Abstract
Very long-chain fatty acids (VLCFAs) possess more than twenty carbon atoms and are the major components of seed storage oil, wax, and lipids. FAE (Fatty Acid Elongation) like genes take part in the biosynthesis of VLCFAs, growth regulation, and stress responses, and are further comprised of KCS (Ketoacyl-CoA synthase) and ELO (Elongation Defective Elongase) sub-gene families. The comparative genome-wide analysis and mode of evolution of KCS and ELO gene families have not been investigated in tetraploid Brassica carinata and its diploid progenitors. In this study, 53 KCS genes were identified in B. carinata compared to 32 and 33 KCS genes in B. nigra and B. oleracea respectively, which suggests that polyploidization might has impacted the fatty acid elongation process during Brassica evolution. Polyploidization has also increased the number of ELO genes in B. carinata (17) over its progenitors B. nigra (7) and B. oleracea (6). Based on comparative phylogenetics, KCS, and ELO proteins can be classified into eight and four major groups, respectively. The approximate date of divergence for duplicated KCS and ELO genes varied from 0.03 to 3.20 million years ago (MYA). Gene structure analysis indicated that the maximum number of genes were intron-less and remained conserved during evolution. The neutral type of selection seemed to be predominant in both KCS and ELO genes evolution. String-based protein-protein interaction analysis suggested that bZIP53, a transcription factor might be involved in the activation of transcription of ELO/KCS genes. The presence of biotic and abiotic stress-related cis-regulatory elements in the promoter region suggests that both KCS and ELO genes might also play their role in stress tolerance. The expression analysis of both gene family members reflect their preferential seed-specific expression, especially during the mature embryo development stage. Furthermore, some KCS and ELO genes were found to be specifically expressed under heat stress, phosphorus starvation, and Xanthomonas campestris infection. The current study provides a basis to understand the evolution of both KCS and ELO genes in fatty acid elongation and their role in stress tolerance.
Similar content being viewed by others
Introduction
Brassicaceae is a vast family of plants including 372 genera and 4,006 species contributing to condiments, biofuel, food, oil, and fulfilling fodder demands for the ecosystem1. This group is also known for polyploidy studies due to diploid parents and allotetraploid species2. Over time U-triangle developed on polyploidization of Brassica’s proved its usefulness in studying the evolution of various genes and phenotypes3. The B. carinata (Ethiopian mustard, BBCC, 2n = 4X = 34) is an important species that contains 17 sets of chromosomes with a genome size of 1.087 Gb. It evolved after natural polyploidization between two diploid species B. nigra (BB, 2n = 2X = 16) and B. oleracea (CC, 2n = 2X = 18) about 0.047 MYA4. The B. carinata carries important agronomic and climate resilience characteristics including lodging, drought, and heat tolerance. Moreover, B. carinata has resistance against powdery mildew and white rust diseases highlighting its usefulness as a resistance resource in crop improvement5. Recently B. carinata has gained popularity as biofuel production in the United States and Canada6. However, its parents are valued for vegetables and condiments7. The B. oleracea being closest to Arabidopsis thaliana is considered an important Brassica species to study polyploidy8.
VLCFAs consist of 20 or more carbon atoms and are the key components of the cell membrane and cuticular lipids in plants9. Almost all plant cell types have VLCFAs and play a variety of crucial roles such as cell morphogenesis, energy storage in seeds, and stress responses10,11. Fatty acids of this group can be found abundantly in the form of suberins, sphingolipids, leaf cuticles, pollen epidermis, and cork cells. Brassica and Jojoba (Simmondsia chinensis) plants store them as a rich source of carbon12.
Biosynthesis of VLCFAs takes place in plastids and their elongation occurs in endoplasmic reticulum. During their biosynthesis, four key enzymes including ketoacyl-CoA synthase (KCS), 3-ketoacyl-CoA reductase, 3-hydroxyacyl-CoA dehydratase, and trans-2,3-enoyl CoA reductase performs elongation of fatty acid carbon chains13,14,15,16. There are four basic steps of chain elongation which includes (1) condensation, (2) l-reduction, (3) dehydration, and (4) ll-reduction. Condensation involves a reactant in the form of an acyl chain and in Brassica’s oleic acid reacts with malonate which is activated in the presence of coenzyme A by adding an extra carbon atom to the acyl chain and yielding β-ketoacyl. Later on, it passes through l-reduction, dehydration, and ll-reduction phases to convert activated β-ketoacyl to n + 217. Two types of condensing enzymes are present in plants and they can be categorized into KCS-like and ELO-like gene families12. The basic mechanism of their interaction is still missing. However, comparative genomics and transcriptomics studies can help to understand their putative roles.
KCS gene family can be categorized into eight groups11. In A. thaliana, 21 KCS genes have been identified18 among which KCS18 has been studied against the erucic acid biosynthesis which determines the edible oil quality19. Out of 21 KCS members, eight are characterized as KCS-1, KCS2 (DAISY), KCS-5 (CER60), KCS-6 (CER6, CUT1), KCS-10 (FDH), KCS-13 (HIC), KCS-18, and KCS-19 (FAE1)11. Members of this family play vital roles in the biosynthesis of epidermal wax20,21,22, root suberin biopolymer23, seed stored triglycerides19 and maintain leaf guard cell density as well24. The ELO gene family was primarily identified in yeast (Saccharomyces cerevisiae). The ELO genes synthesize poly-unsaturated VLCFAs and were found only in lower plants25,26,27. In A. thaliana, only four ELO genes have been reported namely At1g75000 (AtELO1), At3g06460 (AtELO2), At3g06470 (AtELO3), and At4g36830 (AtELO4)28. Until now, only AtELO4 has been functionally characterized29.
Here, we have identified 118 KCS and 31 ELO genes in tetraploid B. carinata and its diploid progenitors B. nigra and B. oleracea. By employing various bioinformatics tools, we report their mode of duplication, phylogenetic reconstruction, genes structural organization, protein–protein interactions (PPI), promotor binding elements, and syntenic relationships during evolution. To broaden their perspective roles, we have also performed their expression profiling using various RNA-seq datasets comprising of heat stress, phosphate starvation, X. campestris inoculation, and during seed development and tissue specificity.
Results
Identification of KCS and ELO gene families in B. carinata and its progenitor species
The genomes of allotetraploid B. carinata and its diploid progenitors B. nigra and B. oleracea were searched against the queries of B. napus KCS and ELO proteins. Initially, 70 KCS proteins in B. carinata, 32 in B. nigra, and 33 in B. oleracea were identified. Later, these proteins were thoroughly scanned for the presence of conserved domains in both KCS and ELO proteins. Finally, 32, 33, and 54 un-truncated KCS proteins from B. nigra, B. oleracea, and B. carinata were retained, respectively. The same criteria opted for identification of ELO proteins and 17 of B. carinata, seven of B. nigra, and six of B. oleracea were found to be untruncated proteins. All these identified genes were renamed according to their chromosomal positions. A comparison of gene numbers among allotetraploid B. carinata and its diploid progenitors revealed that evolution has impacted both KCS and ELO proteins. Counting of gene numbers in B. carinata suggested that its genome might have lost 16 KCS and two ELO genes (Table S1).
Gene lengths of KCS ranged from 1.2 to 2.26 kilo base pairs (kbp) compared to ELO, which ranged from 0.57 to 4.39 kbp with amino acid lengths of 359–969 and 218–297, respectively. The maximum number of KCS and ELO proteins were found to be localized in endomembrane system and least number of proteins were predicted to be localized in nucleus and chloroplast (Table S2).
Chromosomal distribution of KCS and ELO genes
In B. nigra, KCS genes were unevenly scattered on assembled chromosomes. Out of total eight chromosomes, first six (chrB01–chrB06) harbored 32 genes, whereas none of the genes were found to be localized on chrB07 and chrB08. The chrB03 contained maximum (9) genes, followed by chrB02 and chrB04 carrying seven and six genes, respectively (Fig. 1). In B. oleracea only six genes were clustered in paired form on chrC04 and chrC09 and the rest of the chromosomes harbored genes in random manner. The chrC05, chrC07, and chrC08 each contained five genes, whereas chrC01 carried only one gene.
In BB genome of B. carinata, chrB01, chrB06, and chrB08 each carried one KCS gene only and none of the KCS genes were mapped on chrB04 (Fig. 1). The chrB03 contained maximum (10) genes and this chromosome also carried maximum KCS genes in B. nigra genome. In the CC genome of B. carinata, chrC03 had maximum (5) genes, whereas chrC09 did not carry any KCS gene.
Interestingly, unlike the KCS genes, ELO genes were present in clusters indicating that these genes might be part of QTLs. Out of total 17 BcaELO genes, 12 were present in four clusters and the remaining five genes could not be localized on assembled chromosomes (Fig. 1). Moreover, in BB genome, ELO genes were mapped only on chrB04, whereas in CC genome these genes were localized on chrC05, chrC08, and chrC09. The B. nigra chromosomes had seven genes. The chromosomes chrB07 and chrB01 carried three and one genes, respectively, while other genes were identified from unsorted contigs, hence, could not be localized on chromosomes. Similarly, in B. oleracea genome, chrC03, chrC05, chrC06, and chrC07 each carried one ELO gene, whereas chrC01 contained two genes (Table S2).
Intro-exon distribution of KCS and ELO genes
Gene structure was conserved throughout both gene families. Out of 118 KCS genes, 68 were intron-less, 32 genes were with a single intron, 13 were with 2 introns and 5 were with more than 2 introns, respectively (Fig. S1). Out of 32 B. nigra genes, 18 were intron less, whereas 11, and 2 genes had 1, and 2 introns, respectively. However, 9 introns were found in BniKCS14 gene. In B. oleracea BolKCS20 gene had maximum 8 introns. Thirty-three genes were intron-less and 7, and 5 genes contained 1, and 2, introns respectively (Fig. S1). Similarly, in B. carinata 75 genes were intron less and 13 genes were with single intron, whereas 2 genes carried 2 introns, and the remaining 3 genes carried more than 2 introns. While maximum (7) introns were identified in BcaKCS48 gene. Among three genomes the B. nigra ELO genes structure remained conserved during evolution. However, 4 genes of B. oleracea and 1 gene of B. carinata were identified to have introns (Fig. S2).
Phylogenetic classification and motif analysis of KCS and ELO proteins in three Brassica spp.
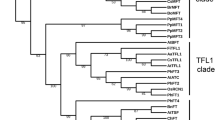
Phylogenetic tree assists in determining the evolution and relationship of genes30. An unrooted comparative phylogenetic tree was constructed by using 50 KCS proteins from B. carinata, 32 from B. nigra, 33 from B. oleracea, 25 from Oryza sativa, and 21 from A. thaliana. Based on phylogenetic clades the KCS proteins grouped into eight groups (α, β, γ, δ, ε, ζ, η, and θ) (Fig. 2a). The θ clade contained maximum (38) members, followed by δ, β, α, γ, ζ, η, and ε which contained 24, 23, 23, 17, 16, 13, and 9 members, respectively (Fig. 2e). The α, β, and θ each had eight KCS orthologous gene pairs, whereas δ and η both contained 5 orthologous gene pairs. The two other remaining groups (ζ and ε) had two and one orthologous gene pairs, respectively. Moreover, constructing phylogeny also assists in assessing gene loss during evolution based on counting members in each clade. It could be inferred that 4 KCS genes from ε, 3 from ζ, and θ each, 2 from β and δ each and 1 from α and η groups each lost during evolution. However, members of γ group remained conserved during evolution.
Phylogenetic classification and motifs distribution among KCS and ELO proteins in B. carinata and its progenitors. Phylogenetic grouping of (a) KCS-like genes and (b) ELO-like genes. Motifs identified among the (c) KCS and (d) ELO proteins, and group-wise distribution of (e) KCS and (f) ELO proteins in the B. carinata, its progenitors, Oryza sativa and A. thaliana, respectively.
Motif analysis was performed through MEME suit server to identify the characteristic regions of KCS and ELO proteins. The 10 and 3 types of motifs were identified in KCS (Fig. 2c) and ELO (Fig. 2d) protein sequences respectively. Interestingly, motifs orientation and numbers were highly conserved throughout the KCS proteins. However, genetic diversification among motifs was observed across members of all eight KCS clades. While seven KCS proteins; 4 from B. oleracea, 2 from B. nigra, and 1 from B. carinata exhibited variable motif structures as compared with other KCS proteins.
Same criteria for the construction of phylogenetic tree of ELO proteins was opted. Seventeen ELO protein sequences from B. carinata, 7 from B. nigra, 6 from B. oleracea, 2 from O. sativa, and 4 from A. thaliana were used for inferring comparative phylogenetic tree (Fig. 2b). The phylogenetic tree classified ELO proteins into four groups (I, II, III, and IV). Group I had 15 members, group II had 11 members, group III had 6, and group IV had 7 ELO protein members (Fig. 2f). Interestingly, group II and IV proteins were only present in Brassica species suggesting these might be Brassica specific groups. Moreover, absence of O. sativa members from these groups indicates that these groups evolved after separation of eudicots from monocots. Similar to KCS proteins, motif structures of ELO proteins were also conserved.
Duplication analysis of KCS and ELO genes
Tandem and segmental types of duplications have their role in gene family’s evolution31. Gene pairs located on same chromosomes having a physical distance of < 50 kbp32 among them are said to be tandemly duplicated, while those pairs localized on different chromosomes are termed as segmentally duplicated33. A total of 6 KCS and 7 ELO paralogous gene pairs were found to be duplicated, of which 2 KCS and 4 ELO pairs were tandemly duplicated, whereas 4 KCS and 3 ELO gene pairs were found to be segmentally duplicated. The segmental duplications among the KCS genes were restricted to α and γ phylogenetic groups. Likewise, segmental duplications among ELO genes were also restricted to II and III phylogenetic groups (Table S1).
Moreover, nucleotide substitutions (Ka and Ks), duplication time, and selection type were also calculated among duplicated gene pairs. The Ka/Ks ratio of all duplicated pairs was < 1 suggesting that these pairs went through purifying selection to maintain their function. Moreover, all the duplicated pairs either tandemly or segmentally duplicated might have duplicated before the polyploidization of allotetraploid B. carinata as indicated from their calculated duplication time. Duplication time of tandemly duplicated pairs ranged from 1.14 to 18.19 million years ago (MYA), whereas duplication time of segmentally duplicated gene pairs was more recent ranging from 0 to 4.49 MYA (Table S1).
Inter chromosomal syntenic relationships of KCS and ELO genes among Brassicas and A. thaliana
The B2 chromosome of B. nigra and B8 of B. carinata were collinear. Chromosomal synteny analysis identified terminal deletions at both arms of B. carinata B2. The orthologs of five B. nigra KCS genes (BniKCS03, BniKCS05, BniKCS06, BniKCS08, and BniKCS09) might have been lost in B. carinata due to this deletion event. While another BniKCS07 gene might be translocated, as its collinear gene was found on chromosome C6 of B. carinata. It could be hypothesized that this translocation might have happened before terminal deletion of B2 chromosome. The C7 of B. oleracea was collinear to C6 of B. carinata and the terminal end of short arm of C6 was showing collinearity with C7 of B. carinata. Interestingly non-collinear chromosomal region of C6 chromosome of B. carinata had three possible translocated ELO genes (BcaELO08, BcaELO10, and BcaELO11) from C9 of B. oleracea. The B5 chromosomes of B. nigra and B. carinata were collinear to each other and deletion in short arm of this chromosome might be responsible for removal of BniKCS25 and BcaELO13 genes, as their orthologs were identified from unassembled contigs in B. carinata. Similar types of possible translocations could also be observed in both KCS and ELO genes (Table S1 and Fig. 3).
The synteny analysis among the AA, BB, and CC genomes of Brassicaceae family. The red bars are depicting BB chromosomes of B. carinata, and B. nigra, the green bars are of A. thaliana (AA), and the blue bars are for CC chromosomes of B. carinata and B. oleracea. The grey strings depict the collinear blocks, red and green lines are indicating orthologues identified in KCS and ELO genes and the blue and black lines are indicating possible translocations of KCS and ELO members.
Prevalence of Cis-regulatory elements in promoters of KCS and ELO genes
Cis-regulatory elements are present in promotor regions and play their role in gene regulation34. Fifteen hundred base pairs upstream regions starting from start codon were used to identify cis-regulatory elements. In promoter regions of KCS genes, 23 types of elements were identified, and we classified them into four major classes: stress, developmental, light, and phytohormone-responsive elements (Table S3). The Myb elements were abundantly found in all KCS promoters and BolKCS30 gene harbored maximum (12) Myb elements (Fig. S3). In ELO genes, 19 types of elements were identified and were classified into same four major groups as in KCS genes.
In ELO genes promoters, only two types of hormone-responsive elements (TGA-element, and DRE-core) were found. Developmental type of elements includes CAAT-box, CAT-box, and HD-zip 3 and playe their roles in endosperm and meristematic expression, and zein metabolism regulation, respectively35. The presence of a significant number of stress-responsive cis-regulatory elements in KCS and ELO promoters marks their involvement in stress responsiveness. MBS, STRE, as-1, DRE core, box s, W box, MYB recognition site, and MYB-like elements were common in both gene families and have roles in drought inducibility, heat stress, pathogenesis-related response, ABA- independent stress response, pathogenesis-related response, and anthocyanin production, respectively. The MYC elements were present in KCS genes only, whereas Myb and Myc were found in ELO genes only. Among phytohormone-responsive elements, only TGA and ABRE were common in both types of genes and were involved in auxin and abscisic acid responses, respectively (Fig. S4).
Predicted protein secondary structures of KCS and ELO proteins
Secondary protein structures give insights into tertiary protein structure and also helps to understand protein functions and relationships36. Secondary proteins contained alpha helix, beta strands, TM helix, and disordered structures. The alpha helix % was dominant in both KCS and ELO proteins. All KCS proteins possessed more than 50% alpha helix, except for BolKCS11, BniKCS08, BolKCS25, and BolKCS44 which contained 39%, 47%, 47%, and 44% alpha-helix, respectively (Table S3). The percentage of beta-strands and TM helix % ranged from 9 to 15%, and 7 to 22%, respectively. Interestingly BolKCS32 protein did not carry a single TM helix. The range of disordered percentages was comparatively low and varied from 7 to 23% (Fig. S5).
Alpha helix was comparatively higher in ELO proteins and the lowest percentage of 69% was observed in BolELO03 protein. Like alpha helix, TM helix was also significantly higher and varied from 51 to 59% for ELO proteins (Fig. S6). However, the disordered structures and beta strands were comparatively lower in ELO proteins.
Protein-protein interaction network analysis of KCS and ELO proteins
The PPI networks of KCS and ELO proteins were constructed through string database to know their potential roles and interactions. KCS proteins of B. oleracea interact with other four BolKCS proteins (BolKCS05, BolKCS20, BolKCS25, and BolKCS33) and showed no interaction with any other protein. Most of the KCS proteins showed a direct interaction with BZIP53 (Fig. 4), a transcription factor involved in regulation of seed maturation genes in A. thaliana37. Moreover, reports also showed that BZIP53 transcription factors have potential roles in biotic and/or abiotic stress responses38,39,40.
Furthermore, KCS proteins also interact with subtilase family proteins (AT3G14067, AT3G14240) galacturonosyltransferase (GAUT10), intracellular copper homeostasis (CCH), phosphatase 2C family proteins (AT4G33500), and PDS-1 (4-hydroxylphenylpyruvate). ELO proteins directly interact with NAD (P) binding rossmann-fold superfamily protein (AT3G50560), CER10, and AT2G25950 protein with unknown functions. In the tetraploid CC genome of B. carinata, KCS and ELO proteins showed the same interaction pattern. However, a new interaction of KCS protein with methylenetetrahydrofolate reductase protein was observed which is not found in PPI of B. oleracea. In BB genome of B. carinata, BcaKCS15 indirectly interacts with two ELO proteins; BcaELO03 and BcaELO02, which was not observed in other genomes. In B. nigra, both KCS and ELO proteins show the same interaction as in B. carinata and B. oleracea (Fig. 4).
Expression patterns of KCS and ELO genes in different tissues under normal conditions
The expression patterns of KCS (Fig. 5a) and ELO (Fig. 5b) genes have been quantified in different tissues and during seed development stages. Among all three species, only eight KCS genes from B. carinata showed higher expression in shoot, root, and leaf tissues. During embryo development, three KCS genes (BcaKCS24, BcaKCS25, and BcaKCS31) showed an abundant expression throughout embryo development stages, whereas the expression of BcaKCS34 and BcaKCS44 decreased at mature seed stage. Most KCS genes exhibited that the transcription rate started decreasing when the embryo reached its maturity stage. Four KCS genes BcaKCS04, BcaKCS23, BniKCS11, and BolKCS09 showed a unique expression rate at the mature embryo stage. However, KCS genes did not show expression in the seed coat. Three genes from B. carinata, (BcaKCS02, BcaKCS22, BcaKCS33), two genes from B. nigra (BniKCS02, BniKCS14), and only one gene from B. oleracea (BolKCS02) showed higher transcription rates during seed coat development. Comparatively B. nigra gene (BniKCS02) showed more transcripts than those of B. carinata genes. The transcription rate significantly decreased at mature seed coat stage. Reduction in transcription rate is obvious from BniKCS02 and BolKCS02 genes at mature seed coat. In silique tissues, only six genes from B. nigra (BniKCS02, BniKCS03, BniKCS10, BniKCS14, BniKCS27, and BniKCS31) got upregulated, whereas none of the genes from B. carinata and B. oleracea showed higher transcription rate. Upregulation of B. nigra KCS genes in silique tissues suggested that these genes have longer transcription cycle than other KCS genes (Table S4).
ELO genes showed different expression patterns in tissues as compared to KCS genes. None of the ELO genes from the three Brassica species were expressed at a higher rate in all tissues. Like KCS genes, expression of ELO genes also decreased in mature seed tissues. The lower expression of ELO genes during embryo development suggested their minor involvement in seed fatty acid biosynthesis. Two ELO genes (BcaELO06 and BcaELO17) expressed at higher rate during seed coat development, whereas their expression was comparatively low during embryo development. Moreover, none of the ELO genes showed higher expression rate at mature embryo stage (Table S4).
Expression patterns of KCS and ELO genes under biotic and abiotic stresses
Expression patterns under biotic and abiotic stresses have also been examined to know the response of KCS and ELO genes against different stresses. Among abiotic stresses, RNA seq data from embryo and endosperm tissues under different temperature regimes were acquired and analyzed. Data of embryonic tissues demonstrated that the majority of the KCS genes are thermo-sensitive, and their transcription rates also changed when tissues were exposed to different temperature regimes. Such patterns can also be observed in endosperm tissue under different temperature treatments. Overall, gene expression gradually decreased in embryonic tissues after 16 days, except at 16 °C where expression remained higher (Fig. 5c). Nine genes (BolKCS03, BolKCS06, BolKCS09, BolKCS13, BolKCS14, BolKCS16, BolKCS17, BolKCS22, and BolKCS23) showed higher expression at 16 °C after 0, 1, 3, 7, 15 and 18 days of embryo development (DAE). Similar upregulation of these genes was also observed at 22 °C after 1, 3, and 7 days of endosperm development. Upregulation of these genes suggested their roles in abiotic stress tolerance.
Eleven KCS genes (BolKCS01, BolKCS04, BolKCS08, BolKCS18, BolKCS19, BolKCS20, BolKCS23, BolKCS24, BolKCS29, BolKCS30, and BolKCS32) got upregulated in root tissues under 4 combinations of zinc and phosphate indicating their possible roles in zinc and phosphate uptake. Five genes (BolKCS04, BolKCS24, BolKCS28, BolKCS29, BolKCS30, BolKCS31, and BolKCS32) upregulated under X. campestris inoculation, zinc, and phosphate stresses, however, these genes did not show up-regulation in endosperm tissues and during seed development stages under heat stress treatments.
ELO genes had more diverse expression patterns than KCS genes. BcaELO03, BcaELO05, and BcaELO06 were upregulated under zinc and phosphate stresses, whereas these genes were downregulated in embryo and endosperm tissues under heat stress treatments. Only one ELO gene (BcaELO04) was upregulated under X. campestris innoculation (Fig. 5d) (Table S5).
Discussion
Due to paramount roles of fatty acid elongation-like genes in biosynthesis of VLCFAs, genome-wide identification of KCS genes has been performed in several plant species such as A. thaliana11 Gossypium hirsutum (cotton)41, Hordeum Vulgare (barley)42, Arachis hypogaea (peanut)43 and Malus domestica (apple)44. Although KCS genes have been reported in B. napus45 and its diploid progenitors, however, genome-wide identification of KCS genes in Brassica triangle is yet to be done. In this study, we identified KCS and ELO genes from whole genomes of B. carinata and its diploid progenitors B. nigra and B. oleracea. The B. nigra, B. oleracea, and B. carinata have 32, 33 and 53 KCS genes, respectively. The results highlighted that difference in number of identified KCS genes between diploid and tetraploid species might be due to terminal deletions in the tetraploid species chromosomes. Moreover, selection type estimates also highlighted that KCS gene went through purifying selections and lost their genomic parts related to KCS functioning. However, after polyploidization, B. carinata gained 4 ELO genes, as we identified a total of 17 ELO genes in tetraploid B. carinata genome, however, the sum of diploid progenitor’s ELO genes was 1345,46.
Phylogenetic analysis revealed eight sub-classes of KCS proteins including α, β, γ, δ, ε, ζ, η, and θ sub-classes. Arabidopsis KCS proteins can be seen in all these sub-classes11. Members of subclass δ shared sequence similarities with AtKCS02 and AtKCS20 which are reported to govern suberin biosynthesis23. Similarly, members of ε sub-class shared sequence homology with AtKCS10 and AtKCS15 which are involved in the growth and development of epidermal cells10. Arabidopsis proteins namely AtKCS05 and AtKCS06 are reported to be responsible for cuticular wax biosynthesis22 and Brassica proteins of related groups might also be involved in similar functions. The AtKCS05 and AtKCS06 genes shared high sequence similarity at protein level and their role in the elongation of carbon chain from C24 to C28 has been confirmed through heterologous expression in yeast and exhibited alike biochemical functions while transformation47. The β sub-class contained three Arabidopsis proteins; AthKCS08, AthKCS16, and AthKCS18 which have already been reported11. AthKCS16 causes elongation of carbon chain 34 (C34) to carbon chain 38 (C38), while AthKCS18 is the first KCS member to be characterized. The AthKCS18 induces the elongation of carbon chain from C20:1 to C22:1 and is predominantly expressed in seeds48, but in contrast to it, AthKCS16 was found to be mainly expressed in cauline leaves and rosette49 and proteins found in this group might be involved in elongation of VLCFAs. Gene structures of ELO and KCS genes varied from species to species.
Expression patterns of KCS and ELO genes were compared in different tissues under biotic and abiotic stresses. The KCS genes expressions were lower at mature seed stages as compared to earlier seed development stages. The KCS1 gene in three Brassica species was expressed at higher rate in vegetative tissues20, while KCS18/ FAE1 gene was mainly expressed in seed tissues and its expression decreased in mature embryos48. Moreover, BnFAE1.1 and BnFAE1.2 genes showed their transcripts in developing seed tissues only50.
In the current study, we have identified KCS and ELO sub-gene families in B. carinata and its progenitors. During polyploidization, tetraploid B. carinata lost its genomic part related to KCS genes, whereas ELO gene family expanded during this process. Gene structures showed that most of the genes were intron-less and BniKCS30 was the longest gene with 22,659bp genomic sequence. RNA-seq analysis highlighted that expression of these seven genes including BolKCS04, BolKCS24, BolKCS28, BolKCS29, BolKCS30, BolKCS31, and BolKCS32 were induced after X. Campestris inoculation and zinc & phosphate stresses. Functional characterization of these genes could provide novel resistant sources.
Materials and methods
Identification of KCS and ELO genes from B. carinata, B. nigra, and B. oleracea
Most updated genomic versions of B. nigra and B. oleracea were acquired from the Brassica database (BRAD) (brassicadb.cn) http://brassicadb.cn, while the most updated version of B. carinata was downloaded from (http://brassicadb.bio2db.com/). Full length proteins of B. napus i.e., BnaKCS (AOS88713.1), and BnaELO (XP_022565432) were acquired from the National Centre for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov) and used as query sequences to do BLASTp in TB tools (Toolbox for Biologist v 1.09832) with an E value < 1e−5 against whole peptides of three Brassica species51. All KCS and ELO proteins were identified by using BLASTp program of TB tools. Moreover, the conserved domain database (CDD) search was carried out to filter out the proteins-containing full-length conserved domains, whereas proteins with incomplete domains were discarded52. Subcellular localization of KCS and ELO proteins was predicted through Bologna Unified Subcellular Component Annotator (http://busca.biocomp.unibo.it).
Gene structure and chromosomal localization of KCS and ELO genes
GFF file of each Brassica species was used to retrieve gene length, gene localization on the chromosomal strand, no. of coding sequences, and chromosome number in TB tools. Gene structures were predicted by putting the gene ids against the GFF file of each species. Moreover, molecular weight (kDa) and iso-electric point (pI) of identified proteins were retrieved from the Expasy compute pI tool available online(https://web.expasy.org/compute_pi/)53.
Relative gene localization on chromosomes was confirmed by the graphics program of TB tools54. The motif analysis was performed by MEME suit (https://meme-suite.org/meme/)55 and presented using iTOL (https://itol.embl.de/).
Phylogenetics and gene divergence analysis
Identified proteins were multiple sequence aligned with Clustal omega56 and opened in MEGA X v 10.2.457 to construct neighbor-joining phylogenetic tree with JTT (Jones-Taylor-Thornton) + G (Gamma distributed) model and 1000 bootstrap repeats58. A comparative tree consisting of three Brassica species, one non-Brassicaceae (O. sativa), and a model plant (A. thaliana) was constructed to explore the evolutionary relationship of KCS and ELO proteins.
Nucleotide substitutions causing amino acid changes are known to be non-synonymous (Ka) and those that do not cause changes are termed synonymous (Ks)59. The nature and magnitude of selection pressure occurring on coding sequences can be estimated through the ratio of Ka and Ks. The mode of selection and divergence time can also be determined through the Ks value60,61. A value of more than 1 (Ka/Ks > 1) indicates adaptive/positive selection62, less than one (Ka/Ks < 1) indicates negative selection63, whereas a value equal to one (Ka/Ks = 1) indicate neutral selection64.
Prediction of secondary protein structure
The peptide sequences were uploaded to the Phyre2 online tool (http://www.sbg.bio.ic.ac.uk/phyre2/)65 and secondary structures were predicted based on homology.
Promoter analysis of KCS and ELO genes
Fifteen hundred bp upstream regions from transcription start site were identified through TBtools by putting genomic FASTA and extracted sequences were submitted to the Plant Care database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) to identify the cis-regulatory elements66.
Synteny analysis and protein-protein interaction
Whole-genome sequences and genome annotation files were put in the MCScanX tool67 to know the syntenic relationship between three Brassica species and model plant A. thaliana. The protein-protein interaction networks were predicted using string database (https://string-db.org/)68 by uploading the peptide sequences of three Brassica species.
Global transcriptome profiling of KCS and ELO genes
To examine the expressions of ELO and KCS genes, tissue-specific RNA-seq data under normal and stressed conditions were retrieved from the European Nucleotide Archive database (https://www.ebi.ac.uk/ena/browser/home). Expression data of seed coat development stages (heart, globular, zygote, octant, bent, torpedo, and mature seed coat) and embryo development stages (heart, globular, zygote, octant, bent, torpedo, unfertilized stage ovule, and mature embryo) under normal conditions were acquired from Bio project PRJNA 641,876. Expression data of Bio project PRJNA 524,852 were used to examine gene expression during seed and embryo development under two different temperatures and day intervals. Root tissue data from Bio project PRJNA 524,852 under zinc and phosphate applications was also explored. Moreover, RNA seq data from Bio project PRJNA 421,190 was used to examine expression patterns under biotic stress of X. campestris.
The expressions of KCS and ELO genes in different tissues were quantified using galaxy Europe server (https://usegalaxy.eu/) and transcripts were evaluated in TPM (Transcripts per kilobase million)69. Moreover, heat maps were constructed using Morpheus software (https://software.broadinstitute.org/morpheus/).
Data availability
Genomes of Brassica species were acquired from (brassicadb.cn)http://brassicadb.cn, and (http://brassicadb.bio2db.com/) databases whereas queries sequences (BnaKCS (AOS88713.1), and BnaELO (XP_022565432) were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/ ). RNA-seq data under normal (PRJNA641876,) and stressed conditions (PRJNA524852, PRJNA524852, PRJNA421190) were downloaded from ENA-EBI database (https://www.ebi.ac.uk/ena/browser/home).
References
Warwick, S. I. Brassicaceae in agriculture. Genet. Genom. Brassicaceae 33–65 (2011).
Nagaharu, U. & Nagaharu, N. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J. Bot. 7, 389–452 (1935).
Zhang, Q. et al. Asymmetric epigenome maps of subgenomes reveal imbalanced transcription and distinct evolutionary trends in Brassica napus. Mol. Plant 14, 604–619 (2021).
Song, X. et al. Brassica carinata genome characterization clarifies U’s triangle model of evolution and polyploidy in Brassica. Plant Physiol. 186, 388–406 (2021).
Raman, R. et al. Molecular diversity analysis and genetic mapping of pod shatter resistance loci in Brassica carinata L. Front. Plant Sci. 1765 (2017).
Ban, Y., Khan, N. A. & Yu, P. Nutritional and metabolic characteristics of Brassica carinata co-products from biofuel processing in dairy cows. J. Agric. Food Chem. 65, 5994–6001 (2017).
Das, S., Roscoe, T., Delseny, M., Srivastava, P. & Lakshmikumaran, M. Cloning and molecular characterization of the Fatty Acid Elongase 1 (FAE 1) gene from high and low erucic acid lines of Brassica campestris and Brassica oleracea. Plant Sci. 162, 245–250 (2002).
Żyła, N., Fidler, J. & Babula-Skowrońska, D. in The Brassica oleracea Genome 1–6 (Springer, 2021).
Wang, X. et al. A β-ketoacyl-CoA synthase is involved in rice leaf cuticular wax synthesis and requires a CER2-LIKE protein as a cofactor. Plant Physiol. 173, 944–955 (2017).
Pruitt, R. E., Vielle-Calzada, J.-P., Ploense, S. E., Grossniklaus, U. & Lolle, S. J. FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc. Natl. Acad. Sci. 97, 1311–1316 (2000).
Joubès, J. et al. The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67, 547–566 (2008).
Haslam, T. M. & Kunst, L. Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210, 93–107 (2013).
Kunst, L. & Samuels, A. L. Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 42, 51–80 (2003).
Bach, L. & Faure, J.-D. Role of very-long-chain fatty acids in plant development, when chain length does matter. C.R. Biol. 333, 361–370 (2010).
Fehling, E. & Mukherjee, K. D. Acyl-CoA elongase from a higher plant (Lunaria annua): metabolic intermediates of very-long-chain acyl-CoA products and substrate specificity. Biochim. Biophy. Acta (BBA)-Lipids Lipid Metab. 1082, 239–246 (1991).
Riezman, H. The long and short of fatty acid synthesis. Cell 130, 587–588 (2007).
Harwood, J. L. in Plant lipids 27–66 (Blackwell, 2020).
Costaglioli, P. et al. Profiling candidate genes involved in wax biosynthesis in Arabidopsis thaliana by microarray analysis. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 1734, 247–258 (2005).
James, D. & Dooner, H. Isolation of EMS-induced mutants in Arabidopsis altered in seed fatty acid composition. Theor. Appl. Genet. 80, 241–245 (1990).
Todd, J., Post-Beittenmiller, D. & Jaworski, J. G. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 17, 119–130 (1999).
Hooker, T. S., Millar, A. A. & Kunst, L. Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129, 1568–1580 (2002).
Fiebig, A. et al. Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12, 2001–2008 (2000).
Lee, S. B. et al. Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 60, 462–475 (2009).
Gray, J. E. et al. The HIC signalling pathway links CO2 perception to stomatal development. Nature 408, 713–716 (2000).
Zank, T. K. et al. Cloning and functional characterisation of an enzyme involved in the elongation of Δ6-polyunsaturated fatty acids from the moss Physcomitrella patens. Plant J. 31, 255–268 (2002).
Kajikawa, M. et al. Isolation and characterization of Δ6-desaturase, an ELO-like enzyme and Δ5-desaturase from the liverwort Marchantia polymorpha and production of arachidonic and eicosapentaenoic acids in the methylotrophic yeast Pichia pastoris. Plant Mol. Biol. 54, 335–352 (2004).
Kajikawa, M. et al. Isolation and functional characterization of fatty acid Δ5-elongase gene from the liverwort Marchantia polymorpha L. FEBS Lett. 580, 149–154 (2006).
Dunn, T. M., Lynch, D. V., Michaelson, L. V. & Napier, J. A. A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana. Ann. Bot. 93, 483–497 (2004).
Quist, T. M. et al. HOS3, an ELO-like gene, inhibits effects of ABA and implicates a S-1-P/ceramide control system for abiotic stress responses in Arabidopsis thaliana. Mol. Plant 2, 138–151 (2009).
Yang, Z. & Rannala, B. Molecular phylogenetics: Principles and practice. Nat. Rev. Genet. 13, 303–314 (2012).
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D. & May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 1–21 (2004).
Zhou, T. et al. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genomics 271, 402–415 (2004).
Panchy, N., Lehti-Shiu, M. & Shiu, S.-H. Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316 (2016).
Wittkopp, P. J. & Kalay, G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69 (2012).
Lohani, N., Babaei, S., Singh, M. B. & Bhalla, P. L. Genome-wide in silico identification and comparative analysis of Dof gene family in Brassica napus. Plants 10, 709 (2021).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen‐bonded and geometrical features. Biopolym. Original Res. Biomol. 22, 2577–2637 (1983).
Alonso, R. et al. A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21, 1747–1761 (2009).
Hwang, I. et al. Genome-wide identification and characterization of bZIP transcription factors in Brassica oleracea under cold stress. BioMed Res. Int. 2016 (2016).
Dubos, C. et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581 (2010).
Alves, M. S. et al. Plant bZIP transcription factors responsive to pathogens: A review. Int. J. Mol. Sci. 14, 7815–7828 (2013).
Xiao, G. H., Wang, K., Huang, G. & Zhu, Y. X. Genome-scale analysis of the cotton KCS gene family revealed a binary mode of action for gibberellin A regulated fiber growth. J. Integr. Plant Biol. 58, 577–589 (2016).
Tong, T. et al. Genome-wide identification and expression pattern analysis of the KCS gene family in barley. Plant Growth Regul. 93, 89–103 (2021).
Huai, D. et al. Genome-wide identification of peanut KCS genes reveals that AhKCS1 and AhKCS28 are involved in regulating VLCFA contents in seeds. Front. Plant Sci. 11, 406 (2020).
Lian, X.-Y. et al. Genome wide analysis and functional identification of MdKCS genes in apple. Plant Physiol. Biochem. 151, 299–312 (2020).
Xue, Y. et al. Genome-wide mining and comparative analysis of fatty acid elongase gene family in Brassica napus and its progenitors. Gene 747, 144674 (2020).
Xue, Y. et al. Genome-wide survey and characterization of fatty acid desaturase gene family in Brassica napus and its parental species. Appl. Biochem. Biotechnol. 184, 582–598 (2018).
Trenkamp, S., Martin, W. & Tietjen, K. Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proc. Natl. Acad. Sci. 101, 11903–11908 (2004).
Rossak, M., Smith, M. & Kunst, L. Expression of the FAE1 gene and FAE1 promoter activity in developing seeds of Arabidopsis thaliana. Plant Mol. Biol. 46, 717–725 (2001).
Hegebarth, D. et al. Arabidopsis ketoacyl-CoA synthase 16 (KCS16) forms C36/C38 acyl precursors for leaf trichome and pavement surface wax. Plant Cell Environ. 40, 1761–1776 (2017).
Chiron, H. et al. Regulation of FATTY ACID ELONGATION1 expression in embryonic and vascular tissues of Brassica napus. Plant Mol. Biol. 88, 65–83 (2015).
Matsuda, F., Tsugawa, H. & Fukusaki, E. Method for assessing the statistical significance of mass spectral similarities using basic local alignment search tool statistics. Anal. Chem. 85, 8291–8297 (2013).
Marchler-Bauer, A. & Bryant, S. H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331 (2004).
Gasteiger, E. et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 (2003).
Chen, C., Chen, H., He, Y. & Xia, R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 289660, 289660 (2018).
Bailey, T. L., Johnson, J., Grant, C. E. & Noble, W. S. The MEME suite. Nucleic Acids Res. 43, W39–W49 (2015).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547 (2018).
Tamura, K. et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Yang, Z. & Nielsen, R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17, 32–43 (2000).
Messier, W. & Stewart, C.-B. Episodic adaptive evolution of primate lysozymes. Nature 385, 151–154 (1997).
Yuan, S. et al. Comprehensive analysis of CCCH-type zinc finger family genes facilitates functional gene discovery and reflects recent allopolyploidization event in tetraploid switchgrass. BMC Genomics 16, 1–16 (2015).
Trabesinger-Ruef, N. et al. Pseudogenes in ribonuclease evolution: A source of new biomacromolecular function?. FEBS Lett. 382, 319–322 (1996).
Anisimova, M., Bielawski, J. P. & Yang, Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol. Biol. Evol. 18, 1585–1592 (2001).
Duret, L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 16, 287–289 (2000).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Lescot, M. et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327 (2002).
Wang, Y. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49–e49 (2012).
Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Rese. gkw937 (2016).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Acknowledgements
This work was supported by Higher Education Commission of Pakistan (HEC) through its grant of Precision Agriculture and Analytics Lab (PAAL) under National Centre in Big Data and Cloud Computing (NCBC). Authors are also thankful to Ministry of Science and Technology for sponsoring National Center for Genome Editing at CAS-AFS, University of Agriculture, Faisalabad. The infrastructural support from the Centre for Advanced Studies in Agriculture and Food Security (CAS-AFS), at University of Agriculture Faisalabad, Pakistan is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
I.A.K., R.M.A. and I.A.R. conceived the idea. I.A.R., U.M.K., R.M. and R.M.A. designed the experiments. U.M.K., N.S. and R.M.A. retrieved and curated the data. U.M.K. and N.S. performed formal analyses and H.M.R. helped them in drafting the manuscript. I.A.R., Q.R., H.M.R., R.M., I.A.K. and R.M.A. reviewed and edited the manuscript. I.A.K., I.A.R. and R.A. provided resources, supervised the experiments, and acquired funding for research. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Khan, U.M., Rana, I.A., Shaheen, N. et al. Comparative phylogenomic insights of KCS and ELO gene families in Brassica species indicate their role in seed development and stress responsiveness. Sci Rep 13, 3577 (2023). https://doi.org/10.1038/s41598-023-28665-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-28665-2
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.