, P., Roszkowska, M., Gawlak, M. & Mioduchowska, M. Integrative redescription of the Minibiotus intermedius (Plate, 1888)—The type species of the genus Minibiotus R.O. Schuster, 1980. Diversity 14, 356. https://doi.org/10.3390/d14050356 (2022).
Londoño, R., Daza, A., Lisi, O. & Quiroga, S. New species of waterbear Minibiotus pentannulatus (Tardigrada: Macrobiotidae) from Colombia. Rev. Mex. Biodivers. 88, 807–814. https://doi.org/10.1016/j.rmb.2017.10.040 (2017).
Vecchi, M. et al. Macrobiotus naginae sp. nov., a new Xerophilous Tardigrade species from Rokua Sand Dunes (Finland). Zool. Stud. 61, e22 (2022).
Stec, D., Dudziak, M. & Michalczyk, Ł. Integrative descriptions of two new Macrobiotidae species (Tardigrada: Eutardigrada: Macrobiotoidea) from French Guiana and Malaysian Borneo. Zool. Stud. 59, e23 (2020).
Stec, D., Roszkowska, M., Kaczmarek, Ł & Michalczyk, Ł. Paramacrobiotus lachowskae, a new species of Tardigrada from Colombia (Eutardigrada: Parachela: Macrobiotidae). N. Z. J. Zool. 45, 43–60. https://doi.org/10.1080/03014223.2017.1354896 (2018).
Sugiura, K., Matsumoto, M. & Kunieda, T. Description of a model tardigrade Paramacrobiotus metropolitanus sp. nov. (Eutardigrada) from Japan with a summary of its life history, reproduction and genomics. Zootaxa 5134, 92–112. https://doi.org/10.11646/zootaxa.5134.1.4 (2022).
Tumanov, D. V. Three new species of Macrobiotus (Eutardigrada, Macrobiotidae, tenuis-group) from Tien Shan (Kirghizia) and Spitsbergen. J. Limnol. 66, 40. https://doi.org/10.4081/jlimnol.2007.s1.40 (2007).
Zawierucha, K., Kolicka, M. & Kaczmarek, Ł. Re-description of the Arctic tardigrade Tenuibiotus voronkovi (Tumanov, 2007 (Eutardigrada; Macrobiotidea), with the first molecular data for the genus. Zootaxa 4196, 498. https://doi.org/10.11646/zootaxa.4196.4.2 (2016).
Stec, D., Tumanov, D. T. & Kristensen, R. M. Integrative taxonomy identifies two new tardigrade species (Eutardigrada: Macrobiotidae) from Greenland. EJT 614, 1–40. https://doi.org/10.5852/ejt.2020.614 (2020).
Fontaneto, D., Flot, J.-F. & Tang, C. Q. Guidelines for DNA taxonomy, with a focus on the meiofauna. Mar. Biodiv. 45, 433–451. https://doi.org/10.1007/s12526-015-0319-7 (2015).
Zhang, J., Kapli, P., Pavlidis, P. & Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29, 2869–2876. https://doi.org/10.1093/bioinformatics/btt499 (2013).
Puillandre, N., Brouillet, S. & Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 21, 609–620. https://doi.org/10.1111/1755-0998.13281 (2021).
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T. & Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34(3), 772–773. https://doi.org/10.1093/molbev/msw260 (2017).
Roszkowska, M., Stec, D., Gawlak, M. & Kaczmarek, Ł. An integrative description of a new tardigrade species Mesobiotus romani sp. nov. (Macrobiotidae: harmsworthi group) from the Ecuadorian Pacific coast. Zootaxa 4450, 550–564. https://doi.org/10.11646/zootaxa.4450.5.2 (2018).
Pilato, G. & Binda, M. G. Definition of families, subfamilies, genera and subgenera of the Eutardigrada, and keys to their identification. Zootaxa 2404, 1–54. https://doi.org/10.11646/zootaxa.2404.1.1 (2010).
Kaczmarek, Ł & Michalczyk, Ł. The Macrobiotus hufelandi group (Tardigrada) revisited. Zootaxa 4363, 101–123. https://doi.org/10.11646/zootaxa.4363.1.4 (2017).
Michalczyk, Ł & Kaczmarek, Ł. A description of the new tardigrade Macrobiotus reinhardti (Eutardigrada: Macrobiotidae, harmsworthi group) with some remarks on the oral cavity armature within the genus Macrobiotus Schultze. Zootaxa 331, 1–24. https://doi.org/10.11646/zootaxa.331.1.1 (2003).
Kaczmarek, Ł, Cytan, J., Zawierucha, K., Diduszko, D. & Michalczyk, Ł. Tardigrades from Peru (South America), with descriptions of three new species of Parachela. Zootaxa 3790, 357–379. https://doi.org/10.11646/zootaxa.3790.2.5 (2014).
Kiosya, Y., Pogwizd, J., Matsko, Y., Vecchi, M. & Stec, D. Phylogenetic position of two Macrobiotus species with a revisional note on Macrobiotus sottilei Pilato, Kiosya, Lisi & Sabella, 2012 (Tardigrada: Eutardigrada: Macrobiotidae). Zootaxa 4933, 113–135. https://doi.org/10.11646/zootaxa.4933.1.5 (2021).
Pilato, G. Analisi di nuovi caratteri nello studio degli Eutardigradi. Animalia 8, 51–57 (1981).
Michalczyk, Ł & Kaczmarek, Ł. The Tardigrada Register: a comprehensive online data repository for tardigrade taxonomy. J. Limnol. 72, e22. https://doi.org/10.4081/jlimnol.2013.s1.e22 (2013).
Bertolani, R. et al. Phylogeny of Eutardigrada: New molecular data and their morphological support lead to the identification of new evolutionary lineages. Mol. Phylogenet. Evol. 76, 110–126. https://doi.org/10.1016/j.ympev.2014.03.006 (2014).
Perry, E., Miller, W. R. & Kaczmarek, Ł. Recommended abbreviations for the names of genera of the phylum Tardigrada. Zootaxa 4608, 145. https://doi.org/10.11646/zootaxa.4608.1.8 (2019).
Degma, P., Michalczyk, Ł & Kaczmarek, Ł. Macrobiotus derkai, a new species of Tardigrada (Eutardigrada, Macrobiotidae, huziori group) from the Colombian Andes (South America). Zootaxa 1731, 1–23. https://doi.org/10.11646/zootaxa.1731.1.1 (2008).
Kaczmarek, Ł, Michalczyk, Ł & Diduszko, D. Some tardigrades from Siberia (Russia, Baikal region) with a description of Macrobiotus garynahi sp. nov. (Eutardigrada: Macrobiotidae: richtersi group). Zootaxa 1053, 35–45. https://doi.org/10.11646/zootaxa.1053.1.3 (2005).
Michalczyk, Ł & Kaczmarek, Ł. Macrobiotus huziori, a new species of Tardigrada (Eutardigrada: Macrobiotidae) from Costa Rica (Central America). Zootaxa 1169, 47–59. https://doi.org/10.11646/zootaxa.1169.1.3 (2006).
Michalczyk, L. & Kaczmarek, L. A new species Macrobiotus magdalenae (Tardigrada: Eutardigrada: Macrobiotidae, richtersi group) from Costa Rican rain forest (Central America). N. Z. J. Zool. 33, 189–196. https://doi.org/10.1080/03014223.2006.9518444 (2006).
Michalczyk, Ł, Kaczmarek, Ł & Węglarska, B. Macrobiotus sklodowskae sp. nov. (Tardigrada: Eutardigrada: Macrobiotidae, richtersi group) from Cyprus. Zootaxa 1371, 45–56. https://doi.org/10.11646/zootaxa.1371.1.4 (2006).
Tumanov, D. V. Notes on the Tardigrada of Thailand, with a description of Macrobiotus alekseevi sp. nov. (Eutardigrada, Macrobiotidae). Zootaxa 999, 1–6. https://doi.org/10.11646/zootaxa.999.1.1 (2005).
Doyère, M. Memoire sur les tardigrades. Ann. Sci. Nat Zool. Ser. 2, 269–362 (1840).
Richters, F. Tardigrada. In Handbuch der Zoologie Vol. 3 (eds Kükenthal, W. & Krumbach, T.) 58–61 (Walter de Gruyter & Co., Berlin and Leipzig, 1926).
Stec, D., Cancellario, T. & Fontaneto, D. Diversification rates in Tardigrada indicate a decreasing tempo of lineage splitting regardless of reproductive mode. Org. Divers. Evol. 22(4), 965–974. https://doi.org/10.1007/s13127-022-00578-4 (2022).
Dellicour, S. & Flot, J.-F. The hitchhiker’s guide to single-locus species delimitation. Mol. Ecol. Resour. 18, 1234–1246. https://doi.org/10.1111/1755-0998.12908 (2018).
Magoga, G., Fontaneto, D. & Montagna, M. Factors affecting the efficiency of molecular species delimitation in a species-rich insect family. Mol. Ecol. Resour. 21, 1475–1489. https://doi.org/10.1111/1755-0998.13352 (2021).
Kaczmarek, Ł, Michalczyk, Ł & McInnes, S. J. Annotated zoogeography of non-marine Tardigrada. Part I: Central America. Zootaxa 3763, 1–62. https://doi.org/10.11646/zootaxa.3763.1.1 (2014).
Kaczmarek, Ł, Michalczyk, Ł & McInnes, S. J. Annotated zoogeography of non-marine Tardigrada. Part II: South America. Zootaxa 3923, 1–107. https://doi.org/10.11646/zootaxa.3923.1.1 (2015).
Kaczmarek, Ł, Michalczyk, Ł & Mcinnes, S. J. Annotated zoogeography of non-marine Tardigrada. Part III: North America and Greenland. Zootaxa 4203, 1–249. https://doi.org/10.11646/zootaxa.4203.1.1 (2016).
Mcinnes, S. J., Michalczyk, Ł & Kaczmarek, Ł. Annotated zoogeography of non-marine Tardigrada. Part IV: Africa. Zootaxa 4284, 1. https://doi.org/10.11646/zootaxa.4284.1.1 (2017).
Michalczyk, Ł, Kaczmarek, Ł & Mcinnes, S. J. Annotated zoogeography of non-marine Tardigrada. Part V: Australasia. Zootaxa 5107, 1–119. https://doi.org/10.11646/zootaxa.5107.1.1 (2022).
Pilato, G., Claxton, S. & Binda, M. G. Tardigrades from Australia. III. Echiniscus marcusi and Macrobiotus peteri, new species of tardigrades from New South Wales. Animalia 16, 43–48 (1989).
Pilato, G., Binda, M. G. & Lisi, O. Eutardigrada from New Zealand, with descriptions of two new species. N. Z. J. Zool. 33, 49–63. https://doi.org/10.1080/03014223.2006.9518430 (2006).
Bartels, P. J., Pilato, G., Lisi, O. & Nelson, D. R. Macrobiotus (Eutardigrada, Macrobiotidae) from the Great Smoky Mountains National Park, Tennessee/North Carolina, USA (North America): Two new species and six new records. Zootaxa 2022, 45–57. https://doi.org/10.11646/zootaxa.2022.1.4 (2009).
Binda, M. G., Pilato, G., Moncada, E. & Napolitano, A. Some tardigrades from Central Africa with the description of two new species: Macrobiotus ragonesei and M. priviterae (Eutardigrada Macrobiotidae). Trop. Zool. 14, 233–242. https://doi.org/10.1080/03946975.2001.10531155 (2001).
Pilato, G., Binda, M. G. & Lissi, O. Notes on tardigrades of the Seychelles with the description of two new species. Ital. J. Zool. 71, 171–178 (2004).
Pilato, G., Binda, M. G. & Lisi, O. Three new species of eutardigrades from the Seychelles. N. Z. J. Zool. 33, 39–48. https://doi.org/10.1080/03014223.2006.9518429 (2006).
Pilato, G., Binda, M. G. & Lisi, O. Notes on tardigrades of the Seychelles with the description of three new species. Ital. J. Zool. 71, 171–178. https://doi.org/10.1080/11250000409356569 (2004).
Pilato, G., Binda, M. G. & Catanzaro, R. Remarks on some tardigrades of the African fauna with the description of three new species of Macrobiotus Schultze 1834. Trop. Zool. 4, 167–178. https://doi.org/10.1080/03946975.1991.10539487 (1991).
Maucci, W. & Durante Pasa, M. V. Tardigradi muscicoli delle Isole Andamane. Boll. Mus. Civ. St. Nat. Verona 7, 281–291 (1980).
Iharos, G. Neuere Daten zur Kenntnis der Tardigraden-Fauna von Neuguinea. Opusc. Zool. Bp. 11, 65–73 (1973).
Binda, M. G. & Pilato, G. Macrobiotus savai and Macrobiotus humilis, two new species of tardigrades from Sri Lanka. Boll. Accad. Gioenia Sci. Nat. Catania 34, 101–111 (2001).
Pilato, G. Macrobiotus centesimus, new species of eutardigrade from the South America. Boll. Accad. Gioenia Sci. Nat. Catania 33, 97–101 (2000).
Daza, A., Caicedo, M., Lisi, O. & Quiroga, S. New records of tardigrades from Colombia with the description of Paramacrobiotus sagani sp. nov. and Doryphoribius rosanae sp. nov. Zootaxa 4362, 29–50. https://doi.org/10.11646/zootaxa.4362.1.2 (2017).
Claps, M. C. & Rossi, G. C. Tardígrados de Uruguay, con descripción de dos nuevas especies (Echiniscidae, Macrobiotidae). Iheringia Sér. Zool. 83, 17–22 (1997).
Iharos, G. Neue tardigraden-arten aus ungarn (neuere beitrage zur kenntnis der tardigraden-fauna ungarns. 6.). Acta Zool. Acad. Sci. Hung. 12(1–2), 111 (1966).
Pilato, G., Kiosya, Y., Lisi, O. & Sabella, G. New records of Eutardigrada from Belarus with the description of three new species. Zootaxa 3179, 39–60. https://doi.org/10.11646/zootaxa.3179.1.2 (2012).
Pasa, D. & Maucci, W. Moss Tardigrada from the Scandinavian Peninsula. in Second International Symposium on Tardigrada, Vol. 79(25). 47–85 (1979).
Lisi, O., Binda, M. G. & Pilato, G. Eremobiotus ginevrae sp. nov. and Paramacrobiotus pius sp. nov., two new species of Eutardigrada. Zootaxa 4103, 344–360. https://doi.org/10.11646/zootaxa.4103.4.3 (2016).
Biserov, V. I. Macrobiotus lorenae sp. n., a new species of Tardigrada (Eutardigrada Macrobiotidae) from the Russian Far East. Arthr Sel. 5, 145–149 (1996).
Biserov, V. I. Tardigrades of the Caucasus with a taxonomic analysis of genus Ramazzottius. Zool. Anz. 236, 139–159 (1997).
Morek, W. et al. Redescription of Milnesium alpigenum Ehrenberg, 1853 (Tardigrada: Apochela) and a description of Milnesium inceptum sp. nov., a tardigrade laboratory model. Zootaxa 4586(1), 35. https://doi.org/10.11646/zootaxa.4586.1.2 (2019).
Morek, W., Surmacz, B., López-López, A. & Michalczyk, Ł. “Everything is not everywhere”: Time-calibrated phylogeography of the genus Milnesium (Tardigrada). Mol. Ecol. 30, 3590–3609. https://doi.org/10.1111/mec.15951 (2021).
Mogle, M. J., Kimball, S. A., Miller, W. R. & McKown, R. D. Evidence of avian-mediated long-distance dispersal in American tardigrades. PeerJ 6, e5035. https://doi.org/10.7717/peerj.5035 (2018).
Vuori, T., Calhim, S. & Vecchi, M. A lift in snail’s gut provides an efficient colonization route for tardigrades. Ecology 103, e3702. https://doi.org/10.1002/ecy.3702 (2022).
Książkiewicz, Z. & Roszkowska, M. Experimental evidence for snails dispersing tardigrades based on Milnesium inceptum and Cepaea nemoralis species. Sci. Rep. 12(4421), 1–10. https://doi.org/10.1038/s41598-022-08265-2 (2022).
Acknowledgements
Studies have been partially conducted in the framework of activities of BARg (Biodiversity and Astrobiology Research group). The PK is scholarship holder of Passport to the future—Interdisciplinary doctoral studies at the Faculty of Biology, Adam Mickiewicz University, Poznań POWR.03.02.00-00-I006/17 and her study was also supported by Minigrant Edition II, contract number POWER6/2021/2ed, Adam Mickiewicz University, POWR.03.02.00-00- I006/17. The work of MM was supported by National Science Centre, Poland, Grant No. 2021/43/D/NZ8/00344 and Grant No. 1220/146/2021 from the Small Grants Programme of the University of Gdansk (i.e., UGrants-first competition). The authors also wish to thank Cambridge Proofreading LLC (http://proofreading.org) for their linguistic assistance.
Author information
Authors and Affiliations
Contributions
Conceptualization, P.K. and Ł.K.; data curation, P.K.; sample collection, Ł.S.; formal analysis, P.K., D.S., M.M. and Ł.K.; investigation, P.K., D.S., Ł.S., M.M., M.G. and Ł.K.; methodology, P.K., D.S., M.M. and Ł.K.; supervision, Ł.K.; validation, P.K., D.S., M.M. and Ł.K.; visualization, P.K. and M.G.; writing—original draft, P.K., D.S., M.M. and Ł.K.; writing—review and editing, All authors reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kayastha, P., Stec, D., Sługocki, Ł. et al. Integrative taxonomy reveals new, widely distributed tardigrade species of the genus Paramacrobiotus (Eutardigrada: Macrobiotidae). Sci Rep 13, 2196 (2023). https://doi.org/10.1038/s41598-023-28714-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-28714-w
This article is cited by
-
The tardigrade Mesobiotus aradasi (Binda, Pilato & Lisi, 2005) is widely distributed along the Antarctic Peninsula
Polar Biology (2024)
-
Morphological and genetic variability in cosmopolitan tardigrade species—Paramacrobiotus fairbanksi Schill, Förster, Dandekar & Wolf, 2010
Scientific Reports (2023)
-
Intraspecific variation of morphological traits backed up with molecular evidence votes for re-appraisal of hitherto distinguished Balaustium species—a case study of Balaustium murorum (Acariformes: Parasitengona, Erythraeidae)
Experimental and Applied Acarology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.